Abstract
The assembly of antigen receptors in developing B-lymphocytes is determined by the spatio-temporal organization of the immunoglobulin heavy chain locus (Igh). In this issue of Immunity, Medvedovicet al. (2013) provide a comprehensive dynamic view of the Igh locus architecture.
The ability of B cellsto recognize an almost infinite array of antigens relies on V(D)J recombination, a process that assembles the variable region genes of antigen receptors (or antibodies) from variable (V), diversity (D) and joining (J) gene segments. V(D)J recombination requires RAG1 and RAG2 endonucleases, which cleave DNA at recognition sites flanking the V, D and J gene segments, and the non-homologous end-joining machinery that ligates the broken DNA ends. The mouseIgH locus, which encodes the heavy chain of antibody molecules, spans 3Mbp in length and is comprised of approximately 200 VH genes dispersed over a 2.5Mbp region divided into distal, central and proximal segments. Downstream of the VH regions are 13-16 DH and 4 JH gene segments, as well as 8 CH regions that encode the Ighconstant regions of the various antibody isotypes(Figure 1) (Perlot and Alt, 2008).
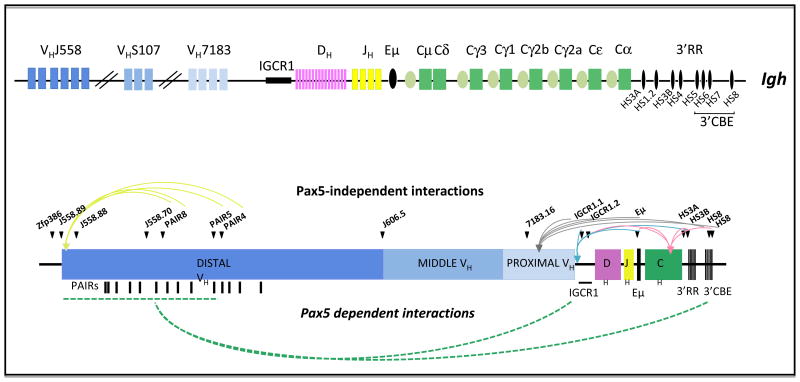
Fig 1. Dynamic architecture of the IgH locus.
Top.Schematic representation of the Ighlocus showing Variable (V), Diversity (D), Joining gene segments (J), Constant regions (C) and the regulatory elements 3′RR, Eμ, IGCR1 and PAIR elements. The CTCF-binding sequence (3′CBE) in the 3′RR region are shown. Bottom.Interactions in the absence or presence of Pax5. Green, grey, pink and blue arrows represent local interactions between the pointed sequences and the corresponding viewpoints. Broken green arrows represent dynamic long-range interactions. Distal VH genes interact mainly with CTCF-binding sites in IGCR1 and 3′CBE. Black triangles indicate the relative position of the 16 viewpoints that were employed for 4C-seq analyses.
In developing pro-B cells, a DH gene segment first recombines with a JH segment to form a DJH junction; subsequent VH to DJH recombination assembles a complete VH(D)JH allele.A salient feature of V(D)J recombination is the unbiased representation of the distal and proximal VH segments in the overall antigen receptor repertoire (Perlot and Alt, 2008). This leads to the central question: what is the mechanism by which each of the ∼200 VH gene segments scattered over a 2.5Mb region have an equal opportunity to establish contact and recombine with the DJH element? Elegant studies employing fluorescence in situ hybridization (FISH) (Fuxa et al., 2004; Kosak et al., 2002; Roldan et al., 2005), combined with painstaking measurement of different contact points throughout the Ighlocus using 3D-FISH and trilateration (Jhunjhunwala et al., 2008)demonstrated that VH to DJH recombination proceeds via large-scale locus contraction occurring through chromatin looping, thus juxtaposing distal VH gene segments close to the DJH element. Several transcription factors, including Pax5, Ezh2, YY1, Ikaros and the CCCTC-binding factor CTCF, interact with various cis- regulatory elements in the Ighlocus to mediate locus contraction (Perlot and Alt, 2008).
The Ighlocus contains four well-characterized regulatory elements (Figure 1). (i) 3′RR: Positioned downstream of the Cα gene, the 3′ regulatory regionis comprised of two distinct modules of DNaseI hypersensitivity (hs) sites- hs3A-hs1.2-hs3B-hs4, and hs5-8 containing a high density of CTCF binding elements (CBEs) interspersed with Pax5-binding sites. (ii) Eμ: Located in the JH-CH intron, Em is required for efficient VH to DJH joining. (iii) PAIR elements: The PAX5-activated intergenic repeat elements are interspersed in the distal VH cluster, and contain binding sites for Pax5 and CTCF, and promote Pax5-dependent anti-sense transcription through theIghlocus in pro-B cells (Ebert et al., 2011). (iv) IGCR1: The intergenic control region is located between the VH and D clusters containing two CBEs. The CBEs in IGCR1 act as insulator elements to V(D)J recombination at the Ighlocus (Guo et al., 2011b).
The DNA-FISH methodology employed thus far has provided an overall idea of the topological architecture of the Ighlocus. However, these studies are of low resolution and do not provide a detailed map of the precise contact points for the myriad long- and short-range interactions that effect locus contraction. To establish a comprehensive map of the interaction domains in the Ighlocus poised to undergo recombination, Medvedovic et al., (2013)used chromosome conformation capture sequencing methodology (4C-seq) that allows unbiased identification of genome-wide interaction partners using defined bait elements or “viewpoints”. In aremarkabletour de forceeffort, Medvedovic et al used 16 distinct viewpoints spanning the entire Ighlocus to compare 3D-chromatin topology between Rag2-/- and Pax5-/- Rag2-/- pro-B cells. Rag2 deficiency “freezes” B cell development at the pro-B stage and thus provides a snapshot of the Pax5-dependent interactions at the IgH locus poised to undergo V(D)J recombination.
The 4C-seq data generated by Medvedovicet alrevealed several novel and interesting aspects of Ightopology that could not be gleaned from the 3D-FISH analysis. First, in pro B-cells, the majority of long-range IgH chromosomal interactions occurred specifically within the Ighlocus, with the region around the 5′ VHJ558 gene acting as the 5′ boundary and the 3′CBE in the hs5-8 module serving as the 3′ limit of the Ighlocus interactions.
Second, even though the regulatory elements 3′RR, Eμ and IGCBR1 form multiple loops across the entire Ighlocus, individual mutations of each of these elements had no effect on locus contraction of the VH genes, leaving open the possibilities that there are either redundant interaction domains and/or the interactions in pro-B cells reflect those that can occur in later stages of B cell differentiation, for example during Ighclass switch recombination (CSR). It is to be noted that the results here differ from a recent report demonstrating the requirement of the Eμ enhancer in Ighlocus contraction using 3D-FISH analyses(Guo et al., 2011a); the reasons behind this discrepancy is not clear at present. Third, local chromatin loops ranging in length from 0.5 Mbp to 1.3 Mbp were observed not only in the absence of Pax5 but also in thymocytes, indicating that these interactions represent the default folding state of the Ighlocus that is largely invariant between different cell types, reminiscent of the topological architecture of the entire mouse chromosome. Fourth, a region between Cγ1 and Cγ2b was found to interact with IGCR1, Eμ, and the 3′RR elements. This novel Cγ1- Cγ2b interaction domain contains two Pax5-dependent DNaseI hypersensitivity sites (DHS) in pro-B cells. Surprisingly, these loops occur even in the absence of both DHS sites in Pax5-deficient pro-B cells. Gene targeting to assess the potential role of this region in regulating loop formation during V(D)J recombination or class switch recombination will be of significant interest. Finally, 4C-seq data obtained with multiple viewpoints across the entire VH cluster in pro-B cells showed that long-range interaction with the VH segments occur in a Pax5-dependent fashion. Significantly, all the viewpoints revealed a continuum of flexible long-range interactions across the entire VH cluster, suggesting that each of the VH genes has a similar probability of being juxtaposed proximal to the rearranged DHJH element during VH-DJH recombination.
In addition to generating a detailed topological map of the Ighinteraction domains, Medvedovicet alelucidated mechanisms by which transcription factors potentially regulate long-range chromatin interactions. Biochemical experiments demonstrated interaction between CTCF and Pax5, suggesting that this association could be relevant for the dynamics of the Ighlocus. This is consistent with the 4C-seq data demonstrating that targeted mutation of CBE elements in IGCR1 abrogated Pax5-dependent interaction between IGCR1 and distal VH gene segments. Medvedovicet al also investigated the mechanism by which the transcription factor YY1 promotes Igh locus contraction. While YY1 could potentially mediate chromosomal interactions through binding sites scattered throughout the IgHlocus, antisense transcripts originating from PAIRelements were found to be severely reduced in YY1-deleted pro-B cells, suggesting that YY1 might control long-range interactions by promoting anti-sense transcription.
In summary, the study by Medvedovicet al not only confirmed genomic interactions predicted from low-resolution 3D-FISH but also identified novel Ighelements that potentially regulate V(D)J recombination. Furthermore, they provided a mechanistic explanation of how the entire complement of VH gene segments could be presented to the DJH element for recombination en route to the generation of an unbiased B cell repertoire. The study also establishes 4C-seq as a bona fide and convenient method to probe local and long-range chromosomal interactions in other processes such as CSR.
Results presented by Medvedovicet allead to additional questions. First, in pro-B cells, the Ighalleles relocate from repressive heterochromatic regions at the nuclear periphery to the center of the nucleus and this movement has been linked to recombination of distal VH genes (Fuxa et al., 2004; Kosak et al., 2002; Roldan et al., 2005). Do locus contraction and nuclear repositioning influence each other or are they independent processes? Second, in pro-B cells, antisense non-coding RNAs traverse across the entire DH-JH region prior to their rearrangement, following which biallelic antisense transcription is initiated across the entire VH gene cluster (Perlot and Alt, 2008). How does anti-sense transcription modulate locus contraction? Does it alter the three dimensional structure of distinct chromatin territories to promote localized accessibility, or do anti-sense transcripts act as molecular scaffolds to recruit factors required for chromatin looping?Third, both Ighalleles undergo homologous pairing in pro-B cells to ensure that VH to DJH recombination occurs on only one Ighallele at a time to effect allelic exclusion (Hewitt et al., 2009). Is there cross-talk between chromosomal loops on individual Ighalleles to “read” the recombination status of the complement? Finally, the study by Medvedovicet al was performed in RAG-deficient B cells. However, RAG1and RAG2bind to active chromatin and localize to the JH segments at the 3′ end of each antigen receptor loci (Perlot and Alt, 2008). How does the topology of the Ighlocus change in the presence of the RAG proteins and following introduction of DNA breaks? These unanswered questions will constitute the next phase of investigation in this complex recombination reaction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes &Development. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Gerasimova T, Hao H, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen R. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011a;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Yoon HS, Franklin A, Jain S, Ebert A, Cheng HL, Hansen E, Despo O, Bossen C, Vettermann C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011b;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, et al. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nature Immunology. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Medvedovic, et al. 2013 This issue. [Google Scholar]
- Perlot T, Alt FW. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Advances in immunology. 2008;99:1–32. doi: 10.1016/S0065-2776(08)00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA. Locus ‘decontraction’ and centromeric recruitment contribute to allelic exclusion of the immunoglobulin heavy-chain gene. Nature immunology. 2005;6:31–41. doi: 10.1038/ni1150. [DOI] [PMC free article] [PubMed] [Google Scholar]