Abstract
Sequestosome 1/p62 (p62) is a scaffold/adaptor protein with multiple functions implicated for neuronal and bone diseases. It carries a ubiquitin binding domain through which it mediates proteasome-dependent proteolysis. In addition, p62 is reported to regulate NF-κB activity in some cells. To date, however, the role of p62 in innate immunity has not been fully elucidated. In this study, we report that IFN-γ plus TLR signaling stimulates late expression of p62 in murine macrophages. Overexpression of p62 inhibited expression of multiple cytokines, IL-12p40, TNF-α, IL-1β, IL-6, and IFN-β, whereas p62 underexpression by small hairpin RNA markedly elevated their expression, indicating that p62 is a broad negative regulator of cytokine expression in stimulated macrophages. We show that p62 interacts with IFN regulatory factor 8 and Ro52, the transcription factor and ubiquitin E3 ligase that are important for IL-12p40 expression. This interaction, detectable at a late stage in stimulated macrophages, led to increased polyubiquitination and destabilization of IFN regulatory factor 8. We also show that upon macrophage stimulation, p62 binds to TNFR-associated factor 6, another E3 ligase important for NF-κB activation, but later this interaction was replaced by the recruitment of the deubiquitinating enzyme, cylindromatosis, an inhibitor of NF-κB activity. Recruitment of cylindromatosis coincided with reduced TNFR-associated factor 6 autoubiquitination and lower NF-κB activation. Our results indicate that p62 orchestrates orderly regulation of ubiquitin modification processes in macrophages to ensure attenuation of cytokine transcription postactivation. Together, p62 may provide a mechanism by which to control excessive inflammatory responses after macrophage activation.
Recognition of pathogen components by TLRs activates macrophages to induce many proinflammatory cytokines, leading to the establishment of innate immunity (1). Macrophages are highly sensitive to IFN-γ, which enables them to elicit enhanced cytokine responses (2, 3). Induction of proinflammatory cytokines, such as IL-1β, IL-6, and TFN-α, depends on the activation of NF-κB family of proteins (4–6). Induction of IL-12p40, in contrast, critically requires IFN regulatory factor (IRF)3 8, a transcription factor of the IRF family (7–11).
A large body of literature documents that ubiquitin modification is a major mechanism of regulating innate immune responses (12, 13). Ubiquitin modification controls NF-κB activation in multiple ways that affect cytokine and chemokine profiles. NF-κB is activated by autoubiquitination of TNFR-associated factor 6 (TRAF6), the RING family of E3 ubiquitin ligase. TRAF6 is a component of the TLR signaling pathways, and its autoubiquitination is required for inhibitor of IκB kinase (IKK) phosphorylation and the subsequent polyubiquitination and proteasome-mediated degradation of IκBα (14, 15). Ubiquitination of a downstream factor, receptor interacting protein, and recognition of ubiquitinated factors by IKKγ (NEMO) are also critical for NF-κB activation (16, 17). E3 ligases of the tripartite motif family, such as TRIM25 and TRIM30α, also act within the pathogen recognition pathways to regulate NF-κB activity (18, 19). Ubiquitination is an integral part of transcription of IL-12p40 as well, a master cytokine important for IFN-γ production (20): IRF8 is ubiquitinated by an IFN-inducible E3 ligase, Ro52, also called TRIM21, following macrophage activation (21). This ubiquitination is associated with enhanced transcription of IL-12p40. Ro52 is also reported to mediate ubiquitination of IRF3 following viral stimulation, which leads to down-regulation of IFN-β expression (22).
Ubiquitination is a widespread event occurring both in the cytoplasm and nucleus. It is a highly dynamic process, reversible by deubiquitinating enzymes. Indeed, some deubiquitinating enzymes are shown to participate in the regulation of innate immunity (13). Among them, cylindromatosis (CYLD), a tumor suppressor in the skin, is shown to negatively regulate NF-κB activity, by reducing TRAF6 autoubiquitination (13, 23–25).
Another family of proteins that regulate ubiquitin modification processes are those that bind to the ubiquitin moiety to facilitate proteasome-dependent clearance of ubiquitinated proteins (26, 27). Among them is the sequestosome 1, also known as p62 or A170 (hereafter p62), which functions as a scaffold/adaptor protein that has many biological activities (28–31). Mutations of p62 are associated with bone diseases (32, 33). Loss of the p62 gene results in impaired bone physiology in mice (34). p62 is also implicated for neuronal diseases, including Alzheimer’s disease (35) and Parkinson’s disease (36). A recent study indicates that p62 may have a role in some tumors (37). The ubiquitin binding domain of p62 binds to proteins that are polyubiquitinated through lysine 63 (K63) of ubiquitin, which leads to protein aggregate formation and eventual proteasome-mediated processing (29, 38, 39). p62 is also known to promote polyubiquitination of TRAF6 and NF-κB activation in nerve growth factor-stimulated PC12 cells and in receptor activator for NF-κB ligand-stimulated osteoclasts (34, 40, 41). In HepG2 cells, p62 is involved in IL-1-stimulated NF-κB activity (42). Interestingly, a more recent study indicates that the p62/ TRAF6 complex interacts with a deubiquitinating enzyme, CYLD, suggesting that p62 assumes a dual role, capable of stimulating and inhibiting NF-κB activity under some conditions (43, 44). As such, p62 may serve as a coordinator of ubiquitin-mediated regulatory processes.
Despite its widespread expression, and the report that p62 participates in Th2-mediated adaptive immune responses (45), little is known as to the role of p62 in innate immunity. Our interest in p62 in innate immunity came from our earlier findings that IFN-γ/TLR stimulation triggers increased ubiquitination and ubiquitin binding of many nuclear proteins in macrophages, including that of p62 (J. Kim, D. E. Anderson, and K. Ozato, submitted for publication). In this study, we show that p62 is an IFN-γ/TLR-inducible protein that negatively regulates expression of many proinflammatory cytokines in activated macrophages. Studies of the underlying mechanisms led to the observations that p62 interacted with Ro52, an E3 ubiquitin ligase for IRF8. Although Ro52 enhanced the transcriptional activity of IRF8 in an early stage, it interacted with p62 in a later stage, which accelerated processing of IRF8, resulting in the inhibition of IL-12p40 expression. Moreover, p62, which interacted with TRAF6 in an early stage, interacted with CYLD, the deubiquitinating enzyme to the p62 complex, in a later stage. The delayed CYLD recruitment coincided with the reduced TRAF6 autoubiquitination and the inhibition of NF-κB activity, a likely basis of reduced cytokine expression. Our results indicate that p62 coordinates orderly regulation of ubiquitination-mediated processes in activated macrophages. The p62 activity observed in this study may represent a previously unrecognized mechanism of attenuating cytokine expression postactivation.
Materials and Methods
Cells and reagents
Raw264.7 (hereafter RAW) cells and 293T cells, both from American Type Culture Collection, were maintained in DMEM with 10% FBS. RAW cells were treated with 150 U/ml IFN-γ (PeproTech) overnight, followed by 100–500 ng/ml CpG DNA 1826 (21, 46). To prepare macrophages, bone marrow mononuclear cells (1 × 107cells) from C57BL/6 mice were incubated in the presence of 20 ng/ml M-CSF (Invitrogen) for 5 days. Nonadherent cells were removed before experiments. Abs to p62, Ro52, TRAF6, CYLD, ubiquitin, and TFIIB Abs were purchased from Santa Cruz Biotechnology; those for phospho-IKK and IKK from Cell Signaling Technology; and anti-hemagglutinin (HA) Ab from Roche.
Plasmids and transfection
To construct an expression vector for HA-tagged p62 (HA-p62), a p62 cDNA was first amplified from the Image clone (ID, 3487289) using the 5′-EcoRI primer, cggaattcatggcgtcgttcacggtgaaggcc, and the 3′-XhoI primer, cgggatcccttggccacagcactatcacaatg, and cloned into pcDNA3-HA, followed by confirmation of correct cloning by sequencing. The plasmid for V5-tagged p62 (V5-p62) was constructed by inserting the p62 cDNA fragment from above into pcDNA3-V5. pcDNA3-Ro52 and pcDNA3-HA-Ub were described (21). RAW cells were transfected with pcDNA3-HA-p62 or the empty vector (pcDNA3-HA) using LipofectaminePlus, and were selected with G418 (1 mg/ml; Invitrogen) for 4 wk. More than 10 stable clones were isolated and maintained in the presence of G418 (500 µg/ml). Two days before the IFN-γ/CpG stimulation, G418 was removed from the medium. V5-p62, Flag-tagged IRF8 (Flag-IRF8), Ro52, and HA-Ub were transiently transfected in 293T cells with SuperFect (Qiagen) for 24 h, according to the manufacturer’s protocol. Some samples were treated with 20 µM MG132 (Sigma-Aldrich) for a final 6 h.
p62 small hairpin RNA (shRNA) vector and transduction
p62 shRNA and control shRNA were cloned into pSUPER retro vector (Oli-goengine). The shRNA sequence used in this study was from +762 to +782 relative to the transcription start site, as follows: 5′-GGTTGACATTGATGT GGAACA-3′. The sequence for control shRNA was 5′-TGTAGATGGGTA CGATGCGTA-3′. Viral supernatants were prepared by transfection of the above vector into 293ET cells using LipofectaminePlus (Invitrogen) (47). Supernatants were collected 48 h later and used for transduction into RAW cells. The effect of p62 shRNA was monitored by quantitative RT-PCR (qRT-PCR) and by immunoblot analysis before each experiment.
RNA extraction and qRT-PCR
Total RNA was extracted from RAW cells with the TRIzol reagent (Invitrogen), reverse transcribed by using Superscript II with the random primers (Invitrogen), and amplified by PCR in a final reaction volume of 20 µl using the SYBR Green mixture (Applied Biosystems) with 3 pmol primers. qRT-PCR was performed using primers designed by Primer Express software (Applied Biosystems) in the ABI Prism 7500 Sequence Detection System (Applied Biosystems). The primer sequences for p62 were 5’-AGCTGCCCTCAGCCCTCTA-3’ and 5’-GGCTTCTCTTCCCTCCAT GTT-3’, and data were normalized by Gapdh transcript levels using the primers, 5’-GTGTTCCTACCCCCAATGT-3’ and 5’-TGTCATCATACT TGGCAGGTTTC-3’. Primer sequences or other genes are available upon request.
Immunoprecipitation and immunoblot analysis
Cells were washed with PBS and lysed with the lysis buffer containing 20 mM HEPES, 150 mM NaCl, 20% glycerol, 0.1% Nonidet P-40, 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 mM N-ethyl-maleimide, and the mixture of protease inhibitor (Roche) at 4°C for 30 min with shacking. For coimmunoprecipitation (Co-IP), lysates (0.5–1 mg) were precleared by protein G, and then incubated with 1 µg of Ab (against p62, IRF8, Ro52, CYLD, and HA) at 4°C overnight, followed by incubation with protein G for 6 h at 4°C Precipitates were washed three times in lysis buffer and eluted in 2× sample buffer. For the detection of TRAF6 autoubiquitination, cells were lysed in the modified radioimmunoprecipi-tation assay buffer (50 mM HEPES, 100 mM NaCl, 4 mM EDTA, 0.1% sodium deoxycholate, 0.5% Triton X-100, 10 mM NaF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 mM N-ethylmaleimide, and the mixture of protease inhibitor), and lysates (500 µl) were incubated with 1 µg of Ab overnight at 4°C, followed by incubation with protein G for 6 h at 4°C Precipitates were washed three times in radioimmunoprecipitation assay buffer and eluted in 2× sample buffer. Eluted materials were resolved on a Nu-PAGE gel (Invitrogen), transferred onto polyvinylidene difluoride membrane (Millipore), and immunoblotted with indicated Abs, and Ab binding was detected using the ECL kit (SuperSignal West Dura Extended Duration Substrate; Pierce).
IL-12p40 promoter and ELISA analysis
A quantity amounting to 1 µg of plasmids containing murine IL-12p40 luciferase reporter (from −413 to +12) (48), Flag-IRF8, Ro52, HA-p62, and Renilla luciferase plasmid was transiently transfected into 293T cells using SuperFect, as above. Luciferase activity was measured 24–36 h after transfection with the Dual-Luciferase Reporter Assay System (Promega). RAW cells expressing p62 shRNA or HA-p62 and corresponding control cells were stimulated with IRF-γ/TLR for 24 h, and supernatants were tested by IL-12p40 protein by ELISA using the kit (Pierce).
Results
Induction of p62 by IFN-γ in macrophages
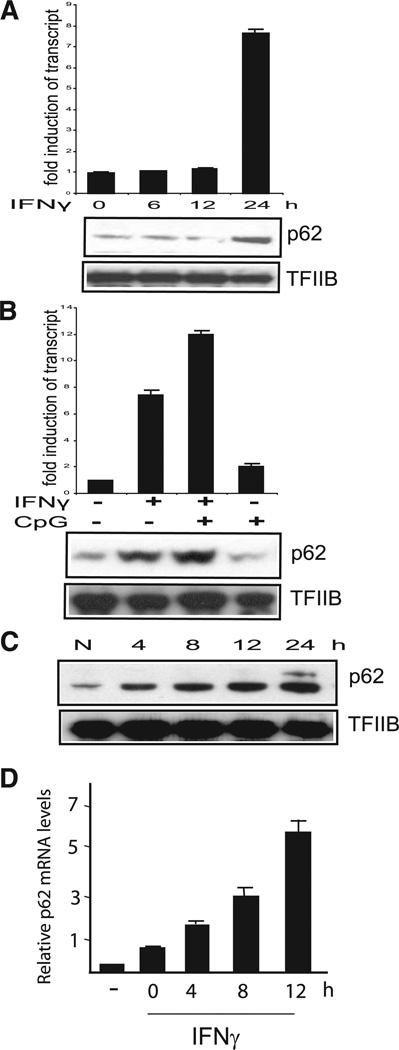
Our previous proteomic analysis identified p62 as one of ubiqui-tinated/ubiquitin-binding proteins that are increased in stimulated macrophages (J. Kim, D. Anderson, and K. Ozato, submitted for publication). In light of the paucity of information regarding the expression of p62 in macrophages, we examined whether p62 is induced in response to IFN-γ and TLR stimulation in RAW macrophages. As shown in Fig. 1A, p62 transcripts and the protein were expressed in unstimulated RAW cells at moderate levels. The p62 transcripts and proteins were markedly increased 24 h after IFN-γ treatment, indicating that p62 is a late responder to IFN-γ. Many other IFN-γ-responsive genes, including IRF8 and Ro52, were shown to be expressed much earlier, within 4 h (21, 49–51), suggesting that the function of p62 may be linked to a late event in macrophage activation. We found that when cells were pretreated with IFN-γ for 20 h, the subsequent treatment with CpG for 4 h further increased p62 expression, although CpG treatment alone did not stimulate p62 expression (Fig. 1B). Immunoblot data in Fig. 1C show that p62 levels increased as IFN-γ/CpG treatment was extended up to 24 h. The upper p62 band seen at 24 h most likely represents ubiquitinated p62 (52). Similarly, p62 was induced by IFN-γ with and without CpG in bone marrow-derived macrophages, and another macrophage line J774A.1 (Fig. 1D; data not shown). We also found that bacterial polysaccharides (LPS) similarly increased p62 expression when added following IFN-γ treatment (data not shown). These results led us to conclude that p62 is an IFN-γ-inducible gene whose expression is further increased by TLR signaling in macrophages. Thus, in all experiments below, RAW cells were primed with IFN-γ overnight, followed by CpG stimulation.
FIGURE 1.
Induction of p62 by IFN-γ and CpG stimulation in macrophages. A, RAW cells were stimulated with IFN-γ (150 U/ml) for indicated times. p62 transcripts and p62 protein levels were measured by qRT-PCR and immunoblot, respectively. Values represent the average of three determinations ± SD. B, Cells were pretreated with IFN-γ for 20 h and/or stimulated with CpG (150 ng/ml) for 4 h, and p62 expression was detected as above. C, Cells were pretreated with IFN-γ overnight and then stimulated with CpG (150 ng/ml) for indicated times, and p62 protein expression was tested by immunoblot analysis. N, Indicates neither IFN-γ nor CpG treatment. D, Bone marrow-derived macrophages were treated with IFN-γ (100 U/ml) overnight and then stimulated with CpG (150 ng/ml) for indicated times, and p62 transcript levels were measured as above. –, Denotes no IFN-γ treatment.
p62 attenuates IFN-γ/TLR-stimulated IL-12p40 expression
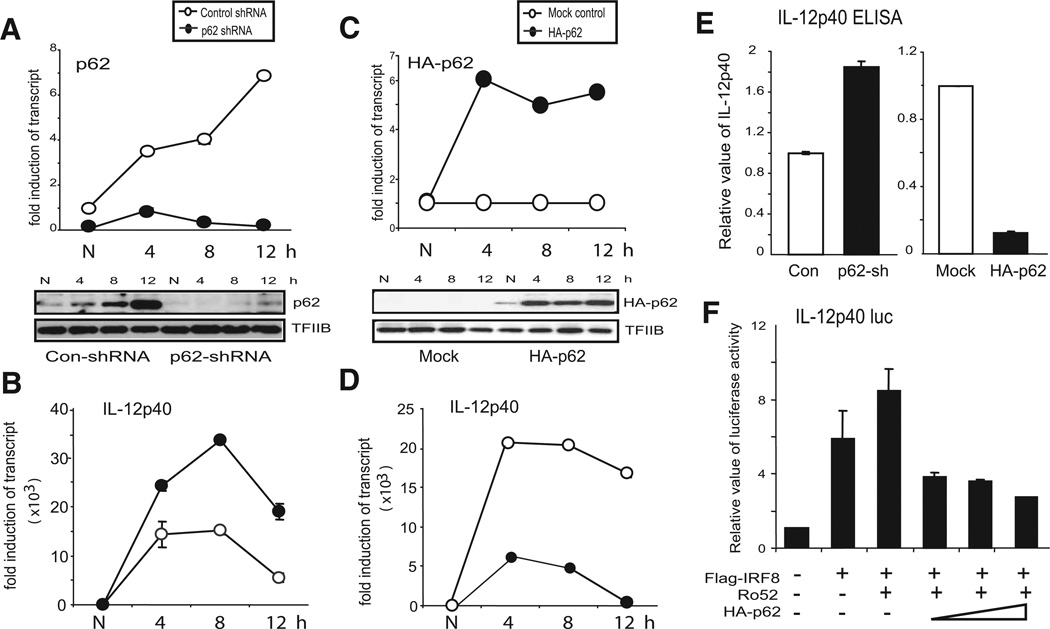
To address the biological function of p62 in cytokine expression in macrophages, we introduced a p62 shRNA vector into RAW cells. As shown in Fig. 2A (upper panel), p62 mRNA levels were markedly reduced in cells with p62 shRNA both before and after IFN-γ/CpG treatment compared with cells with control shRNA. Cells with p62 shRNA showed little p62 induction after IFN-γ/TLR stimulation, indicating effective p62 knockdown by this shRNA. In agreement, p62 protein expression was strongly inhibited in knockdown cells (Fig. 2A, lower panel). TFIIB expression was similar in control and knockdown cells before and after stimulation, supporting specificity of p62 knockdown. We observed ~90% inhibition of p62 expression in RAW cells using this p62 shRNA vector. IL-12p40, a subunit of the master cytokine IL-12, is one of the cytokines induced in macrophages by IFN-γ/TLR (20). Therefore, we tested IL-12p40 transcript expression in p62 knockdown cells. As shown in Fig. 2B, IFN-γ/CpG induced IL-12p40 mRNA by more than 1000-fold both in control and in p62 knockdown cells. However, the IL-12p40 mRNA levels were significantly higher in knockdown cells at all three time points after stimulation, indicating that p62 negatively regulates IL-12p40 expression in macrophages. These results led us to predict that p62 overexpression might, conversely, inhibit IL-12p40 expression. To test this prediction, we stably transfected RAW cells with a plas-mid vector for a HA-tagged p62 (HA-p62), and generated a number of clones stably overexpressing HA-p62. Fig. 2C (upper panel) shows results of one such clone, in which p62 transcripts and protein levels were strongly increased after IFN-γ/CpG stimulation. IFN-γ/CpG-stimulated expression of exogenous HA-p62 is not surprising, because this vector is driven by the CMV promoter that responds to IFNs (53). Immunoblot and qRT-PCR analyses indicated that total p62 levels were increased in HA-p62-transfected clones over mock-transfected cells by ~2-fold (data not shown). As shown in Fig. 2D, IL-12p40 mRNA induction in cells with HA-p62 was substantially lower than in cells with vector alone. Among 10 stable clones tested, those expressing significant HA-p62 all showed reduced IL-12p40 expression compared with control cells. In Fig. 2E, we measured the amount of IL-12p40 protein by ELISA. Consistent with the transcript data above, IL-12p40 protein levels were significantly higher in p62 knockdown cells, and lower in HA-p62-expressing cells, relative to their respective control cells.
FIGURE 2.
p62 inhibits IFN-γ/CgG-stimulated IL-12p40 expression. A, Knockdown of p62 by shRNA. RAW cells were transduced with a retroviral vector for p62 shRNA or control shRNA and stimulated with IFN-γ overnight, followed by CpG for indicated period of time. Upper panel, Represents the levels of p62 transcripts; lower panel, shows p62 protein expression. B, Enhanced expression of IL-12p40 in p62 knockdown cells. Cells with p62 shRNA (●) or control shRNA (○) were stimulated with IFN-γ/CpG, as in A, and IL-12p40 mRNA levels were quantified by qRT-PCR. The values represent the average of three determinations ± SD. C, Overexpression of HA-tagged p62. RAW cells were stably transfected with HA-p62, and a selected clone was tested for expression of HA-p62 transcripts and proteins, as above. D, Reduced expression of IL-12p40 transcripts by p62 overexpression. Cells expressing HA-p62 or empty vector (Mock) were stimulated with IFN-γ/CpG, and IL-12p40 mRNA levels were measured, as in B. E, Cells with p62 knockdown cells, cells expressing HA-p62, and respective control cells were stimulated with IFN-γ/CpG for 24 h, and IL-12p40 in supernatants was measured by ELISA. The amounts of IL-12p40 were normalized to respective control cells, which produced 350–425 pg/ml proteins. F, Inhibition of IL-12p40 reporter activity by ectopic HA-p62 expression. Cells were transiently transfected with IL-12p40 luciferase reporter along with expression vectors for IRF8, Ro52, and p62 (150, 300, and 550 ng) for 30 h. pcDNA-HA was added to adjust the total amount of plasmids to be 850 ng in all samples. Reporter activity was normalized by Renilla luciferase activity. Values represent the average of three assays ± SD.
IL-12p40 expression is a critical event in macrophage innate immunity (20). It has previously been shown that IRF8 is required for IFN-γ- and TLR-stimulated IL-12p40 induction in macrophages and dendritic cells (7–10). A more recent study showed that ubiquitination of IRF8 contributes to enhanced IL-12p40 transcription (21). It was shown that Ro52, a TRIM family member of E3 ubiquitin ligases, was induced after IFN-γ/CpG stimulation, and interacted with IRF8, resulting in IRF8 ubiquitination, which coincided with enhanced IL-12p40 production in RAW cells. To assess whether p62 inhibition of IL-12p40 is mediated by IRF8, we next examined the effect of p62 on IRF8-dependent IL-12p40 promoter activity (Fig. 2F). As expected, transfection of IRF8 alone enhanced IL-12p40 promoter activity, which was further augmented by cotransfection of Ro52. However, additional cotrasfection of p62 reduced the promoter activity to ~50% in a Ro52 dose-dependent manner. These results indicate that p62 regulates IL-12p40 expression by inhibiting the activity of IRF8 and Ro52.
p62 interacts with the IRF8/Ro52 complex
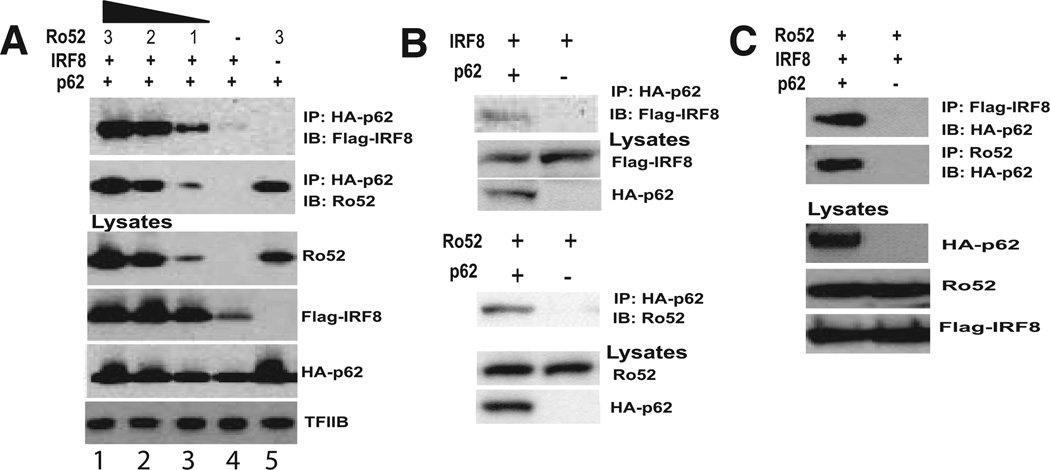
Above data indicated that p62 may regulate the activity of IRF8/ Ro52. To address this possibility, we next examined whether p62 could directly interact with these proteins. Co-IP assays were performed with 293T cells transfected with HA-p62, Flag-IRF8, and Ro52. In Fig. 3A, lysates were immunoprecipitated with anti-HA Ab, and immunoblotted for IRF8 and Ro52. Both IRF8 and Ro52 were coprecipitated along with HA-p62. Interestingly, when IRF8 was transfected without Ro52, only a small amount of IRF8 was coprecipitated with HA-p62. In contrast, when Ro52 was transfected without IRF8, a large amount of Ro52 was coprecipitated with HA-p62 coprecipitated with HA-p62 (lane 5), indicating that p62 bound to Ro52 more strongly than IRF8, and that IRF8 was brought down by p62 partly through its interaction with Ro52. To ascertain whether coprecipitation of IRF8 and Ro52 was due to their interaction with p62, rather than cross-reactivity of HA Ab, Co-IP experiments were performed in the presence or absence of HA-p62. In Fig. 3B, IRF8 (upper panel) and Ro52 (lower panel) were precipitated only in the presence of HA-p62, but not in the absence. Reciprocal Co-IP experiments in Fig. 3C showed that anti-Flag Ab and anti-Ro52 Ab coprecipitated HA-p62, further supporting the interaction of p62 with IRF8 and Ro52.
FIGURE 3.
Ro52-mediated interaction of p62 with IRF8. A, 293T cells were transfected with expression vectors for Ro52 (3, 2, and 1 µg) along with HA-p62 (3 µg) and Flag-IRF8 (3 µg). Lysates were precipitated with anti-HA Ab, and the eluted materials were blotted against anti-Flag Ab (IRF8, upper panel) and anti-Ro52 Ab (lower panel). The bottom four panels represent immunoblot data to confirm the expression of transfected proteins. B, Cells were transfected with Flag-IRF8 (upper panel) or Ro52 (lower panel) along with HA-p62 or empty vector. Lysates were precipitated with anti-HA Ab and blotted for Flag-IRF8 or Ro52. C, Cells were transfected with vectors for Flag-IRF8, Ro52, along with HA-p62 or empty vector. Lysates were precipitated with anti-Flag (upper panel) or anti-Ro52 Ab and blotted for HA-p62.
p62 captures polyubiquitinated IRF8 and facilitates its degradation
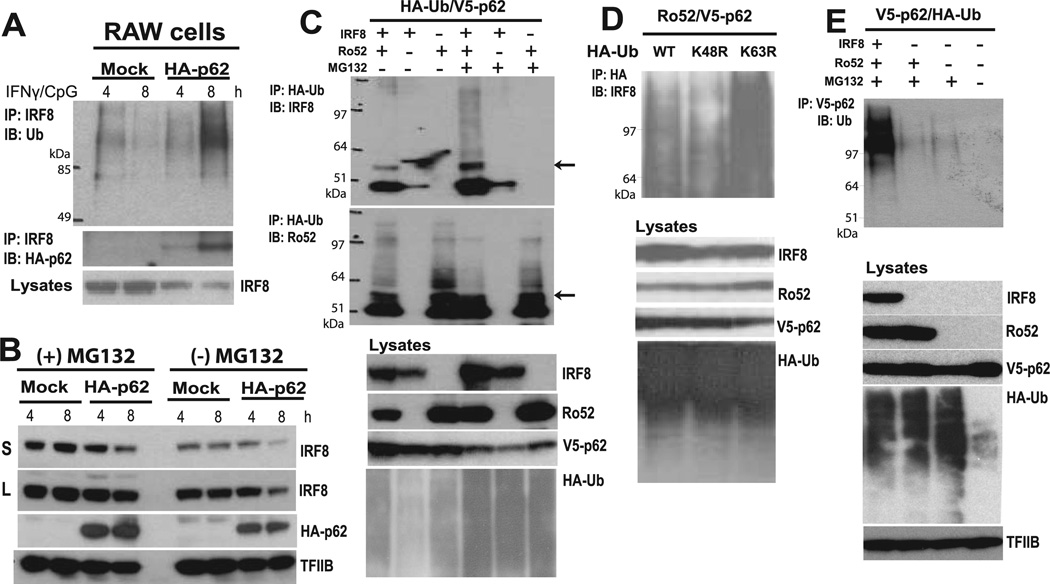
We next sought to assess whether p62 influenced processing of ubiquitinated IRF8 after IFN-γ/CpG stimulation. Because expression of p62 was delayed relative to that of Ro52 and IRF8 after IFN-γ/CpG stimulation, we wondered whether p62 may exert its function in a delayed fashion. In Fig. 4A, control RAW cells or RAW cells expressing HA-p62 were stimulated with IFN-γ/CpG for 4 or 8 h; endogenous IRF8 were imunoprecipitated with anti-IRF8 Ab; and precipitates were tested for IRF8 ubiquitination by anti-ubiquitin Ab. In HA-p62-expressing cells, the IRF8 immune precipitates showed extensive ubiquitination at 8 h, although only a modest amount of ubiquitinated IRF8 was detected at 4 h. However, in control cells, the amounts of ubiquitinated IRF8 were much lower both at 4 and 8 h. Importantly, we noted that the total IRF8 protein levels were substantially lower in HA-p62-expressing cells than in control cells, which was particularly noticeable at 8 h, indicating that HA-p62 accelerated IRF8 processing (Fig. 4A, lower panel). To establish that p62 affects the stability of IRF8, immunoblot analysis was performed in the presence or absence of proteasome inhibitor, MG132. Data in Fig. 4B showed that IRF8 protein levels were reduced in HA-p62-expressing cells both in the presence and absence of MG132, and that IRF8 levels were higher when cells were incubated with MG132 than without. These data support the idea that p62 facilitates ubiquitination of IRF8 in stimulated macrophages and promotes IRF8 destabilization through proteasome-mediated degradation. To verify that these effects were dependent on Ro52, we next tested IRF8 ubiquitination in cells cotransfected with Ro52 and HA-ubiquitin. In Fig. 4C (upper panel), anti-HA Ab precipitated mono- and polyubiquitinated IRF8. The amounts of ubiquitin-conjugated IRF8 were greater when coexpressed with Ro52, than without Ro52 (see an arrow for monoubiquitinated IRF8). Furthermore, broad polyubiquitinated IRF8 species, migrating above monoubiquitinated species, were more visible in the presence of MG132. As reported, Ro52 itself also showed extensive autoubiquitination, whose levels did not seem to differ in the presence or absence of MG132 (lower panel) (54). These data are consistent with the notion that polyubiquiti-nated IRF8 is escorted to proteasome-dependent degradation. As seen in Fig. 4C (bottom panel), MG132 stabilized other ubiquitinated proteins, as would have been expected. p62 has been shown to interact with TRAF6 ubiquitinated through K63 (30, 41), although in other papers p62 was shown to bind to proteins ubiqui-tinated through K48 as well (33, 55). To assess whether IRF8 ubiquitination was through a linkage by K48 or K63, Co-IP analysis was performed with cells transfected with wild-type or mutant ubiquitins in which K at position 48 or 63 was replaced by R. In Fig. 4D, all three ubiquitins were coprecipitated with IRF8, although the K48R mutant showed slightly less ubiquitination, and K63R slightly more ubiquitination than wild-type ubiquitin. These data suggest that IRF8 is ubiquitinated both through K48 and K63, although K48-mediated ubiquitination appears somewhat more predominant. To examine proteins other than IRF8 that may be targeted by p62 for ubiquitination, cells were transfected with V5-p62, and proteins precipitated by anti-V5 Ab were blotted for ubiquitin (Fig. 4E). V5-p62 precipitated a large amount of ubiquitinated materials when cells were transfected with IRF8 and Ro52. However, p62 precipitated a very low amount of ubiquitinated materials when transfected with Ro52 alone without IRF8, indicating that IRF8 itself was the major substrate of ubiquitination under these conditions. Again, ubiquitination of other proteins was equivalent regardless of the presence of IRF8 and Ro52, indicating that many other proteins were ubiquitinated independently of p62 (bottom panel). Together, these results indicate that p62 interacts with the IRF8/Ro52 complex, facilitates polyubiquitination of IRF8, and subsequently shepherds it to the proteasome system for degradation.
FIGURE 4.
p62 binds to polyubiquitinated IRF8. A, RAW cells transfected with empty vector (Mock) or HA-p62 were stimulated with IFN-γ/CpG for indicated periods. MG132 was added to the culture for the final 4 h. Lysates were precipitated by anti-IRF8 Ab and tested for for ubiquitin (Ub, upper panel) and p62 (lower panel) by immunoblot analysis. B, RAW cells transfected with empty vector (Mock) or HA-p62 were stimulated with IRFγ/CpG for 4 and 8 h in the presence or absence of MG132 for the final 4 h. Lysates were immunoblotted for indicated proteins. IRF8 levels were tested after a short (S) or a long (L) exposure. C, 293T cells were transfected with expression plasmids for HA-ubiquitin, V5-p62, along with Flag-IRF8 and Ro52 in the presence or absence of MG132. Lysates were precipitated with anti-HA Ab and blotted for IRF8 (upper panel) or Ro52 (lower panel). Middle panel, Total lysates were immunoblotted for indicated proteins to confirm proper transfection. Bottom panel, Indicates total HA-ubiquitin-conjugated proteins. D, 293T cells were transfected with Flag-IRF8, Ro52, V5-p62, and HA wild-type ubiquitin (Ub, WT) or HA-tagged ubiquitin mutants in which lysine (K) residue at 48 or 63 was replaced by arginine (R) (K48R, K63R). Lysates were precipitated by anti-HA Ab and blotted for IRF8. MG132 was added for the final 6. E, Cells were transfected with V5-p62 and HA-ubiquitin along with Flag-IRF8 and Ro52 in the presence or absence of MG132 for the final 6 h. Lysates were precipitated with anti-V5 Ab and blotted for HA-ubiquitin by HA Ab. Middle panels, Lysates were immunoblotted for indicated proteins to monitor expression of transfected proteins and total ubiquitin-conjugated proteins.
p62 inhibits induction of other proinflammatory cytokines
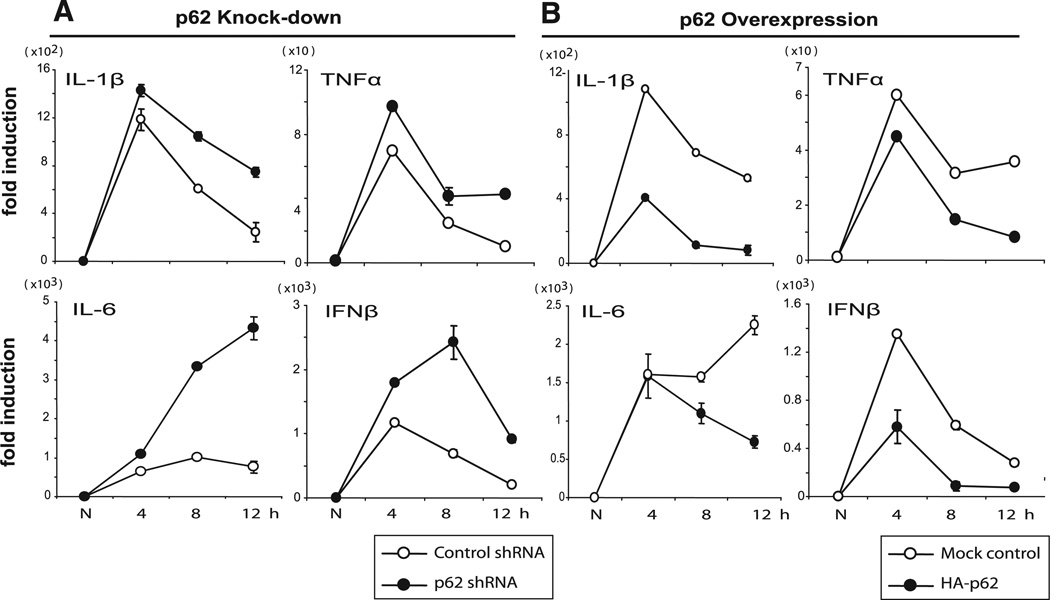
IFN-γ/TLR signaling triggers induction of not only IL-12p40, but other proinflammatory cytokines, many of which are induced in a manner dependent on activation of NF-κB (4, 5). In light of the previous reports that p62 has a positive role in NF-κB activation in neuronal and bone cells, it was of importance to test whether p62 stimulates NF-κB-dependent cytokine gene expression in macrophages. In Fig. 5, we examined the effect of p62 knockdown and overexpression on the expression of IL-1β, TNF-α, IL-6, and IFNβ transcripts. Similar to the results for IL-12p40, expression of all of these cytokines was substantially elevated in p62 shRNA cells (Fig. 5A). The effect was more pronounced at later time points, namely 8 and 12 h after stimulation, compared with an early time point at 4 h. Conversely, expression of these cytokines was universally reduced in cells with HA-p62 expression, compared with control cells (Fig. 5B). In all cases, transcript expression fell faster and more precipitously after stimulation in p62-overexpressing cells relative to control cells, although there was a clear initial peak at 4 h. These results suggested that p62 inhibits expression of NF-κB-dependent cytokine genes in IFN-γ/CpG-stimulated macrophages.
FIGURE 5.
The opposite regulation of proinflammatory cytokine gene expression by p62 knockdown and p62 overexpression. A, RAW cells expressing control or p62 shRNA were stimulated with IFN-γ/CpG for indicated times (h), and transcripts for the indicated cytokine were measured by qRT-PCR. Values represent the average of three assays ± SD. B, RAW cells overexpressing HA-p62 were tested for expression of the cytokines, as above. Three other clones expressing HA-p62 tested showed similar inhibition.
p62 interacts with the deubiquitinating enzyme, CYLD, in IFN-γ/CpG-stimulated macrophages
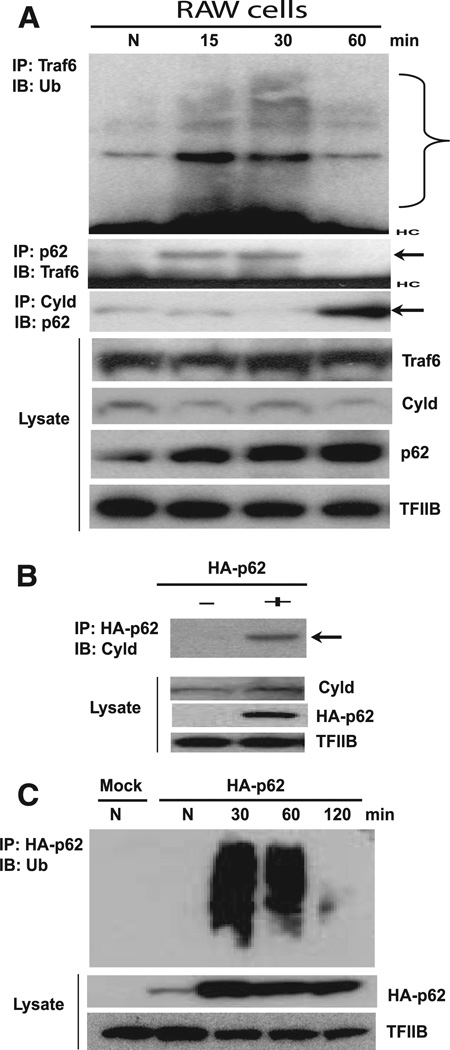
The above data pointed to the possibility that p62 inhibited expression of these cytokines by regulating NF-κB activity. Previously, p62 has been shown to promote NF-κB activation in various cells by enhancing TRAF6 ubiquitination (34, 40–42). TRAF6 is a ubiquitin E3 ligase that is activated by TLR ligands among other stimuli, and its K63-mediated autoubiquitination is required for activation of IKKβ and degradation of IκB that leads to the release of active NF-κB (5). Adding complexity to the issue, recent studies showed that a deubiquitinating enzyme, CYLD, a known negative regulator of NF-κB activity, interacts with p62 as well as TRAF6 to dysregulate NF-κB activity under certain conditions (43, 44). CYLD is shown to negatively regulate TLR- and TNFR-mediated innate immunity as well (13, 23, 24). These studies raised the possibility that p62 may have a dual activity capable of both up-and down-regulating NF-κB activity in macrophages. Thus, we examined the interaction of CYLD with p62 and autoubiquitination of TRAF6 following IFN-γ/CpG stimulation in RAW cells. In Fig. 6A, endogenous TRAF6 showed strong ubiquitination 30 min after stimulation, but ubiquitination levels were diminished subsequently, at 60 min (upper panel), indicating a rapid fall in TRAF6 autoubiquitination after stimulation. TRAF6 was coprecipitated with p62 at 15 and 30 min after stimulation, but levels of coprecipitated TRAF6 diminished thereafter (60 min; Fig. 6A, middle panel). Moreover, p62 coprecipitated a large amount of CYLD at 60 min, although before this time, only a low level of CYLD was precipitated (Fig. 6A, middle panel). These data indicated that CYLD was recruited to the p62 complex subsequent to the interaction of p62 with TRAF6, which led to reduced TRAF6 autoubiquitination. Given that TRAF6 interacts with CYLD in some cells (43, 44), it is possible that CYLD recruitment was mediated by the initial interaction with TRAF6. In Fig. 6B, the interaction of p62 with CYLD was also seen in a reciprocal Co-IP after IFN-γ/ CpG stimulation. To gain functional insight into the late interaction of p62 with CYLD, we examined ubiquitination of total p62- associated proteins following stimulation. As seen in Fig. 6C, the amounts of ubiquitinated and p62-bound proteins increased in an early stage after stimulation, but then declined sharply by 2 h, consistent with the idea that p62-bound proteins were processed later by CYLD.
FIGURE 6.
Interaction of p62 with TRAF6 and CYLD in stimulated macrophages. A, Parental RAW cells were stimulated with IFN-γ/CpG for indicated times. Lysates were precipitated by anti-TRAF6 Ab and blotted for ubiquitin by immunoblot (upper panel). The range of ubiquitinated proteins is bracketed. Lower panels, Lysates were precipitated with Ab against p62 or CYLD, and blotted for TRAF6 or p62 (marked by arrows). Bottom panels, Total lysates were tested for the expression of indicated proteins. B, RAW cells transfected with HA-p62 were stimulated without (−) or with (+) IFN-γ/CpG, and HA-p62 immune precipitates were blotted for the endogenous CYLD. Bottom panels, Indicate the expression of indicated proteins in total lysates. C, RAW cells transfected with empty vector (Mock) or HA-p62 vector were stimulated with IFN-γ/CpG for indicated periods, and HA-p62 immune precipitates were tested for reactivity with ubiquitin (Ub).
p62 overexpression inhibits TRAF6 ubiquitination and IKK phosphorylation
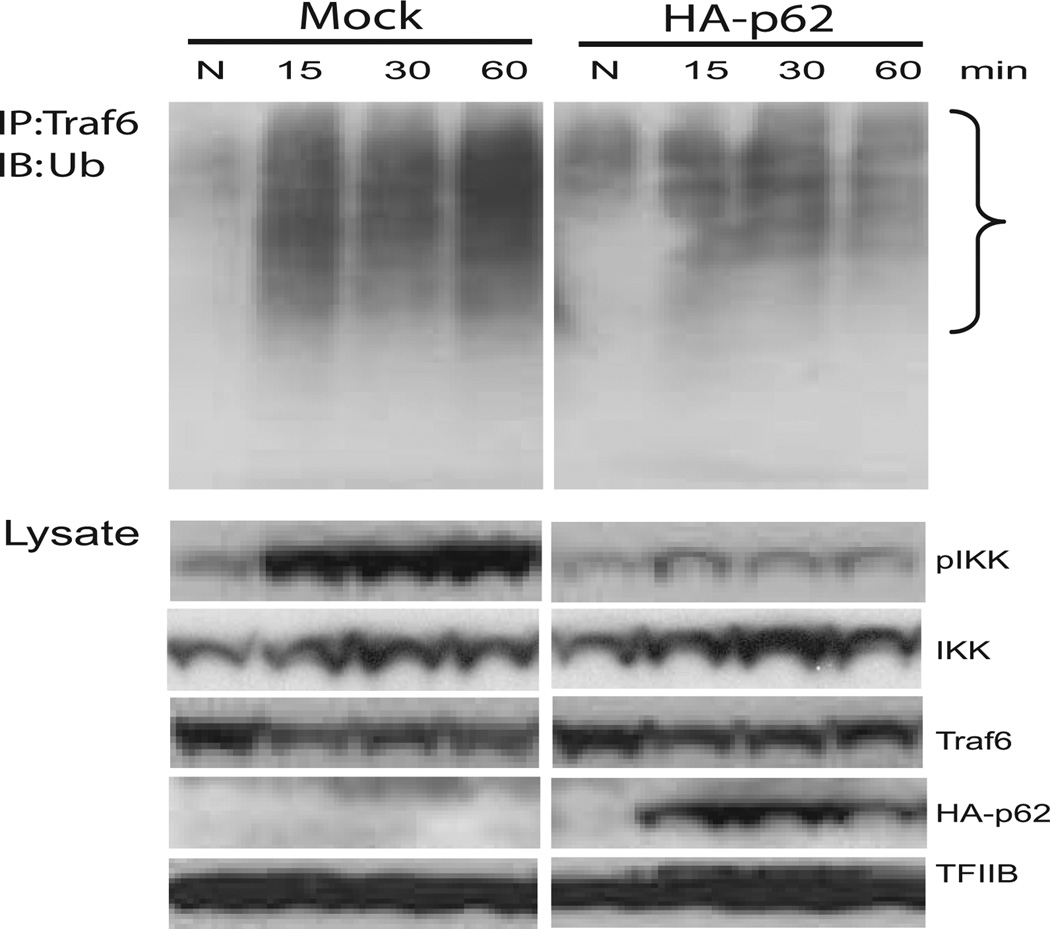
It is possible that in macrophages, p62 negatively regulates NF-κB activity by inhibiting the activation of TRAF6 and IKKs, required for NF-κB activation. To test this possibility, we examined TRAF6 ubiquitination and IKK phosphorylation in RAW cells expressing HA-p62. Data in Fig. 7 showed that the amounts of ubiquitinated TRAF6 were significantly lower in HA-p62-expressing cells compared with control cells, although TRAF6 ubiquitination was increased after stimulation in both cells (upper panel). Supporting the idea that ubiquitination, rather than TRAF6 expression, was affected by p62, total TRAF6 protein levels were similar in these cells and throughout the stimulation (lower panel). Furthermore, phosphorylated IKK levels were significantly lower in HA-p62-expressing cells than control cells, although total IKK levels were similar between control and HA-p62 expression cells (see the lower, lysates panel). These results reinforce the notion that p62, upon IFN-γ/CpG induction, interacted with CYLD and inhibited TRAF6 autoubiquitination, leading to the negative regulation of NF-κB activity in macrophages.
FIGURE 7.
Reduced autoubiquitination of TRAF6 by p62 overexpression. Cells transfected with empty vector (Mock) or HA-62 were stimulated with IFN-γ/CpG for indicted times, and TRAF6 immune complexes were blotted for ubiquitin (upper panel). Total lysates were tested for phosphorylated IKK, total IKK, and indicated proteins (lower panel).
Discussion
p62 is a ubiquitin-binding protein with multiple biological activities, including the removal of polyubiquitinated proteins from the cells (30, 31, 38, 56). The present study revealed that p62 is induced by IFN-γ/TLR stimulation and acts as an important regulator of cytokine expression in activated macrophages. Overexpression and underexpression of p62 led to clear opposing effects pointing to the inhibitory role of p62 in cytokine expression. Subsequent studies to delineate underlying mechanisms revealed that p62 regulates ubiquitin modification steps that affect two distinct transcriptional pathways involving IRF8 and NF-κB.
The role of p62 in regulating IRF8
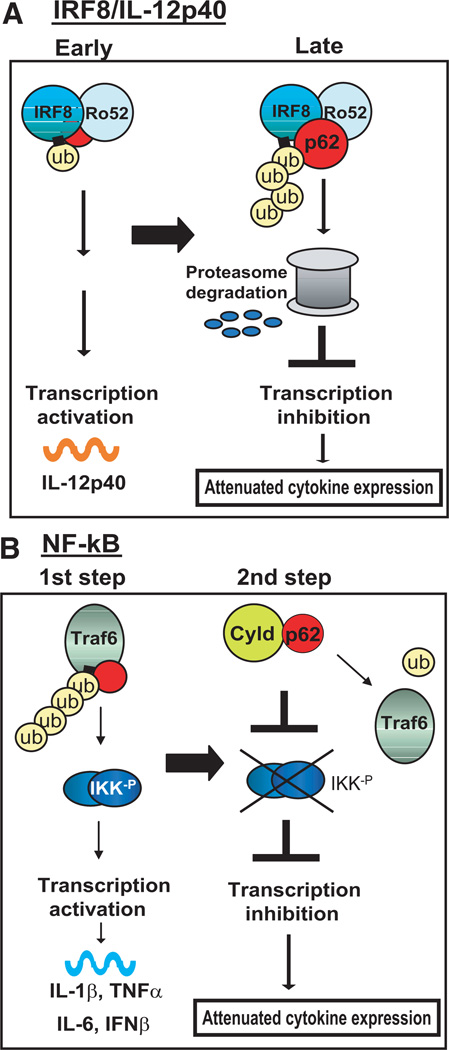
Inhibition of IL-12p40 expression by p62 was linked to increased polyubiquitination of IRF8 upon p62 expression, and the subsequent proteolysis. Increased IRF8 polyubiquitination was presumably mediated by Ro52, an IFN-inducible ubiquitin ligase that interacted with IRF8 (diagrammed in Fig. 8A). By interacting with p62, Ro52 may play two distinct roles in regulating IRF8. Induced by IFN-γ/TLR earlier than p62, Ro52 most likely stimulates the transcriptional activity of IRF8 in an early stage when p62 levels are too low to engage in an extensive interaction with Ro52 (21). In a later stage, when p62 expression is increased, Ro52 may promote greater ubiquitination of IRF8, eventually driving it to proteasome-mediated proteolysis. The notion that IRF8 is subjected to the activity of the ubiquitin/26S proteasome system is in line with the earlier report that this factor interacted with CSN2 (Trip15, the subunit of the COP9 signalosomes, a regulatory component of the proteasome) (57, 58). Our results indicate that p62 is a previously unsuspected regulator of Ro52-IRF8 interactions: p62 may be critically required for the transition of IRF8 from an active to an inactive state during macrophage stimulation. It should be noted in this study that whereas Ro52 was found in the p62 complex, another E3 ligase, Cbl, may also be involved with the activity of p62, based on the previous report that Cbl interacts with and ubiquitinates IRF8 (47).
FIGURE 8.
A model for a dual mode of p62 action. A, IRF8 and Ro52, transcription factors important for IL-12p40 expression, are induced by IFN-γ/TLR early, whereas p62 is induced later in macrophages. In an early stage, Ro52-mediated ubiquitination of IRF8 enhances IL-12p40 transcription. In a later stage as the p62 expression increases, the p62-Ro52 interaction increases, facilitating polyubiquitination of IRF8 and the subsequent degradation. B, p62 interacts with another E3 ubiquitin ligase, TRAF6, upon IFN-γ/TLR stimulation, causing TRAF6 autoubiquitination and NF-κB activation, leading to the induction of proinflammatory cytokines (the first step). Subsequently, the interaction of p62 with TRAF6 is replaced by that with CYLD to reduce TRAF6 autoubiquitination, weakening NF-κB activation (the second step). In this model, p62 orchestrates timely attenuation of cytokine expression by coordinating ubiquitin modification of signaling molecules and transcription factors.
The role of p62 in the regulation of NF-κB activity
It was evident that p62 has a negative effect on NF-κB-dependent cytokine expression in RAW cells. A series of cytokines, IL-1β, TNF-α, IL-6, and IFN-β, important for host resistance and inflammation, were all down-regulated by p62 overexpression and up-regulated by p62 knockdown. These results were at odds with the previous reports that p62 stimulates NF-κB activity in several other systems (34, 37, 40–42). The apparent dichotomy with the previous reports may be reconciled by the new insight into the dual role of p62 in NF-κB activity revealed recently. It was found that the deubiquitinating enzyme, CYLD, is recruited to the p62-TRAF6 complex, and that this recruitment results in dysregulation of TRAF6 ubiquitination and NF-κB activity (43, 44). CYLD has an inhibitory activity on NF-κB activation, and is shown to specifically remove polyubiquitin chains from TRAF6 (13). Disruption of the Cyld gene in the mouse results in increased production of NF-κB-dependent cytokines and heightened responses to TLR and TNFR (13, 23–25). What we found in macrophages was a time-dependent, ordered interaction of p62 with TRAF6 and CYLD. As diagrammed in Fig. 8B, whereas the p62-TRAF6 interaction was found immediately after IFN-γ/TLR stimulation, this interaction was largely replaced by the p62-CYLD interaction in a later stage, indicating that TRAF6 ubiquitination, triggered upon stimulation, was reversed at a later time, coinciding with the recruitment of CYLD. In light of constitutive p62 expression, one can surmise that the negative role of p62, mediated by its interaction with CYLD, is the predominant, net effect brought on the NF-κB activity in stimulated macrophages. Given that this was seen within 1–2 h after stimulation, much earlier than increased p62 expression, the ordered interaction of p62 with TRAF6 and CYLD during macrophage activation is not likely to be entirely attributed to inducible p62 expression, but may involve posttrans-lational modification of p62, such as ubiquitination (52) and phosphorylation (59). It is possible that CYLD inhibits NF-κB, not only through the regulation of TRAF6, but through the regulation of IKKγ (17, 23, 24). Furthermore, there remains the possibility that p62 interacts with other deubiquitinating enzymes such as A20, also known to regulate NF-κB activity (13, 60).
p62 as a coordinator of ubiquitin-mediated innate immune responses in macrophages
Together, p62 functions in two main pathways of cytokine transcription, by exerting complex, time-dependent regulation on transcription. It interacts with at least two ubiquitin ligases, Ro52 and TRAF6, through which it negatively regulates the activity of their targets and substrates, i.e., IRF8 and NF-κB. This negative regulation may have a greater impact on a later stage of macrophage activation, because p62 expression is markedly increased after IFN/TLR simulation. Collectively, our results place p62 as a central coordinator of cytokine gene regulation in macrophage postactivation. It seems reasonable to envisage that p62 provides a mechanism by which to quench a burst of inflammatory responses triggered by IFN-γ and pathogen signals.
Multiple layers of mechanisms exist to reverse cytokine gene expression in activated macrophages (61, 62). These mechanisms help prevent excessive inflammatory responses following pathogen/stress signaling. Proteins of the classical suppressor of cytokine signaling family and those of the protein inhibitor of activated STAT family are important players of this type of regulation. Recent reports that ubiquitination of IRF3 and modification of IRF7 by the small, ubiquitin-like molecules (Sumo) after stimulation indicate the presence of additional mechanisms to insure rapid down-regulation of inflammatory responses (22, 63). In addition to p62, there may be additional molecules that facilitate timely extinction of pathogen-activated gene expression.
Acknowledgments
We thank Drs. H. Kong, H. Xiong, and R. Yoshimi, and members of the Ozato laboratory for reagents, suggestions, and critical discussions of this work.
Footnotes
This work was supported by the intramural program of National Institute of Child Health and Human Development and in part by the Intramural AIDS Targeted Antiviral Program of the National Institutes of Health.
Abbreviations used in this paper: IRF, IFN regulatory factor; Co-IP, coimmunoprecipitation; CYLD, cylindromatosis; HA, hemagglutinin; IKK, inhibitor of IκB kinase; qRT-PCR, quantitative RT-PCR; shRNA, small hairpin RNA; TRAF6, TNFR-associated factor 6.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Kawai T, Akira S. TLR signaling. Semin. Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 3.O’Shea JJ, Murray PJ. Cytokine signaling modules in inflammatory responses. Immunity. 2008;28:477–487. doi: 10.1016/j.immuni.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor κBrecruitment to target promoters. J. Exp. Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayden MS, Ghosh S. Shared principles in NF-κB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 6.Sethi G, Sung B, Aggarwal BB. Nuclear factor-κB activation: from bench to bedside. Exp. Biol. Med. 2008;233:21–31. doi: 10.3181/0707-MR-196. [DOI] [PubMed] [Google Scholar]
- 7.Wang IM, Contursi C, Masumi A, Ma X, Trinchieri G, Ozato K. An IFN-γ-inducible transcription factor: IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J. Immunol. 2000;165:271–279. doi: 10.4049/jimmunol.165.1.271. [DOI] [PubMed] [Google Scholar]
- 8.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J. Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 9.Aliberti J, Schulz O, Pennington DJ, Tsujimura H, Reis e Sousa C, Ozato K, Sher A. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 10.Tsujimura H, Tamura T, Gongora C, Aliberti J, Reis e Sousa C, Sher A, Ozato K. ICSBP/IRF-8 retrovirus transduction rescues dendritic cell development in vitro. Blood. 2003;101:961–969. doi: 10.1182/blood-2002-05-1327. [DOI] [PubMed] [Google Scholar]
- 11.Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J. Biol. Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- 12.Liu YC, Penninger J, Karin M. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 2005;5:941–952. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun SC. Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 2008;8:501–511. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen ZJ. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, Akira S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Kobayashi M, Blonska M, You Y, Lin X. Ubiquitination of RIP is required for tumor necrosis factor α-induced NF-κB activation. J. Biol. Chem. 2006;281:13636–13643. doi: 10.1074/jbc.M600620200. [DOI] [PubMed] [Google Scholar]
- 17.Israel A. NF-κB activation: nondegradative ubiquitination implicates NEMO. Trends Immunol. 2006;27:395–397. doi: 10.1016/j.it.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 19.Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, et al. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- 20.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 21.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse III HC, Ozato K. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 22.Higgs R, Ni Gabhann J, Ben Larbi N, Breen EP, Fitzgerald KA, Jefferies CA. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 2008;181:1780–1786. doi: 10.4049/jimmunol.181.3.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumor suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumor suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Stirling B, Temmerman ST, Ma CA, Fuss IJ, Derry JM, Jain A. Impaired regulation of NF-κB and increased susceptibility to colitis-associated tumorigenesis in Cyld-deficient mice. J. Clin. Invest. 2006;116:3042–3049. doi: 10.1172/JCI28746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem. J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moscat J, Diaz-Meco MT, Rennert P. NF-κB activation by protein kinase C isoforms and B-cell function. EMBO Rep. 2003;4:31–36. doi: 10.1038/sj.embor.embor704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell. Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC. Signaling: polyubiquitination, trafficking, and inclusions: sequestosome 1/p62’s role in neurodegenerative disease. J. Biomed. Biotechnol. 2006:62079. doi: 10.1155/JBB/2006/62079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Laurin N, Brown JP, Morissette J, Raymond V. Recurrent mutation of the gene encoding sequestosome 1 (SQSTM1/p62) in Paget disease of bone. Am. J. Hum. Genet. 2002;70:1582–1588. doi: 10.1086/340731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavey JR, Ralston SH, Hocking LJ, Sheppard PW, Ciani B, Searle MS, Layfield R. Loss of ubiquitin-binding associated with Paget’s disease of bone p62 (SQSTM1) mutations. J. Bone Miner. Res. 2005;20:619–624. doi: 10.1359/JBMR.041205. [DOI] [PubMed] [Google Scholar]
- 34.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev. Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 35.Ramesh Babu J, Lamar Seibenhener M, Peng J, Strom AL, Kemppainen R, Cox N, Zhu H, Wooten MC, Diaz-Meco MT, Moscat J, Wooten MW. Genetic inactivation of p62 leads to accumulation of hyperphosphorylated tau and neurodegeneration. J. Neurochem. 2008;106:107–120. doi: 10.1111/j.1471-4159.2008.05340.x. [DOI] [PubMed] [Google Scholar]
- 36.Nakaso K, Yoshimoto Y, Nakano T, Takeshima T, Fukuhara Y, Yasui K, Araga S, Yanagawa T, Ishii T, Nakashima K. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 2004;1012:42–51. doi: 10.1016/j.brainres.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 37.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cells. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J. Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 39.Geetha T, Seibenhener ML, Chen L, Madura K, Wooten MW. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 2008;3741:33–37. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J. The atypical protein kinase C-interacting protein p62 is a scaffold for NF-κB activation by nerve growth factor. J. Biol. Chem. 2001;276:7709–7712. doi: 10.1074/jbc.C000869200. [DOI] [PubMed] [Google Scholar]
- 41.Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J. The p62 scaffold regulates nerve growth factor-induced NF-κB activation by influencing TRAF6 polyubiquitination. J. Biol. Chem. 2005;280:35625–35629. doi: 10.1074/jbc.C500237200. [DOI] [PubMed] [Google Scholar]
- 42.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J. Biol. Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 44.Jin W, Chang M, Paul EM, Babu G, Lee AJ, Reiley W, Wright A, Zhang M, You J, Sun SC. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J. Clin. Invest. 2008;118:1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin P, Diaz-Meco MT, Moscat J. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. EMBO J. 2006;25:3524–3533. doi: 10.1038/sj.emboj.7601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsujimura H, Tamura T, Kong HJ, Nishiyama A, Ishii KJ, Klinman DM, Ozato K. Toll-like receptor 9 signaling activates NF-κB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J. Immunol. 2004;172:6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 47.Xiong H, Li H, Kong HJ, Chen Y, Zhao J, Xiong S, Huang B, Gu H, Mayer L, Ozato K, Unkeless JC. Ubiquitin-dependent degradation of interferon regulatory factor-8 mediated by Cbl down-regulates interleukin-12 expression. J. Biol. Chem. 2005;280:23531–23539. doi: 10.1074/jbc.M414296200. [DOI] [PubMed] [Google Scholar]
- 48.Masumi A, Tamaoki S, Wang IM, Ozato K, Komuro K. IRF-8/ ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 2002;531:348–353. doi: 10.1016/s0014-5793(02)03556-1. [DOI] [PubMed] [Google Scholar]
- 49.Politis AD, Ozato K, Coligan JE, Vogel SN. Regulation of IFN-γ-induced nuclear expression of IFN consensus sequence binding protein in murine peritoneal macrophages. J. Immunol. 1994;152:2270–2278. [PubMed] [Google Scholar]
- 50.Lehtonen A, Matikainen S, Julkunen I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J. Immunol. 1997;159:794–803. [PubMed] [Google Scholar]
- 51.Brucet M, Marques L, Sebastian C, Lloberas J, Celada A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon γ is mediated by STAT1 and IRF-1. Genes Immun. 2004;5:26–35. doi: 10.1038/sj.gene.6364035. [DOI] [PubMed] [Google Scholar]
- 52.Aono J, Yanagawa T, Itoh K, Li B, Yoshida H, Kumagai Y, Yamamoto M, Ishii T. Activation of Nrf2 and accumulation of ubiquitinated A170 by arsenic in osteoblasts. Biochem. Biophys. Res. Commun. 2003;305:271–277. doi: 10.1016/s0006-291x(03)00728-9. [DOI] [PubMed] [Google Scholar]
- 53.Ramanathan M, Hasko G, Leibovich SJ. Analysis of signal transduction pathways in macrophages using expression vectors with CMV promoters: a cautionary tale. Inflammation. 2005;29:94–102. doi: 10.1007/s10753-006-9005-z. [DOI] [PubMed] [Google Scholar]
- 54.Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, Horvath L, Wallerskog T, Oukka M, Nyberg F, et al. The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J. Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- 55.Ciani B, Layfield R, Cavey JR, Sheppard PW, Searle MS. Structure of the ubiquitin-associated domain of p62 (SQSTM1) and implications for mutations that cause Paget’s disease of bone. J. Biol. Chem. 2003;278:37409–37412. doi: 10.1074/jbc.M307416200. [DOI] [PubMed] [Google Scholar]
- 56.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62: more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen H, Azriel A, Cohen T, Meraro D, Hashmueli S, Bech-Otschir D, Kraft R, Dubiel W, Levi BZ. Interaction between interferon consensus sequence-binding protein and COP9/signalosome subunit CSN2 (Trip15): a possible link between interferon regulatory factor signaling and the COP9/ signalosome. J. Biol. Chem. 2000;275:39081–39089. doi: 10.1074/jbc.M004900200. [DOI] [PubMed] [Google Scholar]
- 58.Huang X, Hetfeld BK, Seifert U, Kahne T, Kloetzel PM, Naumann M, Bech-Otschir D, Dubiel W. Consequences of COP9 signalosome and 26S proteasome interaction. FEBS J. 2005;272:3909–3917. doi: 10.1111/j.1742-4658.2005.04807.x. [DOI] [PubMed] [Google Scholar]
- 59.Yanagawa T, Yuki K, Yoshida H, Bannai S, Ishii T. Phosphory-lation of A170 stress protein by casein kinase II-like activity in macrophages. Biochem. Biophys. Res. Commun. 1997;241:157–163. doi: 10.1006/bbrc.1997.7783. [DOI] [PubMed] [Google Scholar]
- 60.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J. Biol. Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 61.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat. Rev. Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 62.Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat. Rev. Immunol. 2007;7:454–465. doi: 10.1038/nri2093. [DOI] [PubMed] [Google Scholar]
- 63.Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, Kato A, Ozato K. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 2008;283:25660–25670. doi: 10.1074/jbc.M804479200. [DOI] [PMC free article] [PubMed] [Google Scholar]