Abstract
Coronary heart disease (AHD) is the leading cause of death and disability worldwide. In patients with acute coronary syndromes (ACS), timely and effective myocardial reperfusion by percutaneous coronary intervention (PCI) is the primary treatment of choice to minimize the ischemic injury and limit MI size. However, reperfusion can itself promote cardiomyocyte death which leads to cardiac dysfunction via reperfusion injury. The molecular mechanisms of ischemia/reperfusion (I/R) injury are not completely understood and new drug targets are needed. Recently we reported that cardiac troponin I-interacting protein kinase (TNNI3K), a cardiomyocyte-specific kinase, promotes I/R injury via profound oxidative stress, thereby promoting cardiomyocyte death. By using novel genetic animal models and newly developed small-molecule TNNI3K inhibitors, we demonstrate that TNNI3K-mediated I/R injury occurs through impaired mitochondrial function and is in part dependent on p38 MAPK. Herein we discuss the emerging role of TNNI3K as a promising new drug target to limit the I/R-induced myocardial injury. We will also examine the underlying mechanisms that drive the profoundly reduced infarct size in mice in which TNNI3K is specifically deleted in cardiomyocytes. Since TNNI3K is a cardiac-specific kinase, it could be an ideal molecular target since inhibiting it would have little or no effect on other organ systems, a serious problem associated with the use of kinase inhibitors targeting kinases that are more widely expressed.
Introduction
Acute myocardial infarction (AMI), with subsequent left ventricular dysfunction and heart failure, continues to be a major cause of morbidity and mortality worldwide. Rapid advances in the treatment of AMI, mainly through timely reperfusion, have substantially improved outcomes but at the same time promoting cardiomyocyte death and cardiac dysfunction via reperfusion injury.1 A number of preclinical and clinical studies have been published on various pharmacological agents to prevent myocardial cell death during the time of ischemia and subsequent reperfusion.2–4 Many of these agents have failed in the translational phase largely because they were inefective or they produced adverse side effects related to both on- or off-target toxicity in various organs.2,3,5 Since most kinases are ubiquitously expressed, it is not surprising that their systemic administration leads to harmful on-target side effects. Given the fact that localized delivery or gene therapy is still relatively far from clinical reality, it would be a significant step forward if we could identify a cardiac specific drug target with the ability to limit infarct size and I/R mediated injury post AMI.
Recently, we report that inhibition of TNNI3K, a cardiomyocyte specific kinase, limits oxidative stress, infarct size and adverse ventricular remodeling post-MI suggesting that inhibition of TNNI3K could be an attractive cardiac-specific therapeutic target for AMI.6 Studies to test this hypothesis are ongoing.
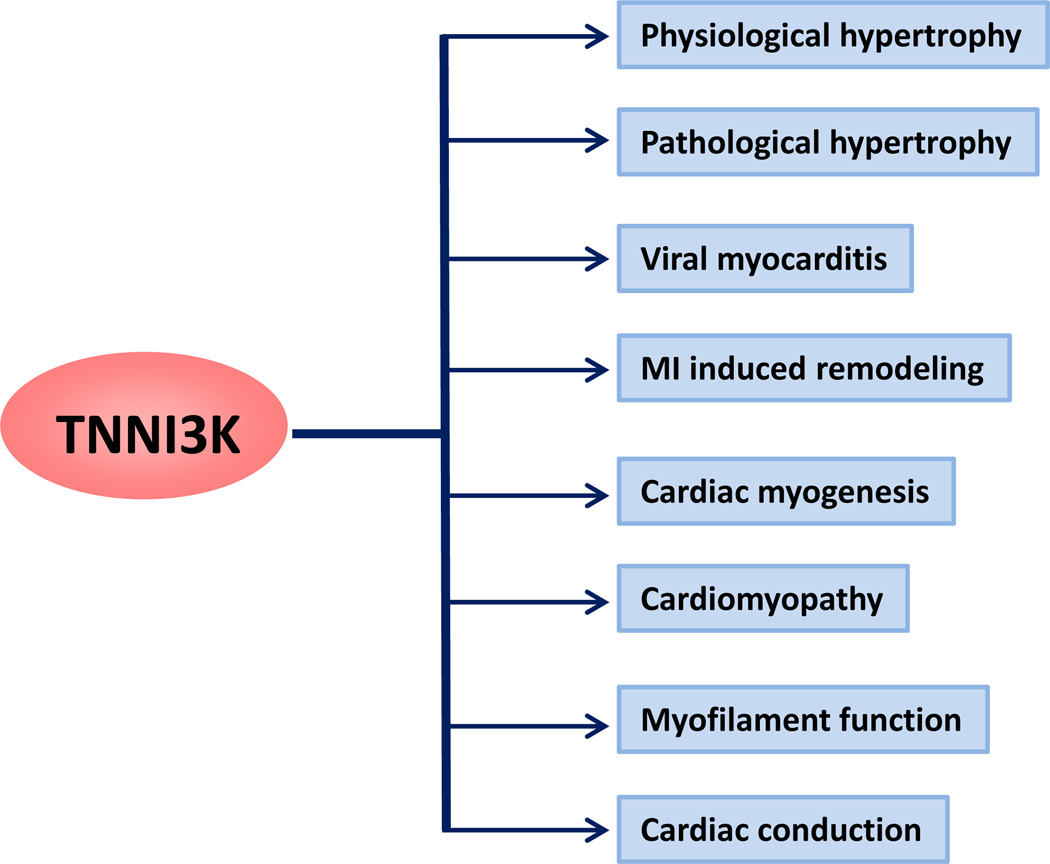
More recent evidence suggests that TNNI3K may have a vital role in several important aspects of cardiac biology including viral myocarditis, cardiac conduction, cardiomyopathy, obesity and metabolic disorders, and pathological and physiological hypertrophy7–9 (Fig. 1). However, most of these observations were either made with transgenic mouse models (gain of function) or from large scale genetic screens (Table 1). Thus we need further studies employing loss of function strategies (KOs and pharmacological inhibition). Here, we review the current findings regarding the role of TNNI3K in different aspect of cardiac biology. We discuss how the inhibition of TNNI3K has been proposed to ameliorate many facets of cardiac injury and metabolic function.
Fig. 1. Schematic of TNNI3K structure.
Three distinct domains are shown. The N-terminal ankyrin repeat, a central kinase domain and the Serine-rich domain at C-terminal. It shares a domain structure similar to that of ILK. ANK, Ankyrin; Ser-rich, Serine-rich; ILK, integrin-linked kinase.
Table.
List of TNNI3K loss of function and gain of function studies with genetically modified animal models.
| SN | Genetic model | HF Model | Phenotype/ outcome |
Mechanism/characteristic | Reference |
|---|---|---|---|---|---|
| 1 | Cardiac specific-TG | I/R | Detrimental | p38 activation, Mitochondrial dysfunction, ROS overproduction, Impaired cardiomyocytes bioenergetics | 6 |
| 2 | Cardiac specific-KO | I/R | Protective | Reduced p38 activation, ROS production and cardiomyocyte death, | 6 |
| 3 | Cardiac specific-TG | - | Physiological hypertrophy | Increased heart mass with enhanced cardiac function, no necrosis or apoptosis | 8 |
| 4 | Cardiac specific-TG | TAC | Detrimental | Cardiac remodeling, Increased expression of ANP and BNP | 9 |
| 5 | Cardiac specific-kinase dead TG | TAC | Protective | Attenuated remodeling, decreased expression of ANP and BNP | 9 |
| 6 | Intra-myocardial injection of TNNI3K-overexpressing P19CL6 cells | MI | Protective | Improved LV function and remodeling post-MI. | 10 |
| 7 | Double TG of TNNI3K and Calsequestrin | Cardiomyopathy | Detrimental | Severely impaired systolic function and increased mortality. | 11 |
Cloning and characterization
TNNI3K was initially cloned in 2003 by investigators at Peking University Union Medical College in Beijing.12 They identified the kinase via a bioinformatics approach and found that it was highly expressed in the heart but not expressed in any other tissues. Subsequently, the full-length TNNI3K mouse mRNA sequence was cloned, and the basal promoter regions were characterized (GenBank accession no. NM015978).10,13
Multiple fetal and adult Northern blot experiments, as well as gene arrays, confirmed the cardiac specific expression of TNNI3K.6,10 Within the heart, TNNI3K was variably expressed in all heart regions with highest levels in the interventricular septum and apex. Immunohistochemical analysis detected TNNI3K predominantly localized to perinuclear or nuclear regions of fetal and adult cardiac myocytes.6,12 TNNI3K has a full-length cDNA with 3,420 bp and contains a continuous open reading frame of 2,505 bp, which encodes a protein of 835 amino acids and a molecular mass of 93 kDa.10
TNNI3K contains a central kinase domain, flanked by an ankyrin repeat domain in the amino terminus and a serine-rich domain in the C-terminus (Fig. 2). BLAST analysis of TNNI3K sequence identifies integrin-linked kinase (ILK), a four-ankyrin-repeat kinase, as its closest relative. Integrin-linked kinase (ILK) is a highly evolutionarily conserved intracellular protein that was originally identified as an integrin-interacting protein. ILK plays a vital role in embryonic development and tissue homeostasis.14,15 Importantly, ILK also has central roles in cardiac and smooth-muscle contractility, and ILK dysregulation causes cardiomyopathies in humans.15,16 The kinase domain of TNNI3K contains primary sequence motifs conserved in both serine/threonine and tyrosine protein kinases. This assigns TNNI3K to a family of protein kinases, the mixed lineage kinase (MLK) family.
Fig. 2.
TNNI3K has been implicated in regulation of several essential myocardial pathophysiological conditions.
Mechanism of cardiac specific expression
Wang et al13 cloned the full-length mRNA sequence and characterized the basal promoter region of the mouse TNNI3K gene. By using a bioinformatics approach, five potential conserved transcription factor binding sites were identified in the core promoter region of the mouse TNNI3K gene. These include Tbx5, SRE, M-CAT box, MEF2 and GATA4. MEF2C plays a critical role in regulating TNNI3K transcriptional activity as mutations in MEF2 binding sites caused a drastic decrease in transcriptional activity.13 MEF2C transcripts are largely restricted to the cardiac and skeletal muscle lineages.17,18 In mice homozygous for a null mutation of MEF2C, heart development arrests at the looping stage, the right ventricle did not form, and a subset of cardiac muscle genes were not expressed, suggesting that MEF2C is an essential regulator of cardiac myogenesis and right ventricular development.19 In the adult heart, overexpression of MEF2 induces dilatative cardiomyopathy in transgenic mice and activates a genetic program promoting LV chamber dilatation and mechanical dysfunction in a heart failure model.20,21 Thus, considering the known function of MEF2 family members in cardiac development and disease processes, it was not surprising that the cardiac-enriched transcription factor Mef2c is the most critical for cardiac-specific expression of TNNI3K.
TNNI3K binding partners
TNNI3K was named on the basis of its interaction with cardiac troponin I (cTnI).12 A yeast two-hybrid screen using TNNI3K as bait identified cardiac troponin I as an interacting protein and, therefore, a possible substrate. Feng et al.22 identified peroxiredoxin 3 (PRDX3) (Previously named “antioxidant protein 1” (AOP-1)) as another TNNI3K interacting partner. The PRDX3 belongs to thiol-specific antioxidant protein family.23,24 PRDX3 interacts with TNNI3K to down regulates its kinase activity.22 A yeast two-hybrid screen and an in-vitro binding assay confirmed a direct interaction of PRDX3 and TNNI3K. Studies with deletion mutants of TNNI3K demonstrated that PRDX3 could not only bind to the ankyrin motif but also to the protein kinase domain of TNNI3K, suggesting that PRDX3 exerts its inhibitory effect through its interaction with the protein kinase domain of TNNI3K. Studies with deletion mutants of PRDX3 demonstrated that the binding site located within the C-terminal 60–256 amino acids of PRDX3, that is, the thiol-specific antioxidant domain. Further studies are warranted to determine the molecular details and physiological significance of the PRDX3 and TNNI3K interaction in-vivo. Several other potential TNNI3K interacting partners have been proposed which includes, fatty acid binding proteins 3, aryl hydrocarbon receptor-interacting protein, adult skeletal muscle actin and cardiac myosin binding protein C.10,12 However, further investigation is required to confirm these potential interactions and their biological significance.
TNNI3K as a potential target of cardiac remodeling
a) Role of TNNI3K in cardiomyopathy and MI-induced remodeling
Initial in vitro studies suggested a protective role of TNNI3K in cell death, as its overexpression led to increased beating mass which was associated with reduced apoptosis.10 This study utilized the pluripotent P19CL6 cells with or without transfection by pcDNA6-TNNI3K plasmid to determine its role in cardiac myogenesis. The authors concluded that TNNI3K promotes the differentiation process based on the increased beating mass and number of α-actinin-positive cells after TNNI3K overexpression. Intramyocardial administration of TNNI3K-overexpressing P19CL6 cells in mice post-MI improved cardiac performance and attenuated ventricular remodeling compared with injection of wild-type P19CL6 cells. Thus early data suggested that TNNI3K had a protective role after myocardial injury.
However, several high profile studies with genetically modified animals challenged the protective role of TNNI3K in cardiac disease processes and strongly suggested that activation of TNNI3K leads to a detrimental phenotype and conversely its inhibition leads to significant protection both at baseline and post-injury.6,7,9,11 Wheeler et al.11 demonstrated that both normal expression and TNNI3K overexpression had a detrimental effect on cardiac function. This was demonstrated by utilizing different inbred and congenic mouse stains, which have significant heterogeneity in the expression level of endogenous TNNI3K. Interestingly, some strains represent natural loss of function models as they harbor 100 fold less TNNI3K expression than others. Mice not expressing TNNI3K were resistant to Calsequestrin-induced cardiomyopathy, whereas normal or high expressers were highly susceptible to two different models of heart failure. TNNI3K overexpression greatly accelerates cardiac dysfunction in mouse models of cardiomyopathy, indicating an important role of TNNI3K in modulating cardiac disease progression.11
By using cardiac-specific TNNI3K transgenic, conditional knockout, and novel pharmacological inhibitors, we demonstrated that TNNI3K plays an important role in development of I/R injury and thus its inhibition could be a novel drug target to treat myocardial I/R injury.6 Genetic overexpression of TNNI3K leads to larger infarct size in comparison to wilt-type littermates after I/R injury. Consistently, TNNI3K transgenic mice had elevated plasma levels of cTnI. Kinase activity of TNNI3K was responsible for the larger infarct size as kinase dead transgenic mice had significantly smaller infarcts and reduced plasma cTnI levels. Surprisingly, we found that TNNI3K regulates several essential functions of mitochondrial biology including mitochondrial ROS production (mROS), mitochondrial membrane potential and mitochondrial calcium flux. Thus, TNNI3K-dependent cell death and I/R-induced cardiac dysfunction is mediated through overproduction of mROS and subsequent mitochondrial impairment. Furthermore, we identified p38αMAPK as a key downstream effector of TNNI3K that is, in part, responsible for its detrimental effect (Fig. 3). Consistent with the gain of function model, cardiomyocyte specific deletion of TNNI3K led to smaller infarct size and decreased production of injury markers in comparison to wild-type. Thus, genetic gain- and loss of function studies suggest that inhibition of TNNI3K could be beneficial in the mouse heart after I/R injury.6
Fig. 3. Molecular signaling mechanism of TNNI3K in response to cardiac injury.
After cardiac injury, TNNI3K induces p38 phosphorylation by an unknown mechanism. Activation of p38 results in increased mitochondrial reactive oxygen species generation, resulting in cell death which leads to increased infarct size and adverse ventricular remodeling. The role of TNNI3K-dependent mitochondrial dysfunction or oxidative stress in development of cardiac hypertrophy is not known. Overexpression studies have suggested that TNNI3K also increases the velocity of diastolic depolarization of phase 4 of spontaneous action potential through suppressing phosphorylation of cTnI, resulting in increased contractile force. TNNI3K may increase the contractile force by modulating the ryanodine receptor-mediated intracellular Ca2+ concentration. MI, Myocardial infarction; I/R, Ischemia-reperfusion injury; TAC, Transverse aortic constriction; SR, Sarcoplasmic reticulum, RyR, Ryanodine receptor; cTnI, Cardiac troponin I.
To begin to translate these preclinical observations to the clinic, we developed two small molecule TNNI3K inhibitors and tested their efficacy in a myocardial I/R model in vivo. Pharmacological inhibition of TNNI3K reduced mROS, p38 activation, and infarct size when delivered at reperfusion to mimic clinical intervention. Treatment with TNNI3K inhibitor also limits chronic adverse remodeling and preserves cardiac function in the re-perfused heart.6 Thus taken together our recent preclinical studies indicate that TNNI3K inhibition could represent a therapeutic strategy for acute coronary syndromes. This hypothesis is being tested in studies to be done at NHLBI via the CAESAR Program.
b) Physiological and pathological hypertrophy
Pathological cardiac hypertrophy is associated with increased interstitial fibrosis, cell death and cardiac dysfunction, and is a key risk factor for heart failure. Wheeler et al11 were the first to demonstrate that overexpression of TNNI3K accelerates disease progression in a pressure- overload-induced model of pathological hypertrophy. The TNNI3K transgenic mice showed increased adverse dilatative remodeling in response to thoracic aortic constriction (TAC) surgery, as reflected by increased left-ventricular end diastolic diameter (LVEDD) and left-ventricular end systolic diameter (LVESD). This dilatative remodeling was associated with a reduction of ventricular function as reflected by reduced fractional shortening, confirming that TNNI3K expression has a detrimental effect in a model of pathological hypertrophy.11 Wang et al25 demonstrated that adenovirus-mediated overexpression of TNNI3K promotes cardiomyocyte cellular hypertrophy. Ad-TNNI3K led to an increase in sarcomere organization, cell surface area, H-leucine incorporation and β-MHC re-expression.25 Tang et al9 used transgenic kinase-dead mice to determine whether TNNI3K kinase activity is responsible for TNNI3K mediated pathological hypertrophy. TAC-induced hypertrophic responses were significantly attenuated in the transgenic kinase dead mice confirming that TNNI3K kinase activity plays an essential role in pro-hypertrophic responses driven by biomechanical stress-induced pathologic cardiac hypertrophy.
TNNI3K has also been implicated in physiological cardiac hypertrophy.8 In contrast to pathological hypertrophy, physiological cardiac hypertrophy is reversible and is characterized by normal cardiac morphology (i.e. no fibrosis or apoptosis) and enhanced cardiac function. Wang et al.8 reported that TNNI3K transgenic mice develop a phenotype of concentric hypertrophy with enhanced cardiac function as revealed by echocardiography and hemodynamic assessments. The authors concluded that it was physiological hypertrophy as no necrosis or myocyte apoptosis was observed in the heart of TNNI3K transgenic mice. However, we do not observed physiological or pathological hypertrophy in the TNNI3K transgenic or KOs mice at baseline. Taken together the available evidence suggests that TNNI3K plays an important role in mediating pathological hypertrophy, however, its role in physiological hypertrophy needs further confirmation.
C) Role of TNNI3K in viral myocarditis
Myocarditis is an inflammatory disease of the heart frequently resulting from viral infections and/or subsequent immune- responses. Evidence of viral infection as a cause of heart failure has been recognized for >50 years, but it is still a challenging disease to diagnose and treat.26,27 Several viruses have been associated with myocarditis in humans. Of the virus genomes identified, adenovirus, enterovirus and cytomegalovirus are the most common groups of viruses. However, coxsackievirus B3 is still considered the dominant etiological agent.28 In a multicenter analysis of 624 patients in the United States with histologically-proven myocarditis, the presence of various virus genomes was confirmed in 239 of biopsy samples (38%).26,29 At present, no diagnostic gold standard is available for viral myocarditis, however, even after proper diagnosis, management of viral myocarditis represents a difficult challenge as there is no clinically proven treatment to inhibit the development of subsequent dilated cardiomyopathy.27,30 This is primarily due to lack of known molecular drug candidates for viral myocarditis susceptibility.31 Wiltshire et al32 identified TNNI3K as a candidate gene for viral myocarditis in an unbiased screen based on quantitative trait locus analysis, pathway analysis and consomic mapping. A combination of analyses revealed very strong evidence for the existence and location of viral myocarditis susceptibility 1 (Vms1) locus on chromosome 3. Further microarray and candidate gene analysis identified TNNI3K as a likely candidate for Vms1. Further study is warranted to investigate the pathophysiology and mechanistic details of this association.
E) Role of TNNI3K in cardiac conduction
Atrioventricular (AV) conduction disease is characterized by a prolonged PR interval on the surface electrocardiogram (ECG). Importantly, a prolonged PR interval is a strong predictor of atrial fibrillation (AF). AF is the most commonly observed arrhythmia which is associated with an increased risk of stroke, heart failure, and sudden cardiac death. The association of TNNI3K and cardiac conduction was first observed in a microarray screening in human failing heart samples from patients with arrhythmogenic right ventricular cardiomyopathy.33 Arrhythmogenic cardiomyopathy was associated with upregulation of TNNI3K regardless of cardiomyopathic etiology. Further regression analysis showed a positive correlation between TNNI3K and ProANP in Arrhythmogenic failing hearts.33 Recently Lodder et al.34 provided strong evidence that increased expression of TNNI3K is significantly correlated with increased PR interval. Measurement of PR interval duration and TNNI3K levels in six inbred lines identified a positive correlation between the levels of TNNI3K and PR interval duration.34 In the same study investigators performed in vivo functional studies using TNNI3K transgenic mice to confirm the role of TNNI3K in regulating PR interval in atrioventricular conduction.34,35 The molecular mechanism underlying TNNI3K mediated atrioventricular conduction is not known and requires further in-depth investigation.
F) Role of TNNI3K in obesity
Several recent human genetic studies with diverse populations have suggested an important role of TNNI3K in regulation of BMI and development of obesity.36–38 Zhao et al39 were the first to find an association between common childhood obesity and a single-nucleotide polymorphism (SNP) in TNNI3K in a study of European Americans, aged between 2 and 18 years old. Subsequently, the observation from the Look AHEAD trial suggested a significant association of risk allele at TNNI3K with lower percentage of energy from protein sources.38 A GWAS analysis of BMI in adolescents and young adults reveals that TNNI3K had a larger effect on BMI during adolescence and young adulthood compared with older adults.37 Another recent GWAS analysis suggests that TNNI3K is positively associated with emotional and uncontrolled eating.36 Thus, although a strong correlation between TNNI3K and obesity has been indicated by population based genetic studies, the mechanistic basis of this association is lacking and needs further investigation.
Conclusions and future perspectives
Infarct size is a major determinant of cardiac remodeling. To date, multiple experimental interventions have been reported to protect the ischemic myocardium in experimental animals. Unfortunately, with the exception of timely reperfusion, none have translated into clinical practice. As most of these therapeutic interventions are based on targeting of ubiquitously expressed kinases, one can speculate that harmful side-effects could arise from systemic administration of one or more compounds that modulate kinase activity. Our recent findings that TNNI3K inhibition can limit infarct size and adverse cardiac remodeling after I/R injury, is especially promising as TNNI3K inhibition would be cardiac-specific, and thus limiting adverse effects of systemic kinase inhibition. However, in order to provide effective translation of TNNI3K inhibition and cardio-protection in the clinical setting, it is fundamental to first understand, in pre-clinical animal models, not only the pharmaco-kinetics but also the signaling pathways involved. Further investigation of TNNI3K binding partners and downstream targets would bring clarity to TNNI3K mediated-cellular responses in contexts of interest. Defining the molecular link between TNNI3K and the immune response to viral pathogens may reveal new principles for the management of viral myocarditis. Several genome wide analyses have suggested that TNNI3K may regulate insulin resistance and obesity, but, further investigation is warranted to establish this link. Finally, by unravelling the molecular details of TNNI3K biology, identifying binding partners and downstream targets, and establishing their roles in various aspect of cardiac biology, would benefit the field tremendously. Finally, another patient population that has a profound unmet therapeutic need are patients with frequent and recurrent ischemic events, but they are not candidates for surgery. The development of enhanced TNNI3K inhibitor-based therapeutics could protect these patients from the common scenario of gradually declining contractile function.
Acknowledgements
This work was supported by grants from the NHLBI to T. Force (HL061688, HL091799) and American Heart Association Scientist Development Grant (13SDG16930103) to H.L.
Relationship with Industry disclosure:
Dr. Force received research funding, as well as consultancy fees from GlaxoSmithKline.
References
- 1.Hausenloy DJ, Yellon DM. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J Clin Invest. 2013;123:92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerczuk PZ, Kloner RA. An update on cardioprotection: A review of the latest adjunctive therapies to limit myocardial infarction size in clinical trials. J Am Coll Cardiol. 2012;59:969–978. doi: 10.1016/j.jacc.2011.07.054. [DOI] [PubMed] [Google Scholar]
- 3.Kloner RA. Current state of clinical translation of cardioprotective agents for acute myocardial infarction. Circ Res. 2013;113:451–463. doi: 10.1161/CIRCRESAHA.112.300627. [DOI] [PubMed] [Google Scholar]
- 4.Minamino T. Cardioprotection from ischemia/reperfusion injury: Basic and translational research. Circ J. 2012;76:1074–1082. doi: 10.1253/circj.cj-12-0132. [DOI] [PubMed] [Google Scholar]
- 5.Frohlich GM, Meier P, White SK, Yellon DM, Hausenloy DJ. Myocardial reperfusion injury: Looking beyond primary pci. Eur Heart J. 2013;34:1714–1722. doi: 10.1093/eurheartj/eht090. [DOI] [PubMed] [Google Scholar]
- 6.Vagnozzi RJ, Gatto GJ, Jr, Kallander LS, Hoffman NE, Mallilankaraman K, Ballard VL, et al. Inhibition of the cardiomyocyte-specific kinase tnni3k limits oxidative stress, injury, and adverse remodeling in the ischemic heart. Sci Transl Med. 2013;5:207ra141. doi: 10.1126/scitranslmed.3006479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abraham DM, Marchuk DA. Inhibition of the cardiomyocyte-specific troponin i-interacting kinase limits oxidative stress, injury, and adverse remodeling due to ischemic heart disease. Circ Res. 2014;114:938–940. doi: 10.1161/CIRCRESAHA.113.303238. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Wang J, Su M, Wang C, Chen J, Wang H, et al. Tnni3k, a cardiac-specific kinase, promotes physiological cardiac hypertrophy in transgenic mice. PLoS One. 2013;8:e58570. doi: 10.1371/journal.pone.0058570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Xiao K, Mao L, Rockman HA, Marchuk DA. Overexpression of tnni3k, a cardiac-specific mapkkk, promotes cardiac dysfunction. J Mol Cell Cardiol. 2013;54:101–111. doi: 10.1016/j.yjmcc.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai ZF, Chen YZ, Feng LP, Meng XM, Ding JF, Wang LY, et al. Overexpression of tnni3k, a cardiac-specific map kinase, promotes p19cl6-derived cardiac myogenesis and prevents myocardial infarction-induced injury. Am J Physiol Heart Circ Physiol. 2008;295:H708–H716. doi: 10.1152/ajpheart.00252.2008. [DOI] [PubMed] [Google Scholar]
- 11.Wheeler FC, Tang H, Marks OA, Hadnott TN, Chu PL, Mao L, et al. Tnni3k modifies disease progression in murine models of cardiomyopathy. PLoS Genet. 2009;5:e1000647. doi: 10.1371/journal.pgen.1000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y, Meng XM, Wei YJ, Zhao XW, Liu DQ, Cao HQ, et al. Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin i. J Mol Med (Berl) 2003;81:297–304. doi: 10.1007/s00109-003-0427-x. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Chen C, Song X, Chen J, Zhen Y, Sun K, et al. Mef2c is an essential regulatory element required for unique expression of the cardiac-specific cark gene. J Cell Mol Med. 2008;12:304–315. doi: 10.1111/j.1582-4934.2007.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannigan GE, McDonald PC, Walsh MP, Dedhar S. Integrin-linked kinase: Not so 'pseudo' after all. Oncogene. 2011;30:4375–4385. doi: 10.1038/onc.2011.177. [DOI] [PubMed] [Google Scholar]
- 15.Hannigan GE, Coles JG, Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ Res. 2007;100:1408–1414. doi: 10.1161/01.RES.0000265233.40455.62. [DOI] [PubMed] [Google Scholar]
- 16.Lal H, Verma SK, Foster DM, Golden HB, Reneau JC, Watson LE, et al. Integrins and proximal signaling mechanisms in cardiovascular disease. Front Biosci (Landmark Ed) 2009;14:2307–2334. doi: 10.2741/3381. [DOI] [PubMed] [Google Scholar]
- 17.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 18.Olson EN, Perry M, Schulz RA. Regulation of muscle differentiation by the mef2 family of mads box transcription factors. Dev Biol. 1995;172:2–14. doi: 10.1006/dbio.1995.0002. [DOI] [PubMed] [Google Scholar]
- 19.Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor mef2c. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, Gong NL, Bodi I, Aronow BJ, Backx PH, Molkentin JD. Myocyte enhancer factors 2a and 2c induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281:9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 21.van Oort RJ, van Rooij E, Bourajjaj M, Schimmel J, Jansen MA, van der Nagel R, et al. Mef2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation. 2006;114:298–308. doi: 10.1161/CIRCULATIONAHA.105.608968. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Liu DQ, Wang Z, Liu Z, Cao HQ, Wang LY, et al. Aop-1 interacts with cardiac-specific protein kinase tnni3k and down-regulates its kinase activity. Biochemistry (Mosc) 2007;72:1199–1204. doi: 10.1134/s0006297907110053. [DOI] [PubMed] [Google Scholar]
- 23.Immenschuh S, Baumgart-Vogt E. Peroxiredoxins, oxidative stress, and cell proliferation. Antioxid Redox Signal. 2005;7:768–777. doi: 10.1089/ars.2005.7.768. [DOI] [PubMed] [Google Scholar]
- 24.Rigoulet M, Yoboue ED, Devin A. Mitochondrial ros generation and its regulation: Mechanisms involved in h(2)o(2) signaling. Antioxid Redox Signal. 2011;14:459–468. doi: 10.1089/ars.2010.3363. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Wang H, Ye J, Xu RX, Song L, Shi N, et al. Adenovirus-mediated overexpression of cardiac troponin i-interacting kinase promotes cardiomyocyte hypertrophy. Clin Exp Pharmacol Physiol. 2011;38:278–284. doi: 10.1111/j.1440-1681.2011.05499.x. [DOI] [PubMed] [Google Scholar]
- 26.Yajima T, Knowlton KU. Viral myocarditis: From the perspective of the virus. Circulation. 2009;119:2615–2624. doi: 10.1161/CIRCULATIONAHA.108.766022. [DOI] [PubMed] [Google Scholar]
- 27.Corsten MF, Schroen B, Heymans S. Inflammation in viral myocarditis: Friend or foe? Trends Mol Med. 2012;18:426–437. doi: 10.1016/j.molmed.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Dennert R, Crijns HJ, Heymans S. Acute viral myocarditis. Eur Heart J. 2008;29:2073–2082. doi: 10.1093/eurheartj/ehn296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowles NE, Ni J, Kearney DL, Pauschinger M, Schultheiss HP, McCarthy R, Hare J, et al. Detection of viruses in myocardial tissues by polymerase chain reaction. Evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003;42:466–472. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 30.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–155. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 31.Kindermann I, Barth C, Mahfoud F, Ukena C, Lenski M, Yilmaz A, et al. Update on myocarditis. J Am Coll Cardiol. 2012;59:779–792. doi: 10.1016/j.jacc.2011.09.074. [DOI] [PubMed] [Google Scholar]
- 32.Wiltshire SA, Leiva-Torres GA, Vidal SM. Quantitative trait locus analysis, pathway analysis, and consomic mapping show genetic variants of tnni3k, fpgt, or h28 control susceptibility to viral myocarditis. J Immunol. 2011;186:6398–6405. doi: 10.4049/jimmunol.1100159. [DOI] [PubMed] [Google Scholar]
- 33.Wei YJ, Cui CJ, Huang YX, Zhang XL, Zhang H, Hu SS. Upregulated expression of cardiac ankyrin repeat protein in human failing hearts due to arrhythmogenic right ventricular cardiomyopathy. Eur J Heart Fail. 2009;11:559–566. doi: 10.1093/eurjhf/hfp049. [DOI] [PubMed] [Google Scholar]
- 34.Lodder EM, Scicluna BP, Milano A, Sun AY, Tang H, Remme CA, et al. Dissection of a quantitative trait locus for pr interval duration identifies tnni3k as a novel modulator of cardiac conduction. PLoS Genet. 2012;8:e1003113. doi: 10.1371/journal.pgen.1003113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curran J, Mohler PJ. Defining the pathways underlying the prolonged pr interval in atrioventricular conduction disease. PLoS Genet. 2012;8:e1003154. doi: 10.1371/journal.pgen.1003154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cornelis MC, Rimm EB, Curhan GC, Kraft P, Hunter DJ, Hu FB, et al. Obesity susceptibility loci and uncontrolled eating, emotional eating and cognitive restraint behaviors in men and women. Obesity (Silver Spring) 2013 doi: 10.1002/oby.20592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graff M, Ngwa JS, Workalemahu T, Homuth G, Schipf S, Teumer A, et al. Genome-wide analysis of bmi in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. Hum Mol Genet. 2013;22:3597–3607. doi: 10.1093/hmg/ddt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, et al. Obesity susceptibility loci and dietary intake in the look ahead trial. Am J Clin Nutr. 2012;95:1477–1486. doi: 10.3945/ajcn.111.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Bradfield JP, Zhang H, Sleiman PM, Kim CE, Glessner JT, et al. Role of bmi-associated loci identified in gwas meta-analyses in the context of common childhood obesity in european americans. Obesity (Silver Spring) 2011;19:2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]