SUMMARY
Neural stem cells (NSCs) in the subventricular zone (SVZ) rely on environmental signals provided by the neurogenic niche for their proper function. However, little is known about the initial steps of niche establishment, as embryonic radial glia transition to postnatal NSCs. Here, we identify Gli3 repressor (Gli3R), a component of the Sonic hedgehog (Shh) pathway, as a critical factor controlling both cell type specification and structural organization of the developing SVZ. We demonstrate that Gli3R expressed in radial glia temporally regulates gp130/STAT3 signaling at the transcriptional level to suppress glial characteristics in differentiating ependymal cells. In addition, Gli3R maintains the proper level of Numb in ependymal cells to allow localization of cell adhesion molecules such as VCAM and E-cadherin. Thus, our findings reveal a novel role for Gli3R as a mediator of niche establishment and provide insights into the conditions required for proper SVZ neurogenic niche formation.
INTRODUCTION
Neurogenesis persists in the subventricular zone (SVZ) of the lateral ventricle in the postnatal rodent forebrain (Kriegstein and Alvarez-Buylla, 2009). In the SVZ, neural stem cells (NSCs) are in close contact with ependymal cells, transit-amplifying cells, neuroblasts and endothelial cells (Doetsch, García-Verdugo and Alvarez-Buylla, 1997). Collectively, these cells constitute the neurogenic niche. The ventricular surface of the SVZ is covered with ependymal cells that surround NSCs in pinwheel-like arrangements (Mirzadeh et al., 2008). These stereotypical arrangements of distinct cell types in the SVZ appear important for providing NSCs with the necessary environmental signals for proper production of interneurons of the olfactory bulb (Ihrie and Alvarez-Buylla, 2011; Tavazoie et al., 2008; Shen et al., 2008).
The neurogenic SVZ in postnatal animals is derived from the embryonic ventricular zone (VZ) of the lateral ganglionic eminence (LGE) (Young et al., 2007). Around birth, radial glial cells (RGCs) in the VZ of the LGE transform into postnatal NSCs and ependymal cells (Kriegstein and Alvarez-Buylla, 2009). Very little is known about how adult NSCs form from embryonic radial glia, and understanding the differential signaling events directing NSC and ependymal cell formation would give us an idea of how adult NSCs are initially established and regulated.
One critical environmental signal, Sonic hedgehog (Shh), has been known to control the maintenance and proliferation of NSCs (Machold et al., 2003; Ahn and Joyner, 2005) and other neural progenitors (Corrales et al., 2006) in adults. Interestingly, Shh-responding NSCs do not emerge until late embryonic stages in the SVZ and are not fully directing neurogenesis until after birth (Ahn and Joyner, 2005). However, prior to Shh signaling activation, the RGCs in the VZ do express downstream components of the pathway, including Gli2 (Allen Brain Atlas) and Gli3 (Wang et al., 2011). It is well known that Gli3, in particular, plays a critical role as a repressor (Gli3R) in brain patterning in embryonic development in the absence of Shh signaling (Theil et al., 1999). The presence of Shh effectors in the absence of active Shh signaling raises the question as to whether Gli3 in the VZ (the future SVZ) plays any role prior to Shh signaling activation in the neurogenic niche development.
Shh signaling is not the only signaling pathway critical for formation of the neurogenic niche. Postnatal SVZ NSCs inherit glial cell features from RGCs and turn on GFAP expression around birth as well (Kriegstein and Alvarez-Buylla, 2009). gp130/JAK-STAT signaling promotes glial fate and activates the GFAP promoter during normal gliogenesis (Bonni et al., 1997; Nakashima et al., 1999a; Nakashima et al., 1999b; Takizawa et al., 2001). gp130/JAK-STAT signaling is shared by the IL-6 family cytokines, including CNTF, OSM, IL-6, LIF and CT-1 (Nakashima and Taga, 2002). Although CNTF is the major in vivo glial fate promoting cytokine during development, all five signals have the ability to induce GFAP positive glial cells in vitro due to the common downstream signal pathway involving activation of the STAT family transcription factors (Nakashima and Taga, 2002). However, how STAT signaling is specifically activated in postnatal NSCs and how it interacts with Shh signaling remains unclear.
Not all RGCs become postnatal NSCs, however. They also form ependymal cells (Spassky et al., 2005), another important cell population in homeostasis of postnatal neurogenesis in the SVZ. Instead of taking on glial characteristics, these cells differentiate into cuboid, multi-ciliated cells that are required for circulating cerebrospinal fluid (Spassky et al., 2005). Very little is understood about the nature of the signals that instruct ependymal development, but it has been shown that Numb and Numblike are required for the formation and maintenance of ependymal cells in the SVZ (Kuo et al., 2006). Numb is a well-known negative regulator of the Notch signaling pathway that degrades Notch intracellular domain (Di Marcotullio et al., 2011). In addition, Numb also plays a role in maintaining RGC polarity and apical adhesion during development in the SVZ as an endocytic adaptor (Rasin et al., 2007; Zhou et al., 2011). These apical adhesions between NSCs and ependymal cells are required for proper NSC function (Karpowicz et al., 2009; Paez-Gonzalez et al., 2011). For example, disruption of several cell adhesion molecules including Ankyrin-3, vascular cell adhesion molecule-1 (VCAM), and E-cadherin has been shown to impair the SVZ structure and NSC function (Paez-Gonzalez et al., 2011; Kokovay et al., 2012; Karpowicz et al., 2009). Furthermore, E-cadherin localization is also regulated through Numb (Rasin et al., 2007), emphasizing the importance of Numb in the neurogenic SVZ.
While the Shh, Notch, and gp130/STAT signaling pathways all have been implicated in the maintenance of the adult neurogenic niche, less is known about the way in which the niche is originally formed. Since previous work in our lab identified the importance of Gli3R, the repressor of the Shh pathway, in specifying neural progenitor fates (Wang et al., 2011), we asked whether Gli3R plays a role in fate specification of NSCs in the SVZ. We also probed whether Gli3R interacts with other pathways to establish as well as maintain the neurogenic niche. We found that Gli3 acts as a repressor in the developing neurogenic niche to promote fate specification of both NSCs and ependymal cells and in establishment of the SVZ niche structure.
RESULTS
The establishment of postnatal NSC niche is disrupted in the absence of Gli3
Gli3, which serves as a context-dependent regulator of the Shh pathway, is expressed in the ventricular zone (VZ)/SVZ of the brain, including the developing LGE (Schimmang et al., 1992; Fotaki et al., 2006; Wang et al., 2011). Gli3 expression was also detected in both NSCs and ependymal cells of the SVZ in the adult brain (Figure S1C) (Lee et al., 2012). In contrast, Shh positive signaling as measured by Gli1 expression (Bai et al., 2002) is only expressed in a very limited fashion embryonically and is not required for olfactory bulb neurogenesis until E18.5 (Ahn and Joyner, 2005). In adults, Shh signaling activation was only detected in cells capable of proliferation (NSCs and transit-amplifying cells) in the postnatal SVZ (Ahn and Joyner, 2005 and Figure S1B). Together, the expression data suggests that Gli3 protein likely functions as a repressor (Gli3R) in radial glial cells during development and in postnatal ependymal cells (Figure S1A).
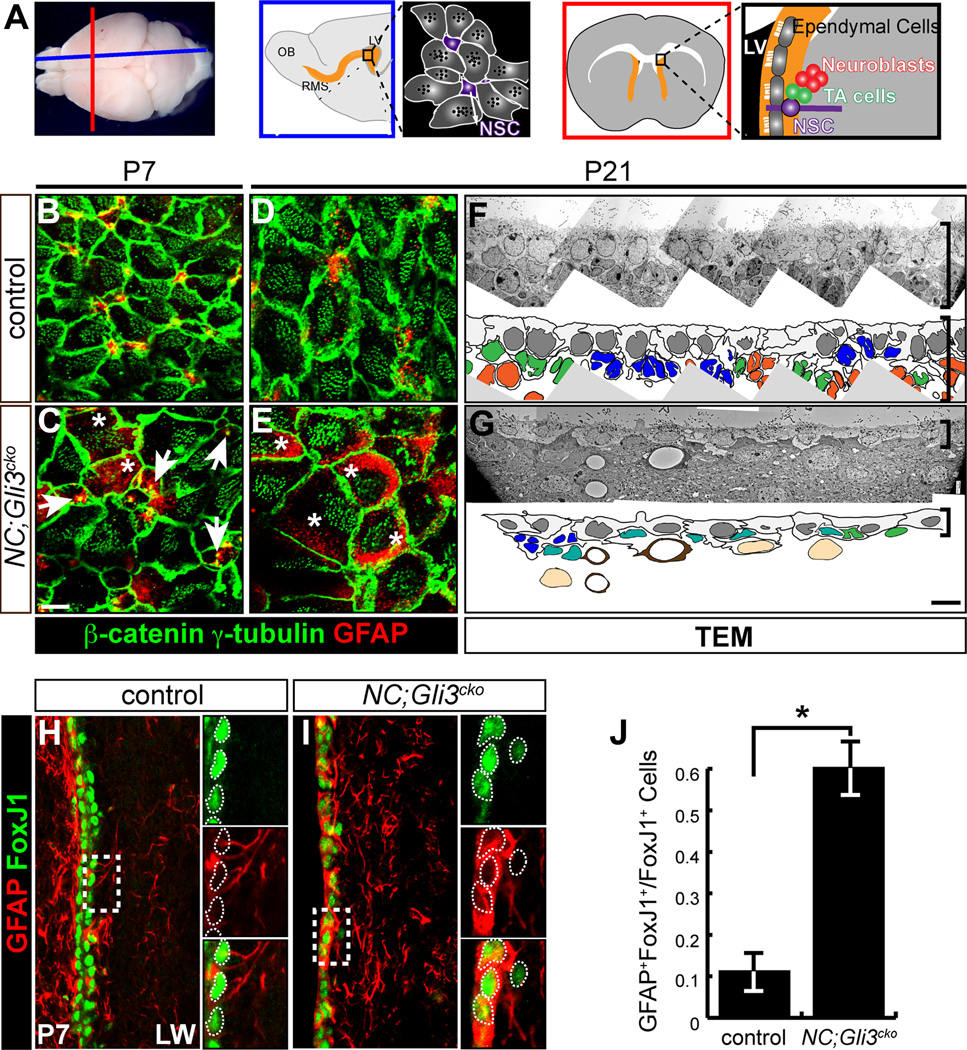
We investigated the earlier role of Gli3 (as Gli3R) by conditionally removing it in radial glia cells (RGCs) with Nestin-Cre (NC) mice (Tronche et al., 1999) before active Shh signaling emerges in the ventral SVZ (Ahn and Joyner, 2005), but after the embryonic patterning is established (Wang et al., 2011). We first analyzed the structure of the developing neurogenic niche on the lateral wall of the lateral ventricles (Figure 1A). Whole mount analysis of the SVZ showed a pinwheel-like arrangement of NSCs (GFAP+) and ependymal cells (β-catenin+ for cellular morphology and γ-tubulin+ for basal bodies of cilia) (Mirzadeh et al., 2008) in the control brain by postnatal day (P)7 (Figure 1B). NSCs were located in the center of intercellular spaces between ependymal cells (Figure 1B and 1D). In contrast, the NC;Gli3cko mutants did not show any apparent pinwheel-like structure (Figure 1C and 1E). Expression of the NSC marker, GFAP, was generally upregulated in RGC-like cells as identified by a single prominent basal body in NC;Gli3cko mutants (Figure 1C, arrows). These RGC-like cells are only found in the mutant samples, suggesting a delay in niche maturation in NC;Gli3cko mutants (Figure S1D). At P21, GFAP expression persisted in mutant cells exhibiting numerous γ-tubulin+ ciliary basal bodies, a hallmark of mature ependymal cells (Figure 1E). This ectopic GFAP expression indicates that a clear distinction between NSCs and ependymal cells fails to develop when Gli3 is absent.
Figure 1. Gli3 is required for proper establishment of the neurogenic niche in the subventricular zone.
(A) Schematics of mouse brain cut along the sagittal (blue) and coronal (red) orientation show the arrangement of cell types found in the neurogenic subventricular zone (SVZ) including neural stem cells (NSCs), transit-amplifying (TA) cells, ependymal cells, and neuroblasts. LV, lateral ventricle; OB, olfactory bulb; RMS, rostral migratory stream.
(B–E) en face view of the lateral wall of the ventricle shows that the pinwheel-like arrangements of ependymal cells (β-catenin+, green cell border; γ-tubulin+, green dots) and neural stem cells (GFAP+, red) are established by P7 in the controls (B and D). NC;Gli3cko mutants that lack Gli3 function in embryonic radial glia cells show persistence of radial glia cells with a single basal body (white arrows) and no clear pinwheel arrangements and ectopic expression of GFAP within β-catenin+ γ-tubulin+ cells (C and E, white asterisks). Scale bar, 5µm.
(F and G) Transmission Electron Micrograph (TEM) analysis of the control (F) and NC;Gli3cko mutants (G). Disrupted cytoarchitecture of the SVZ is shown in the mutants. The SVZ cell types are indicated in colors: blue (NSC); green (TA cell); orange (neuroblast); gray (ependymal cell); teal (atypical cells); light brown (striatal neurons). Brackets indicate the thickness of the SVZ. Scale bar, 5µm.
(H–J) Immunohistochemistry of FoxJ1 (green) and GFAP (red) staining in control (H) and NC;Gli3cko (I) SVZ at P7. FoxJ1 expression is similar between control and NC;Gli3cko SVZ, but many more cells co-express GFAP and FoxJ1 in the NC;Gli3cko mutant (I) as compared to the control (H, quantified in J). Insets reveal co-localization of FoxJ1 and GFAP, with white dashed lines encircling FoxJ1+ cells. Scale bar, 10µm. *, p < 0.05.
See also Figure S1.
In order to confirm the lack of proper cell fate specification in our NC;Gli3cko mutants, we performed immunohistochemical staining for FoxJ1, a marker for mature ependymal cells (Jacquet et al., 2009). We found that fewer FoxJ1+ cells were present in the NC;Gli3cko mutant SVZ at P4 (Figure S1F), compared to control (Figure S1E, quantified in S1G). By P7, the NC;Gli3cko mutant SVZ expressed FoxJ1 at equivalent levels to the control, but the majority of FoxJ1+ cells co-expressed GFAP (Figure 1I), whereas the control showed rarely any FoxJ1+ GFAP+ cells (Figure 1H). Quantification revealed a 6-fold increase in the amount of FoxJ1+ GFAP+ cells in the NC;Gli3cko mutant SVZ (Figure 1J). Together, these results indicate there is a delay in ependymal maturation followed by confusion of cell fate specification that leads to the malformation of the neurogenic niche observed at P21 (Figure 1E).
Since the ability to respond to Shh signaling is one major distinction between NSCs (Ahn and Joyner, 2005) and ependymal cells in the SVZ (Figure S1B), we examined Shh responsiveness in the NC;Gli3cko mutant SVZ. The Gli1nLZ allele allowed us to visualize the cells responding to Shh signaling based on X-gal precipitates identified by TEM analysis. In controls, there was a clear distinction between cells that respond to Shh signaling with strong X-gal precipitates (NSCs) and cells that do not respond (ependymal cells) (Figure S1H). In contrast, it was hard to distinguish the cells with a clear strong presence of X-gal precipitates in the NC;Gli3cko mutant SVZ (Figure S1H). Blind scoring for the strength of Shh-responsiveness showed that instead of clear separation between NSCs and ependymal cells, most of the cells in the NC;Gli3cko mutant SVZ exhibited intermediate Shh-responsiveness (Figure S1I). The lack of differential Shh activation between NSCs and ependymal cells is another indication that these mutant cells are not establishing proper cell identities during niche maturation.
We further confirmed an abnormal SVZ cytoarchitecture in NC;Gli3cko mutants at the ultrastructural level using transmission electron microscopy (TEM). Based on the identification criteria of the SVZ cell types described in Doetsch et al., (1997), we were able to assign cell identities in the control SVZ (Figure 1F). However, in the NC;Gli3cko mutants, there were numerous cells without clear characteristic features of NSCs, transit-amplifying cells, or neuroblasts (Figure 1G). The outermost layer of the SVZ containing ependymal cells was morphologically atypical and did not intercalate extensively in NC;Gli3cko mutants (Figure 1G). In addition, the NC;Gli3cko SVZ was much thinner than the control SVZ (brackets in Figure 1F and 1G), indicating severe structural defects caused by a loss of Gli3 during development.
As a result of this improper fate specification, these Gli3 mutant cells with dual cellular characteristics do not function properly as NSCs or ependymal cells. They are not proliferative (Figure S3D), and thus do not function as NSCs. They also do not circulate cerebrospinal fluid, as evidenced by the severe hydrocephalus in NC;Gli3cko mutant mice (unpublished observation, Ahn lab).
While we thought it likely that Gli3 itself was responsible for niche maturation, we wanted to ensure that the de-repression of the Shh pathway induced by loss of Gli3 was not responsible for our observed findings. In order to assess ectopic Shh activation, we crossed our NC;Gli3cko mice with Gli1nLZ, a reporter that expresses LacZ in any cell responsive to Shh signaling. X-gal staining for LacZ revealed no overall increase in Gli1 expression in the NC;Gli3cko SVZ as compared to the control (Figure S1J), suggesting that activation of Shh signaling is not increased in the absence of Gli3.
In order to confirm Shh activation was not playing a role in maturation of the adult neurogenic niche, we ablated Shh positive signaling directly. First, we ablated Smoothened (Smo), one of the main transducers of Shh activation, via Nestin-Cre. We found that loss of Smo did not result in mixed identity cells nor disorganization of niche structure in the SVZ (Figure S1K). We also used Nestin-Cre to ablate Gli2, the major transcriptional activator of the Shh pathway. In the absence of Gli2, there was no difference in the cytoarchitecture and cell identity between the control and NC;Gli2cko mutant SVZ (Figure S1L). Together, these experiments indicate that niche structure and cell fate are not governed by activation of Shh signaling, but rather the presence of Gli3 repressor.
Loss of Gli3R leads to overexpression of gp130
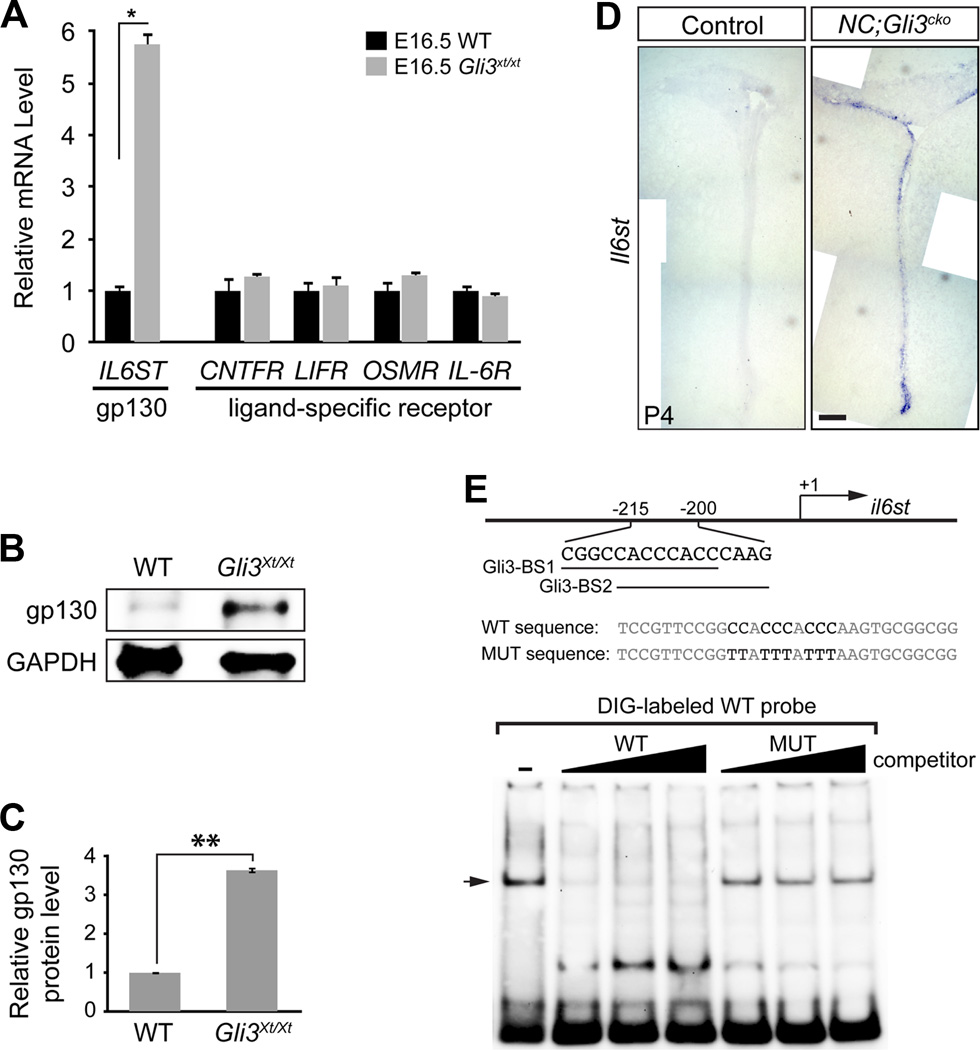
In order to find the downstream events leading to the ectopic expression of GFAP in Gli3 mutant SVZ, we carried out a qRT-PCR-based gene expression analysis for genes that are involved in Shh and/or Notch pathways (Qiagen RT² Profiler™ PCR Array) and were previously identified to contain Gli3 binding sites (Vokes et al., 2008). We compared E16.5 forebrain tissue from wild type embryos with that from Gli3Xt/Xt null mutant animals to observe the maximum difference in gene expression caused by the absence of Gli3. Since active Shh signaling is absent in E16.5 forebrain (data not shown), any phenotypes observed in Gli3Xt/Xt null mutants could be attributed to the loss of Gli3R. Surprisingly, one of the dramatically changed genes was il6st, which encodes a protein named gp130. gp130 is a co-receptor subunit shared by the IL-6 family of cytokines including CNTF, OSM, IL-6, LIF and CT-1 (Nakashima and Taga, 2002). gp130 and the cytokine-specific receptors transduce cytokine signals and activate downstream effectors, including JAK-STAT molecules. In E16.5 forebrain, the mRNA of il6st was increased by 5.75 ± 0.24 fold in Gli3Xt/Xt null mutants compared to wild type controls (Figure 2A). However, the expression levels of all the cytokine-specific co-receptors were unchanged (Figure 2A). Consistent with the mRNA expression level, gp130 protein level was increased by 3.64 ± 0.07 fold in E16.5 mutant forebrain compared to the wild type (Figure 2B and 2C). Based on the Gli3 null mutant embryo results, we next asked whether similar molecular changes occurred in NC;Gli3cko mutants. RNA in situ hybridization revealed that il6st expression was clearly increased in the postnatal SVZ in the NC;Gli3cko mutants (Figure 2D).
Figure 2. Il6st transcription is repressed by Gli3R.
(A) qRT-PCR shows that Il6st (gp130) mRNA is increased in E16.5 forebrain of Gli3Xt/Xt compared to wild type. mRNA level of the IL-6 cytokine family ligand-specific receptors is not different between wild type and Gli3Xt/Xt at E16.5. Error bars represent S.E.M. and * indicates p = 0.016.
(B and C) Western blot shows that gp130 protein is increased in E16.5 forebrain of Gli3Xt/Xt compared to wild type. Error bars indicate S.E.M. **, p < 0.001.
(D) in situ hybridization on control and NC;Gli3cko shows Il6st mRNA is increased in the mutant SVZ at P4. Scale bar, 500µm.
(E) Electrophoresis Mobility Shift Assay (EMSA) shows Gli3R binds to two overlapping putative Gli binding sites located between −215bp and −200bp 5’ of mouse il6st gene. +1 indicates the transcription initiation site of the il6st gene. The arrow indicates the complex of Gli3R and DIG-labeled WT probe.
See also Figure S2.
We then tested whether the ectopic upregulation of il6st was due to the loss of Gli3R, which normally acts as a transcriptional repressor. We identified two putative Gli binding sites sharing the same consensus core sequence between −215bp and −200bp upstream of the il6st transcription initiation site in the mouse genome (Evolutionary Conserved Regions Browser and rVISTA). Electrophoresis Mobility Shift Assay (EMSA) confirmed the direct and specific binding of Gli3R to the identified Gli sites taken from the il6st gene (Figure 2E).
Previous studies indicated that all IL-6 family cytokines have the ability to induce GFAP+ glial cell fate in vitro due to common downstream activation of JAK-STAT3 signaling (Nakashima and Taga, 2002). Thus, we examined the changes in cytokine levels in the Gli3 null mutants using qRT-PCR. Interestingly, at E16.5, OSM and IL-6 were both expressed at significantly higher levels in Gli3Xt/Xt mutants compared to controls (4.03 ± 0.24 fold and 2.77 ± 0.24 fold, respectively) (Figure S2A). We found that OSM mRNA expression was also upregulated in the postnatal SVZ of NC;Gli3cko mutants as expected (Figure S2B). Together, our results indicate that the absence of Gli3R causes overexpression of both ligands and a shared receptor subunit of the IL-6 cytokine family.
Loss of Gli3R induces the sustained activation of STAT3 and overexpression of GFAP in Gli3 mutant SVZ
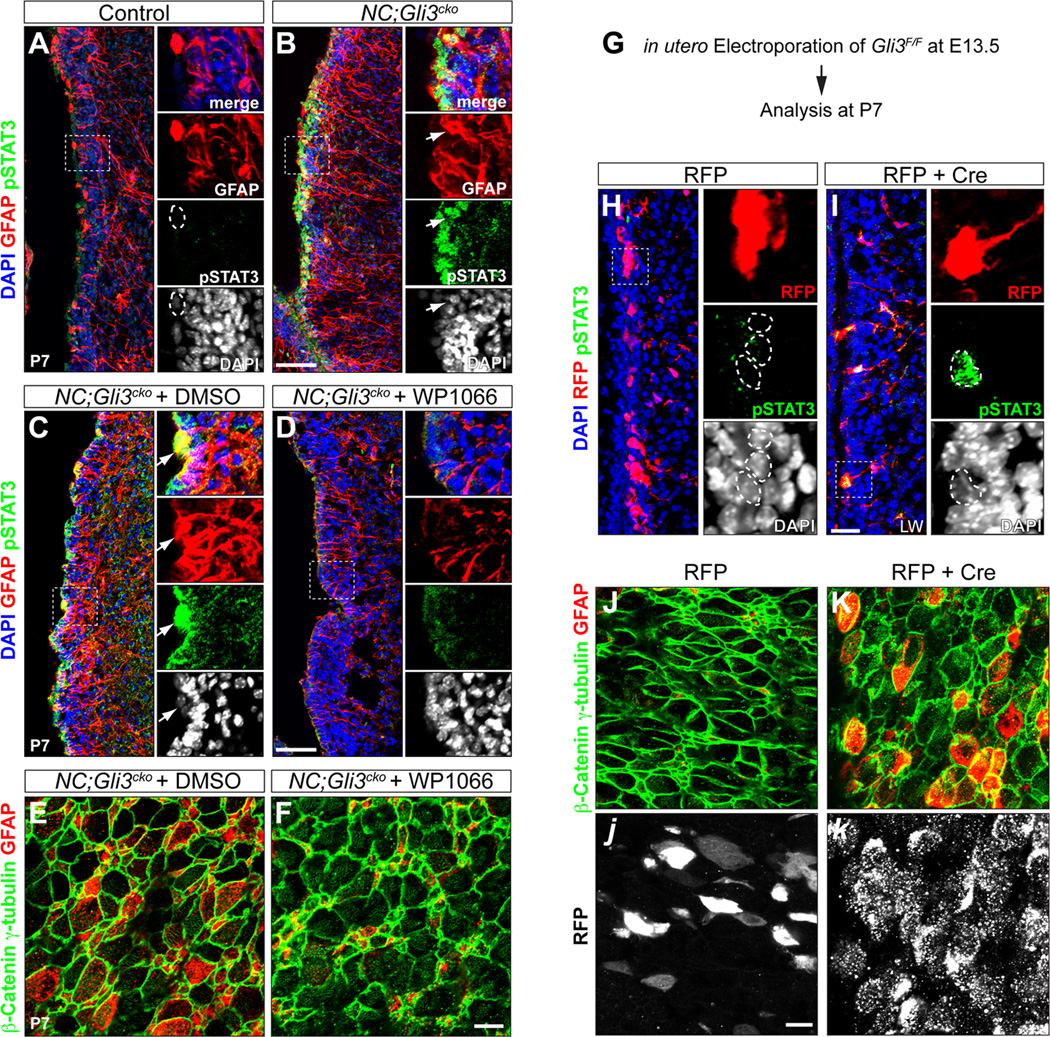
gp130 promotes GFAP expression and glial cell fate through the phosphorylation of the STAT3 (pSTAT3) transcription factor (Nakashima et al., 1999a; Nakashima et al., 1999b; Bonni et al., 1997). In control animals, there was a transient activation of STAT3 in a small population of SVZ cells at P4 (Figure S3A), but pSTAT3 was barely detectible by P7 (Figure 3A) (Herrmann et al., 2008). In contrast, NC;Gli3cko mutants showed widespread presence of pSTAT3 in the SVZ at both P4 and P7 (Figure S3B and Figure 3B). In addition, most of the pSTAT3+ cells also expressed GFAP, indicating that the increased gp130 level induced ectopic activation of STAT3 to induce GFAP overexpression in NC;Gli3cko mutants.
Figure 3. Loss of Gli3 leads to the overactivation of STAT3 and ectopic GFAP expression in NC;Gli3cko mutant SVZ.
(A and B) Immunohistochemistry of phosphorylated STAT3 (pSTAT3) and GFAP in control and NC;Gli3cko SVZ at P7. Both pSTAT3 (green) and GFAP (red) are increased in NC;Gli3cko (B) compared to control (A). Higher magnification images of boxed areas are shown in right panels. GFAP+ cell (outlined by dashed circle) does not express pSTAT3 in the control. Arrows indicate the GFAP+ cell expresses a high level of nuclear pSTAT3 in the mutant. Scale bar, 50µm.
(C and D) NC;Gli3cko embryos received either STAT3 inhibitor (WP1066) or vehicle control DMSO from E16.5 till birth. The animals were analyzed at P7. The nuclear pSTAT3 (green) in GFAP+ (red) cells shown in mutants receiving DMSO (C) is rescued by WP1066 (D). Arrows indicate the GFAP+ cell expresses a high level of nuclear pSTAT3 in the DMSO treated mutant.
(E and F) en face view of P7 NC;Gli3cko SVZ which were treated with either STAT3 inhibitor (WP1066) or vehicle control DMSO prenatally. The ectopic GFAP expression (red) in ependymal-like cells (β-catenin+ γ-tubulin+, green) in NC;Gli3cko treated with DMSO (E) is rescued by WP1066 (F). Scale bar, 50µm.
(G–K) in utero electroporation of either RFP alone or RFP + Cre into Gli3F/F embryos. Electroporation was done at E13.5 and the manipulated pups were sacrificed at P7. The Gli3 mutant cells induced by Cre are labeled by RFP in (I). The electroporated cells received RFP alone (red) does not have nuclear pSTAT3 (green) expression (indicated by dashed circles in H). However, the Gli3 mutant cells (red) express nuclear pSTAT3 (green) (indicated by dashed circles in I). Scale bar, 25 µm. en face view shows that Gli3 mutant cells (white in k) exhibit ectopic GFAP expression (red in K) in ependymal-like cells (β-catenin+ γ-tubulin+, green). The ectopic GFAP expression is absent in cells that received RFP alone (white in j). Scale bar, 50µm.
See also Figure S3.
To further confirm that the sustained activation of STAT3 and overexpression of GFAP in NC;Gli3cko mutants were solely dependent on the loss of Gli3, we acutely induced Gli3 mutant cells in a wild type environment by electroporating Cre-expressing plasmids into Gli3F/F embryos at E13.5 (Figure 3G). We found that by P7, Cre-expressing Gli3 mutant cells in the SVZ showed ectopic activation of STAT3 (Figure 3I) and overexpression of GFAP (Figure 3K), while control electroporated cells never showed such phenotypes (Figure 3H and 3J). We also noticed that the ectopic GFAP+ Gli3 mutant cells in the SVZ formed elongated processes similar to the radial processes of NSCs rather than short processes of typical astrocytes (Figure 3I).
Previous studies showed that pSTAT3 directly activates GFAP gene transcription by binding to its promoter sequence (Takizawa et al., 2001). Thus we tested whether the GFAP overexpression seen in NC;Gli3cko mutants was due to the overactivation of STAT3. We treated NC;Gli3cko embryos with a STAT3 inhibitor (WP1066) between E16.5 and birth via pregnant dams and found that GFAP expression at P7 was largely restored to control levels by the STAT3 inhibitor (Figure 3C-3F).
Together, our results indicate that the loss of Gli3R resulted in an increased level of gp130 and its ligands to ectopically induce STAT3-mediated GFAP expression in the NC;Gli3cko mutant SVZ. This eventually made the mutant SVZ cells maintain both NSC and ependymal cell characteristics and fail to develop into two distinct, functional cell types.
Loss of Gli3 results in alterations in cell adhesion and loss of Numb via LNX2
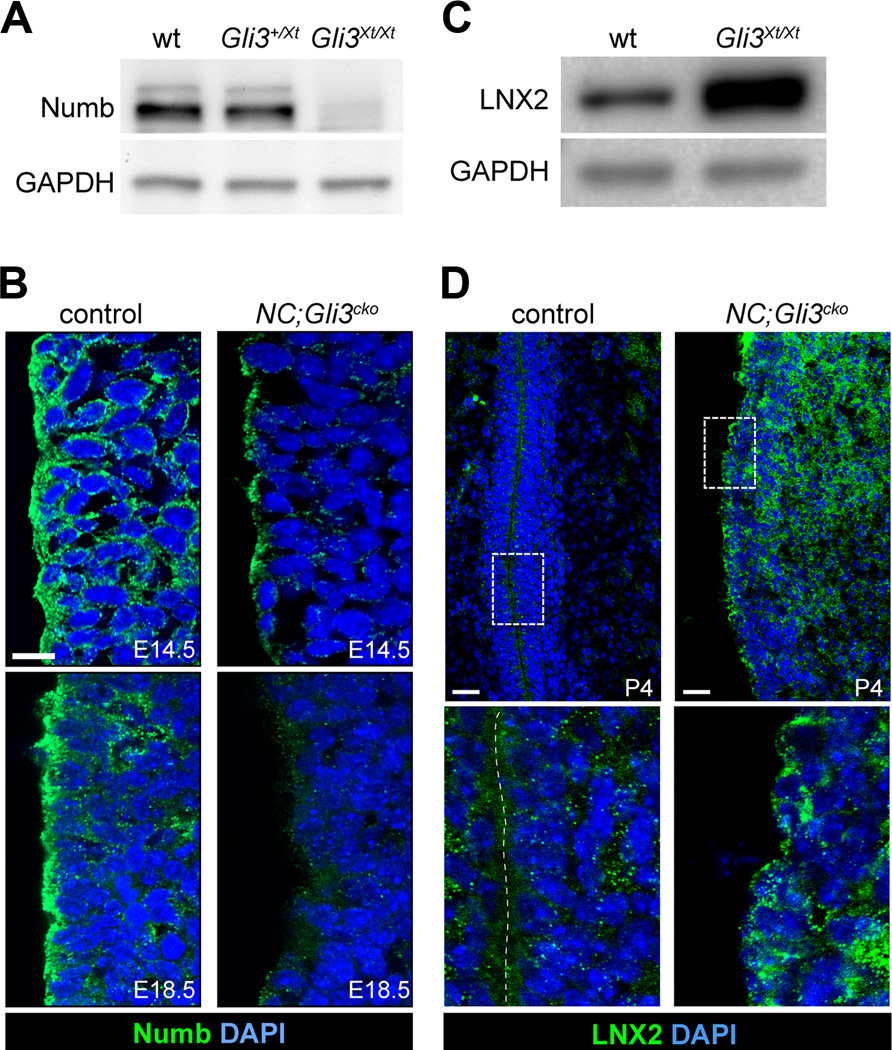
While the STAT3 inhibitor did rescue the ectopic expression of GFAP, it did not rescue the disruption of the pinwheel arrangement of NSCs and ependymal cells in NC;Gli3cko mutants (Figure 1E and Figure 3F). Thus, we assessed an alternative role for Gli3 in establishing the neurogenic niche structure. We found that in the absence of Gli3, the pinwheel organization was not maintained and the ventricular wall integrity was compromised, leading to hydrocephalus. The weakness of the ventricular wall integrity led us to the idea that Gli3 might regulate cell adhesion between ependymal cells and NSCs. Interestingly, our NC;Gli3cko mutant phenotype is very similar to the reported phenotype of Numb;Numblike double conditional mutants, including reduced cortical thickness and hydrocephaly (Kuo et al., 2006; Wang et al., 2011). Thus, we asked whether Numb levels might have been altered in Gli3 mutants. We found that levels of Numb protein were greatly reduced in E18.5 Gli3Xt/Xt null mutants (Figure 4A). The SVZ of NC;Gli3cko mutants also showed a reduction in Numb expression at E14.5 and almost complete loss by E18.5, particularly in the outermost cell layer lining the lateral ventricle (Figure 4B).
Figure 4. Loss of Gli3 results in decreased level of Numb protein.
(A) Western Blot of Numb in E18.5 Gli3Xt/Xt forebrain shows a dose dependent reduction in Numb protein level compared to the wild type.
(B) Immunohistochemistry of Numb shows that Numb (green) is localized to the ventricular surface of RGCs at both E14.5 and E18.5 in the control (left panels). In NC;Gli3cko mice, Numb is significantly reduced at both E14.5 and E18.5 (right panels). Scale bar, 10µm.
(C) Western Blot of E18.5 forebrain samples shows LNX2 protein level is increased in Gli3Xt/Xt compared to wild type.
(D) Immunohistochemistry of LNX2 at P4 shows LNX2 (green) protein level is increased at the ventricular surface and in the basal SVZ in NC;Gli3cko mutant (right panels) as compared to the control (left panels). Higher magnification images of boxed areas are shown below. Scale bar, 10 µm.
To explore the mechanism responsible for this loss of Numb, we next examined the expression level of LNX2, an E3 ubiquitin ligase that is known to target Numb for degradation (Nie, Li and McGlade, 2004). We found that the LNX2 protein level was greatly increased in the forebrain extracts of E18.5 Gli3Xt/Xt null embryos compared to the littermate controls (Figure 4C). NC;Gli3cko mutants also showed elevated LNX2 protein levels as assessed by immunohistochemical staining, particularly in the SVZ (Figure 4D). Our findings suggest that the loss of Gli3 led to an abnormal increase in LNX2 expression that ultimately targeted Numb for degradation in NC;Gli3cko mutants.
Loss of Numb results in loss of proper cell adhesion
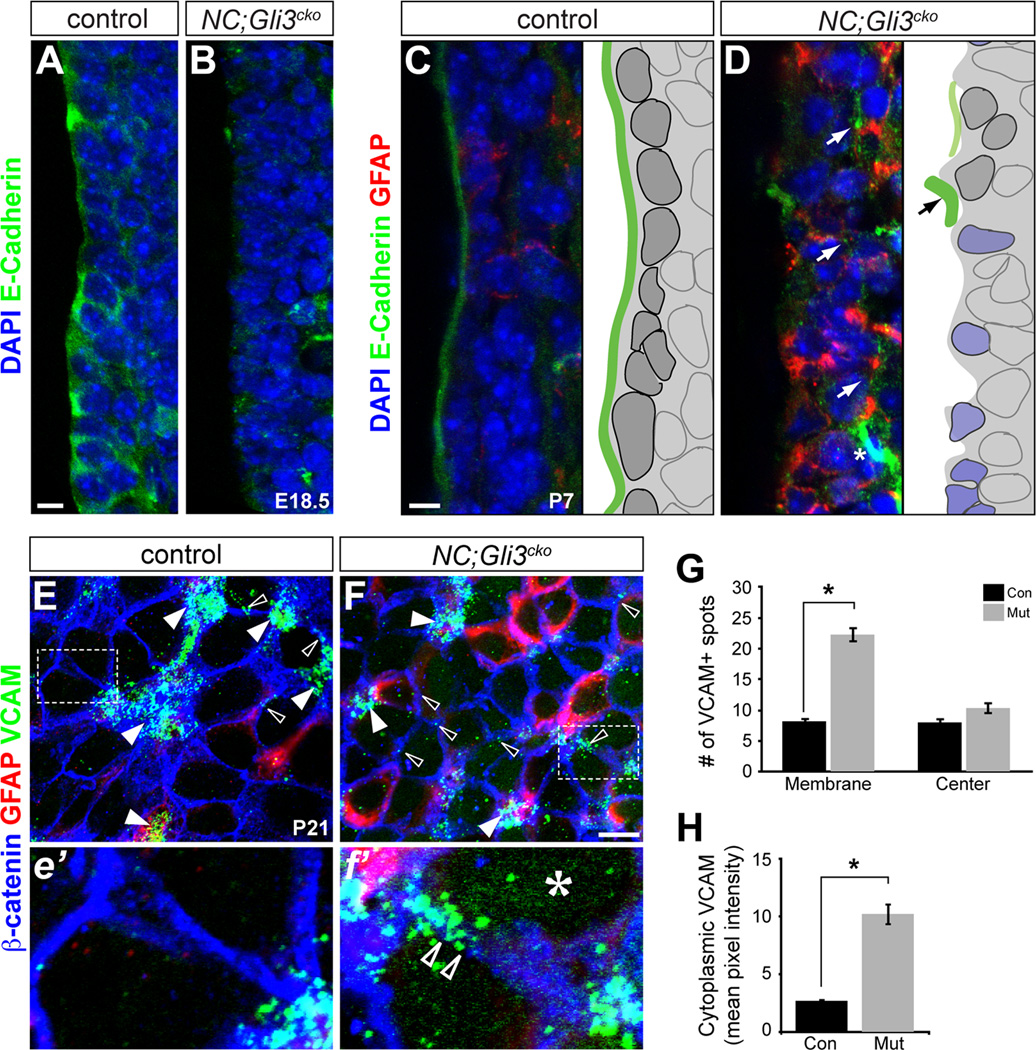
Because the loss of Numb is associated with defects in cell adhesion (Rasin et al., 2007), we next examined changes in cell adhesion molecules in our NC;Gli3cko mutants. In particular, Numb is known to regulate cell adhesion molecules such as E-cadherin, which is critical for proper NSC function and NSC polarity by maintaining adherens junctions (Rasin et al., 2007; Karpowicz et al., 2009; Chenn et al., 1998; Perez-Moreno et al., 2003). When we analyzed changes in E-cadherin expression, we found that the enriched E-cadherin expression seen in the first cell layer of control SVZ was dramatically reduced on the apical surface of the lateral ventricle in NC;Gli3cko mutants at E18.5 (Figure 5A and 5B).
Figure 5. Distribution of adhesion molecules is disrupted in NC;Gli3cko.
(A and B) E-Cadherin protein (green) is highly expressed at the apical surface of RGCs in the control mice at E18.5 (A), but is reduced at the apical surface in NC;Gli3cko mutants (B). Scale bar, 10µm.
(C and D) E-Cadherin protein (green) is expressed just at the ventricular surface by ependymal cells at P7 in control mice (C). NC;Gli3cko mice show reduced E-Cadherin expression and increased GFAP (red) compared to the control (D). Schematics of immunohistochemical results shown to the right of each image. Dark gray cells represent ependymal cells. Blue cells represent atypical mutant cells. Green lines represent E-Cadherin lining. Scale bars, 10µm.
(E and F) Whole mount immunohistochemical staining of the ventricular surface at P21 reveals VCAM1 (green) protein localization in NC;Gli3cko (F, f’) versus controls (E, e’). Higher magnification of inset is shown below low magnification images (e’, f’). GFAP (red) marks NSC. Open arrowheads show membrane localization of VCAM that is not associated with NSC clusters. White arrowheads show VCAM staining clustered at pinwheel centers. The asterisk (f’) shows the upregulation of VCAM in the cytoplasm of mutant cells. Scale bar in (E and F), 10µm.
(G) Quantification of VCAM spots. 10–15 images for each genotype were counted. Areas of staining above background were considered a VCAM+ spot. If the cluster was at the center of a pinwheel, it was deemed a “cluster” spot, and if at the membrane between two cells a “membrane” spot. All were normalized to spots/image. There were significantly more spots at the membrane of mutant animals as compared to controls. Error bars represent S.E.M. *, p < 0.05.
(H) Quantification of cytoplasmic VCAM staining. ImageJ was used to quantify VCAM pixel intensity for each image using a filter based on β-catenin staining to remove VCAM staining localized to the membrane. The mutant images had significantly increased cytoplasmic VCAM as compared to the controls. Error bars represent S.E.M. *, p < 0.05. See also Figure S4.
The reduction in E-cadherin expression was also found in the P7 NC;Gli3cko mutant as in the E18.5 analysis. While there was a clean contiguous E-cadherin expression on the apical surface of the SVZ in controls, there was almost no E-cadherin expression found along the surface of the mutant lateral ventricle (Figure 5C and 5D). Instead, we observed strong E-cadherin expression 2–3 cell layers below the ventricular surface in the NC;Gli3cko mutants as compared to the controls (Figure 5D, white arrow). Thus, E-cadherin is mislocalized and downregulated during early establishment of the postnatal neurogenic niche.
At P7 we also saw that the well-organized ependymal layer observed in control sections (Figure 5C) was not present in the NC;Gli3cko mutants (Figure 5D). Instead, the first layer of cells failed to form a smooth continuous ventricular surface and the majority of them expressed GFAP, directly exposing GFAP+ cells to the apical surface of the lateral ventricle (Figure 5D). Interestingly, we found that the cells with the highest E-cadherin expression (Figure 5D schematic, black arrow) in the NC;Gli3cko mutants were also the cells with very low GFAP expression. Our finding that gross disorganization of the ventricular surface observed in the NC;Gli3cko mutants does not occur until postnatal stages (Figure 5B and 5D) further supports the idea that Gli3R is critical during the transition from RGCs to ependymal cells and NSCs, and that cell adhesion is critical for proper neurogenic niche maturation.
To further assess changes in cell adhesion induced by the loss of Gli3R, we examined vascular cell adhesion molecule-1 (VCAM) expression in the SVZ neurogenic niche. VCAM is an adhesion molecule expressed in both NSCs and ependymal cells, although its expression was found to be 8.87 times higher in NSCs than ependymal cells as assessed by qPCR (Figure S4A). VCAM localizes specifically to NSC-ependymal cell junctions (Kokovay et al., 2012), providing a tool for the assessment of cell adhesion and organization of the SVZ. We found that in P21 NC;Gli3cko mutants, VCAM was diffusely expressed throughout the cytoplasm (Figure 5f’ asterisk and Figure S4B) with a pixel intensity five times greater than that of the control (Figure 5e’ and 5H). Furthermore, there was also a significant upregulation of VCAM staining found in the membrane of the mutant cells (Figure 5F, f’, open arrowheads) as opposed to specifically clustering at NSC-ependymal cell junctions (Figure 5E, F, white arrowheads). When the number of VCAM+ spots were counted, more than double the amount of membrane enriched spots were present in the NC;Gli3cko mutants as compared to the controls (Figure 5G). Thus, there is more VCAM present in the cytoplasm and at non-ependymal cell-NSC junctions in the mutants as compared to the control. This ectopic expression suggests that VCAM may not be properly localized within the cell in the absence of Gli3, suggesting that there are long-term cell adhesion changes when Gli3 is lost during development of the SVZ.
DISCUSSION
NSCs and ependymal cells share a common developmental origin (RGCs) in the embryonic LGE and form highly structured arrangements on the ventricular surface of the postnatal SVZ (Kriegstein and Alvarez-Buylla, 2009; Ihrie and Alvarez-Buylla, 2011). Little is known about the manner by which embryonic RGCs transform into NSCs and ependymal cells, since most studies have focused on the stages after cell fates are already specified. However, it is clear that there are many changes in signaling between embryonic RGCs and postnatal NSCs. Here, we propose to define three stages of postnatal neurogenic niche development and maturation: First, embryonic RGCs in the LGE; Second, transition from embryonic RGCs to postnatal NSCs (~E18.5 to P7); Third, maturation of postnatal NSCs. Our results demonstrate an interesting biphasic requirement of Shh pathway components in these stages for niche maturation. In RGCs, Gli3 (in repressor form) is highly expressed and RGCs are refractory to Shh activation. During the transition between E18.5 and P7, Gli3 repressor plays a critical role in cell fate specification and niche organization, establishing a framework of support cells and stem cells. Finally, once the niche has established (at around P7), Shh responsive NSCs begin active postnatal neurogenesis. Thus, we have found that Gli3 plays a critical role in setting the stage on which Shh can act in the mature niche.
During the establishment phase of the neurogenic SVZ, several molecular changes take place when Gli3 is lost (Figure 6). Among several signaling pathways known to influence postnatal neurogenesis in rodent brains, we found that the coordinated activation and duration of Shh and JAK/STAT3 signaling is critical in determination of NSC and ependymal cell fates. Before birth, RGCs do not actively respond to Shh or cytokine signaling. Just around birth, RGCs that gain responsiveness to both Shh and activated STAT3 become NSCs, while RGCs remaining unresponsive to these signals become ependymal cells. In addition, unlike Shh signaling that remains active in postnatal NSCs, STAT3 signaling is active only transiently for proper development of the SVZ structure. When STAT3 activation is sustained due to the overexpression of il6st (gp130) in NC;Gli3cko mutants, ectopic GFAP expression is widespread across the cell types, resulting in compromised cell fates in the SVZ niche. Thus, while transient STAT3 signaling is necessary for NSC specification, sustained STAT3 signaling is detrimental to the proper specification of cell types generated from RGCs during the establishment period of the neurogenic niche. A recent study reported that adult NSCs in the SVZ express CNTF receptors and respond to exogenous CNTF in vivo (Lee et al., 2013). Since CNTF signaling promotes self-renewal of the SVZ NSCs in vitro (Shimazaki, Shingo and Weiss, 2001; Müller et al., 2009), the fact that NSCs still retain CNTF receptor expression may indicate their potential to respond to exogenously supplied cytokines. Interestingly, the cytokines of IL-6 family are commonly induced by inflammation, suggesting that diseases or injuries in the SVZ might induce changes in NSC behavior through activated STAT3 signaling.
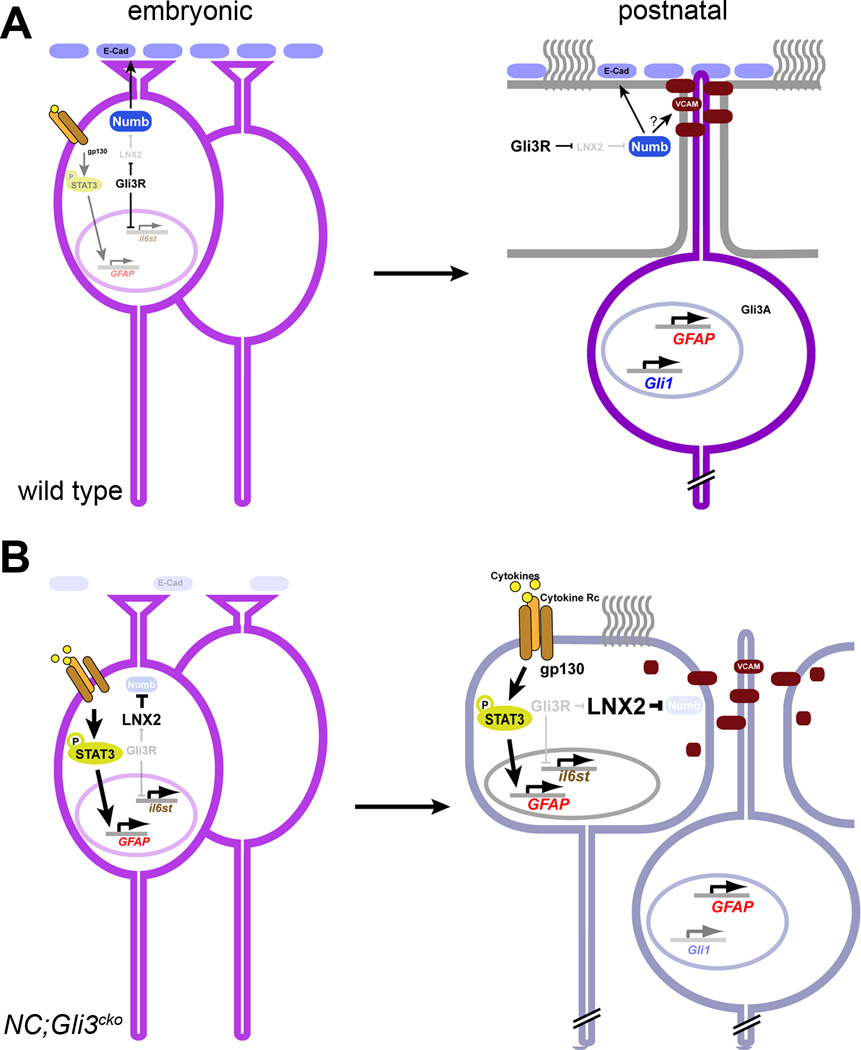
Figure 6. Summary schematic of Gli3 repressor.
(A) In wild type animals, embryonic radial glia express Gli3R and suppress il6st. Thus, gp130-mediated STAT3 signaling is only transiently activated in future NSCs around birth. Due to Gli3R–mediated suppression of LNX2, the Numb level is maintained to regulate E-cadherin at the apical surface of the ventricular zone. In the postnatal brain, Shh-responding NSCs contains Gli activators to induce GFAP and Gli1 expression while non-responsive ependymal cells contain Gli3 as Gli3 repressor. Sustained expression of Numb enables ependymal cells to localize E-cadherin and VCAM on the apical surface to maintain the integrity of the neurogenic niche.
(B) In NC;Gli3cko, il6st is overexpressed due to the loss of Gli3R, resulting in sustained activation of STAT3 to induce GFAP overexpression. Without Gli3R, LNX2 is overexpressed to target Numb for degradation, leading to a loss of E-cadherin on the apical surface of the ventricular zone. In the postnatal brain, cellular characteristics of ependymal cells and NSCs are less distinct and GFAP is overexpressed across the cell types in the SVZ due to the persistent activation of STAT3. In addition, due to the lack of Numb, E-cadherin is missing from the apical lining of the ventricle and VCAM is found also in the cytoplasm. Together, these molecular changes lead to the compromised integrity of the neurogenic SVZ structure.
In addition to its role in the regulation of Shh and STAT3 signaling, our current findings demonstrate a key role for Gli3R in the establishment of the neurogenic niche structure through the maintenance of Numb (Figure 6). Interestingly, we found that in wild type animals, Numb mRNA is expressed at a higher level in ependymal cells than in their neighboring NSCs (Figure S1C). Since Numb is a negative regulator of Notch signaling, our result supports the previously reported phenomenon that positive Notch activity is only observed in NSCs, not in ependymal cells (Imayoshi et al., 2010). However, the absence of Numb did not affect Notch activity in NC;Gli3cko mutants as evidenced by lack of significant change in Hes5 expression, a readout of activated Notch signaling (data not shown). Thus, Numb is not affecting the SVZ structure through alteration of Notch activity as expected. Since Numb is also known to suppress Shh signaling via targeted degradation of Gli1 (Pierfelice, Alberi and Gaiano, 2011; Di Marcotullio et al., 2006), the absence of Numb in NC;Gli3cko mutants could be responsible for a slight induction of Shh response observed in mutant ependymal cells (Figure S1H and S1I), which may contribute to the less distinct cell identities.
While loss of Numb does appear to affect Shh signaling, the primary consequence of its loss in our NC;Gli3cko mutants was the disorganization of the niche structure due to defects in cell adhesion (Kuo et al., 2006). We showed that a loss of Numb in our NC;Gli3cko mutants resulted in changes in adherens junctions between niche cells as evidenced by altered VCAM and E-cadherin expression and localization. Since Numb-mediated loss of E-cadherin is known to change gap junctions between cells (Govindarajan et al., 2010), similar defects in NC;Gli3cko mutants could disrupt the communication between NSCs and ependymal cells for extracellular signaling molecules. One consistent and interesting phenotype we observed from acute deletion of Gli3 using in utero electroporation was that the cellular boundaries of Gli3 mutant cells expressing the reporter protein RFP always appeared to be less clear, raising the possibility that electroporated DNA constructs could be more leaky between mutant cells (Figure 3k). In addition, entanglement of the GFAP+ processes on the apical surface of the SVZ suggests possible defects in polarity, a role in which Numb was implicated previously in RGCs (Rasin et al., 2007). For example, apical processes of NSCs that are critical for sensing signaling molecules in the cerebrospinal fluid of the lateral ventricle (Kokovay et al., 2010; Lehtinen et al., 2011) require proper cell-cell adhesion for their maintenance (Loulier et al., 2009). Thus, structural defects in the SVZ can lead to neurogenic defects when polarity or cell adhesion alterations prevent NSCs from effectively communicating with neighboring cells and factors present in the ventricular lumen/CSF.
Interestingly, we found that there were a few cells in the NC;Gli3cko mutants that still expressed normal levels of E-cadherin at P7, when the stereotypical adult neurogenic niche structure emerges (Figure 5D). These cells did not overexpress GFAP and exhibited normal characteristics of true differentiated ependymal cells, suggesting that they were most likely the ones that escaped Nestin-Cre mediated recombination of the Gli3F conditional allele and were able to maintain normal Gli3R levels. Thus, the expression of E-cadherin appears to be a fundamental feature of proper ependymal cells, and Gli3R is required for its expression via maintenance of Numb.
In summary, we identified a new role for Gli3 repressor (Gli3R) as a critical mediator of cell fate specification and cytoarchitectural organization in the neurogenic niche during the niche establishment phase. We found that Gli3R suppresses glial fates through transcriptional regulation of gp130 and promotes the stable expression and localization of cell adhesion molecules for proper formation of the neurogenic niche cytoarchitecture. Together, our findings provide an interesting biphasic requirement of Shh signaling components in the SVZ neurogenesis: Gli3R shapes the neurogenic niche structure prior to Shh activation, which in turn promotes the neurogenesis. Thus, we have found a crucial role for Gli3R in establishment of the adult neurogenic niche.
EXPERIMENTAL PROCEDURES
Mice and DNA
All procedures in mice followed the guidelines of the Institutional Animal Care and Use Committee of the National Institutes of Health. Gli3 conditional (Gli3F/+) and null (Gli3Xt/+) alleles as well as Nestin-Cre mouse line were previously described (Blaess, Stephen and Joyner, 2008; Hui and Joyner, 1993; Tronche et al., 1999). in utero electroporation was performed on E13.5 embryos following the procedures described in Wang et al., (2011). More detailed information is listed in Supplemental Experimental Procedures.
Histology, immunohistochemistry, RNA in situ hybridization and TEM
Brains were dissected from perfused animals and either frozen sections or vibratome sections were used. Whole mount analysis of the SVZ structure followed the procedure described in (Mirzadeh et al., 2010; Mirzadeh et al., 2008). Antibodies used are listed in Supplemental Experimental Procedures. RNA in situ hybridization followed the procedure described in (Ahn and Joyner, 2004). Tissue processing for TEM and X-gal/TEM was done as described (Wichterle et al., 1999) and cell types were identified according to (Doetsch, García-Verdugo and Alvarez-Buylla, 1997).
Western analysis and qRT-PCR
Western analysis was performed as described (Ahn and Joyner, 2004). qRT-PCR was performed on total RNA obtained from embryonic forebrain tissue or FACS-isolated NSCs and ependymal cells from adult mice as detailed in Supplemental Experimental Procedures.
Electrophoresis Mobility Shift Assay
Oligonucleotide probes were designed based on the Gli binding site located from −215bp to −200bp 5’ of the mouse il6st gene identified by the ECR browser. Wild type probe sequence is 5’-TCCGTTCCGGCCACCCACCCAAGTGCGGCGG-3’. Mutant probe sequence is 5’-TCCGTTCCGGCCATTTATTTAAGTGCGGCGG-3’. Double stranded oligonucleotides were labeled with Dig-ddUTP using Dig Gel Shift Kit (Roche). Gli3R containing nuclear extract was isolated from pGli3R–IRES-nGFP transfected 293T cells and was used for oligonucleotide binding directly. The binding reaction was set up following previous studies (Vortkamp, Gessler and Grzeschik, 1995). A 4–20% gradient precast TBE gel (BioRad) was pre-run for 30 minutes and the electrophoresis was carried out for 90 minutes at 80 volts in 0.5xTBE. Samples were then electro-transferred onto positively charged nylon membrane and subjected to antibody reaction and chemiluminescent detection following the Dig Gel Shift Kit (Roche) protocol.
STAT3 inhibitor treatment
Timed pregnant females carrying NC;Gli3cko mutant embryos were given 600ug/day of either a STAT3 inhibitor, WP1066 (dissolved in DMSO), or DMSO of the same volume starting from E16.5 until birth. Pups were sacrificed at P7 and subjected to section or whole mount immunohistochemistry.
Quantification and statistics
Statistical analysis of qRT-PCR and Western Blotting was performed by student’s t-test. The quantification results are presented as average ± standard error of means (S.E.M) for error bars. n indicates the number of animals analyzed. Quantification of cytoplasmic pixel intensity for VCAM expression was performed using ImageJ. A filter was laid over each image using the threshold tool based on membrane staining by β-catenin. Erode and Dilate were used to fill the cytoplasmic area so that the filter was as accurate as possible, then pixel intensity was measured using Analyze Particles. Pixel intensity measurements were collected in Excel and ANOVA was used to quantify differences between groups. Significance was set at p < 0.05
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank M. Song, M. Zervas and J. Li for critical reading of the manuscript; I. Dawid and M. Won for helpful suggestion on LNX2 study; M. Song for discussion on STAT3; members of the Ahn lab for discussion; G. Ge and E. Karey for help in initial stage of the study. We would also like to thank NICHD Microscopy Imaging Core and NCI Electron Microscopy Core Facilities for confocal microscopy and TEM analysis; NHLBI Flow Cytometry Core Facility for FACS. This study was supported by Intramural Research Program of NIH (1ZIHD008781) to SA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
H.W., A.K., and S.A. designed and performed experiments, analyzed data, and wrote the paper. C.L. performed experiments.
COMPETING INTERESTS STATEMENT
Authors declare they have no competing financial interests.
Supplementary Information accompanies this paper.
REFERENCES
- Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Ahn S, Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–1821. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, Greco A, De Smaele E, Po A, Sico MA, Alimandi M, Giannini G, Maroder M, Screpanti I, Gulino A. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8:1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Greco A, Mazzà D, Canettieri G, Pietrosanti L, Infante P, Coni S, Moretti M, De Smaele E, Ferretti E, et al. Numb activates the E3 ligase Itch to control Gli1 function through a novel degradation signal. Oncogene. 2011;30:65–76. doi: 10.1038/onc.2010.394. [DOI] [PubMed] [Google Scholar]
- Doetsch F, García-Verdugo JM, Alvarez-Buylla A. Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J Neurosci. 1997;17:5046–5061. doi: 10.1523/JNEUROSCI.17-13-05046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. J Neurosci. 2006;26:9282–9292. doi: 10.1523/JNEUROSCI.2673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan R, Chakraborty S, Johnson KE, Falk MM, Wheelock MJ, Johnson KR, Mehta PP. Assembly of connexin43 into gap junctions is regulated differentially by E-cadherin and N-cadherin in rat liver epithelial cells. Mol Biol Cell. 2010;21:4089–4107. doi: 10.1091/mbc.E10-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JE, Imura T, Song B, Qi J, Ao Y, Nguyen TK, Korsak RA, Takeda K, Akira S, Sofroniew MV. STAT3 is a critical regulator of astrogliosis and scar formation after spinal cord injury. J Neurosci. 2008;28:7231–7243. doi: 10.1523/JNEUROSCI.1709-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat Genet. 1993;3:241–246. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Lake-front property: a unique germinal niche by the lateral ventricles of the adult brain. Neuron. 2011;70:674–686. doi: 10.1016/j.neuron.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet BV, Salinas-Mondragon R, Liang H, Therit B, Buie JD, Dykstra M, Campbell K, Ostrowski LE, Brody SL, Ghashghaei HT. FoxJ1-dependent gene expression is required for differentiation of radial glia into ependymal cells and a subset of astrocytes in the postnatal brain. Development. 2009;136:4021–4031. doi: 10.1242/dev.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P, Willaime-Morawek S, Balenci L, DeVeale B, Inoue T, van der Kooy D. E-Cadherin regulates neural stem cell self-renewal. J Neurosci. 2009;29:3885–3896. doi: 10.1523/JNEUROSCI.0037-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E, Goderie S, Wang Y, Lotz S, Lin G, Sun Y, Roysam B, Shen Q, Temple S. Adult SVZ lineage cells home to and leave the vascular niche via differential responses to SDF1/CXCR4 signaling. Cell Stem Cell. 2010;7:163–173. doi: 10.1016/j.stem.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E, Wang Y, Kusek G, Wurster R, Lederman P, Lowry N, Shen Q, Temple S. VCAM1 is essential to maintain the structure of the SVZ niche and acts as an environmental sensor to regulate SVZ lineage progression. Cell Stem Cell. 2012;11:220–230. doi: 10.1016/j.stem.2012.06.016. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Mirzadeh Z, Soriano-Navarro M, Rasin M, Wang D, Shen J, Sestan N, Garcia-Verdugo J, Alvarez-Buylla A, Jan LY, Jan YN. Postnatal deletion of Numb/Numblike reveals repair and remodeling capacity in the subventricular neurogenic niche. Cell. 2006;127:1253–1264. doi: 10.1016/j.cell.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Hu J, Ralls S, Kitamura T, Loh YP, Yang Y, Mukouyama YS, Ahn S. The molecular profiles of neural stem cell niche in the adult subventricular zone. PLoS One. 2012;7:e50501. doi: 10.1371/journal.pone.0050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Batt MK, Cronier BA, Jackson MC, Bruno Garza JL, Trinh DS, Mason CO, Spearry RP, Bhattacharya S, Robitz R, et al. Ciliary neurotrophic factor receptor regulation of adult forebrain neurogenesis. J Neurosci. 2013;33:1241–1258. doi: 10.1523/JNEUROSCI.3386-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulier K, Lathia JD, Marthiens V, Relucio J, Mughal MR, Tang SC, Coksaygan T, Hall PE, Chigurupati S, Patton B, et al. beta1 integrin maintains integrity of the embryonic neocortical stem cell niche. PLoS Biol. 2009;7:e1000176. doi: 10.1371/journal.pbio.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Hayashi S, Rutlin M, Muzumdar MD, Nery S, Corbin JG, Gritli-Linde A, Dellovade T, Porter JA, Rubin LL, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Mirzadeh Z, Han YG, Soriano-Navarro M, García-Verdugo JM, Alvarez-Buylla A. Cilia organize ependymal planar polarity. J Neurosci. 2010;30:2600–2610. doi: 10.1523/JNEUROSCI.3744-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3:265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Chakrapani BP, Schwegler H, Hofmann HD, Kirsch M. Neurogenesis in the dentate gyrus depends on ciliary neurotrophic factor and signal transducer and activator of transcription 3 signaling. Stem Cells. 2009;27:431–441. doi: 10.1634/stemcells.2008-0234. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T. Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci. 1999;19:5429–5434. doi: 10.1523/JNEUROSCI.19-13-05429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Taga T. Mechanisms underlying cytokine-mediated cell-fate regulation in the nervous system. Mol Neurobiol. 2002;25:233–244. doi: 10.1385/MN:25:3:233. [DOI] [PubMed] [Google Scholar]
- Nie J, Li SS, McGlade CJ. A novel PTB-PDZ domain interaction mediates isoform-specific ubiquitylation of mammalian Numb. J Biol Chem. 2004;279:20807–20815. doi: 10.1074/jbc.M311396200. [DOI] [PubMed] [Google Scholar]
- Paez-Gonzalez P, Abdi K, Luciano D, Liu Y, Soriano-Navarro M, Rawlins E, Bennett V, Garcia-Verdugo JM, Kuo CT. Ank3-dependent SVZ niche assembly is required for the continued production of new neurons. Neuron. 2011;71:61–75. doi: 10.1016/j.neuron.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Moreno M, Jamora C, Fuchs E. Sticky business: orchestrating cellular signals at adherens junctions. Cell. 2003;112:535–548. doi: 10.1016/s0092-8674(03)00108-9. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Schimmang T, Lemaistre M, Vortkamp A, Rüther U. Expression of the zinc finger gene Gli3 is affected in the morphogenetic mouse mutant extra-toes (Xt) Development. 1992;116:799–804. doi: 10.1242/dev.116.3.799. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Kokovay E, Lin G, Chuang S-M, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell-cell interactions. Cell stem cell. 2008;3:289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazaki T, Shingo T, Weiss S. The ciliary neurotrophic factor/leukemia inhibitory factor/gp130 receptor complex operates in the maintenance of mammalian forebrain neural stem cells. J Neurosci. 2001;21:7642–7653. doi: 10.1523/JNEUROSCI.21-19-07642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Merkle FT, Flames N, Tramontin AD, García-Verdugo JM, Alvarez-Buylla A. Adult ependymal cells are postmitotic and are derived from radial glial cells during embryogenesis. J Neurosci. 2005;25:10–18. doi: 10.1523/JNEUROSCI.1108-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Ruther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schütz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vortkamp A, Gessler M, Grzeschik KH. Identification of optimized target sequences for the GLI3 zinc finger protein. DNA Cell Biol. 1995;14:629–634. doi: 10.1089/dna.1995.14.629. [DOI] [PubMed] [Google Scholar]
- Wang H, Ge G, Uchida Y, Luu B, Ahn S. Gli3 is required for maintenance and fate specification of cortical progenitors. J Neurosci. 2011;31:6440–6448. doi: 10.1523/JNEUROSCI.4892-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat Neurosci. 1999;2:461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- Young KM, Fogarty M, Kessaris N, Richardson WD. Subventricular zone stem cells are heterogeneous with respect to their embryonic origins and neurogenic fates in the adult olfactory bulb. J Neurosci. 2007;27:8286–8296. doi: 10.1523/JNEUROSCI.0476-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Alfaro J, Chang EH, Zhao X, Porcionatto M, Segal RA. Numb links extracellular cues to intracellular polarity machinery to promote chemotaxis. Dev Cell. 2011;20:610–622. doi: 10.1016/j.devcel.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.