Abstract
Background
Signaling messengers and effector proteins provide an orchestrated molecular machinery to relay extracellular signals to the inside of cells and thereby facilitate distinct cellular behaviors. Formations of intracellular macromolecular complexes and segregation of signaling cascades dynamically regulate the flow of a biological process.
Scope of Review
In this review, we provide an overview of the development and application of FRET technology in monitoring cyclic nucleotide-dependent signalings and protein complexes associated with these signalings in real time and space with brief mention of other important signaling messengers and effector proteins involved in compartmentalized signaling.
Major Conclusion
The preciseness, rapidity and specificity of cellular responses indicate restricted alterations of signaling messengers, particularly in subcellular compartments rather than globally. Not only the physical confinement and selective depletion, but also the intra- and inter-molecular interactions of signaling effectors modulate the direction of signal transduction in a compartmentalized fashion. To understand the finer details of various intracellular signaling cascades and crosstalk between proteins and other effectors, it is important to visualize these processes in live cells. Förster Resonance Energy Transfer (FRET) has been established as a useful tool to do this, even with its inherent limitations.
General Significance
FRET technology remains as an effective tool for unraveling the complex organization and distribution of various endogenous signaling proteins, as well as the spatiotemporal dynamics of second messengers inside a single cell to distinguish the heterogeneity of cell signaling under normal physiological conditions and during pathological events.
Keywords: Förster Resonance Energy Transfer (FRET), Compartmentalized cell signaling, Macromolecular complex
1. Introduction
Cell signaling is a multifaceted process involving numerous signaling messengers and effector proteins [1]. To understand overall cellular behavior, it is important to concentrate on the pattern, duration and preciseness of a signaling cascade in response to cues [2]. Small, yet elegant molecules such as cAMP and cGMP play critical roles as second messengers in carrying out a precise signaling cascade with the right specificity [3–5]. The spatiotemporal regulation of the intracellular dynamics of these cyclic nucleotides can determine the duration and localization of cellular responses and segregates the global network of cell-signaling systems into separate cellular compartments. To understand the localized modulation of cyclic nucleotides, there is a need to focus on the mechanisms by which a cell maintains different cyclic nucleotide environments at different subcellular regions. Together with synthesis and catabolism by different isoforms of phosphodiesterases, nucleotide transporters such as multidrug resistance protein 4 and 5 (MRP4/5) are responsible for determining intracellular concentrations of cAMP and cGMP and the subsequent regulation of different cyclic nucleotide-dependent signalings [2, 6–13]. Cyclic nucleotide-dependent and -independent kinases, such as protein kinases A, G and C, facilitate compartmentalized signaling by restricted activation of specific substrate populations in distinct cellular compartments [14–18]. Other important signaling proteins, including the Ras superfamily of small guanosine triphosphatases (GTPase), also are dynamically regulated in time and space in order to generate discrete and localized cellular effects. The cycles of activation by guanine nucleotide exchange factor (GEF) and inactivation by GTPase-activating protein (GAP) finely control segregated interactions of GTPase with downstream effectors [19–21]. Precise measurement of free cytosolic and organellar Ca2+ is important to understand the highly localized Ca2+ signaling required for allosteric regulation of a myriad of signaling proteins, and cells provide a collection of components for managing the wide range of Ca2+-dependent spatial and temporal signals [22]. Nevertheless, the interactions of receptors either between themselves or with their downstream targets are both indispensable for initiating a signaling cascade and an early hallmark step for a cell response to external cues [23, 24].
The regulation of signaling machineries occur in a highly compartmentalized fashion, and a fine-tuned localized modulation near the target effector rather than a global signaling wave directs the specific signaling flow [2, 25]. For example, although a macromolecular complex containing CFTR and MRP4 at the apical membrane regulates the secretory properties of gut epithelia, this interaction plays an important role for β-adrenergic-stimulated contraction in cardiac myocytes, and both of these occur via the compartmentalized alteration of cAMP dynamics [26, 27]. In the past few decades, efforts have been made to better understand compartmentalized cell signaling and FRET technology, which is a quantum mechanical phenomenon that relies on spectral overlap between donor and acceptor molecules, provides a method to do so [28–30]. After being first theorized by Dr. Theodor Förster in 1948, it was more than 50 years before FRET was used in the field of cell biology for direct visualization and quantification of biological function [29, 31–34]. The spatial and temporal resolution of a signaling flow via modulation of secondary messengers or through the formation of macromolecular complexes provides more-vivid information about complex cell-signaling phenomena in real time. The focus of this review is a comprehensive exploration of FRET technology used to understand compartmentalized cell signaling.
2. Ratiometric and Direct/Sensitized FRET
FRET is the transfer of excitation energy of an electronically excited donor fluorophore to a nearby acceptor chromophore through a long-range dipole-dipole interaction and without the emission of photons [35, 36]. The spectral overlap and the dipole orientations of the donor and acceptor molecules, together with the distance between chromophores, determine the efficiency of FRET and it is inversely proportional to the sixth power of the distance between chromophores [37]. The distance dependency (<10nm) of FRET signal makes it a useful tool for measuring proximity and conformational changes of biological molecules as a spectroscopic ruler with a higher spatial resolution compared to conventional optical microscopy [37–39]. Typically, fluorescence signals are measured for cells expressing donor or acceptor or both molecules, and FRET efficiencies are calculated [40]. Two types of FRET signals can be monitored for this purpose; i) Ratiometric FRET, where the ratio between the intensity of donor and acceptor or FRET and donor signal were measured, and ii) Direct/Sensitized FRET, where the acceptor emission was measured upon the excitation of the donor. Ratiometric FRET is the most-convenient readout when the donor and acceptor are stoichiometrically fixed and fused in a single polypeptide chain. For Ratiometric FRET, the Ratio Numerator and Ratio Denominator determine the channel specificity [26, 41, 42], whereas for Direct/Sensitized FRET, the FRET channel must use a donor exciter and acceptor emitter [43–45]. An alternate intensity-based assay measuring donor dequenching after acceptor bleaching provides specific and accurate static information about FRET efficiency, but this assay is limited to single measurement due to its destructive nature. The nondestructive and fluorescence-decay kinetics-based methods, such as detection of change in the excited-state lifetime of the donor molecule, still need further development [29, 38]. For example using the fluorescence lifetime imaging microscopy (FLIM) technique, the decay kinetics of a chromophore are measured in a nanosecond timeframe. Though this technique can spatially resolve various physiological parameters in live cells independent of the chromophore concentration, temporal resolution still remains as a major limitation associated with FLIM [46, 47].
3. FRET to study compartmentalized cyclic-nucleotide signaling
In recent years, Ratiometric FRET technology has been successfully exploited to monitor the spatially restricted dynamics of a variety of cellular messengers, such as cAMP, cGMP and Ca++, and to visualize the flow of a signaling cascade in real time [48–51]. In this review, we emphasize the development and application of FRET-based sensors to resolve the spatiotemporal dynamics of cyclic nucleotides and cyclic nucleotide-dependent signaling in healthy and diseased cells. In cells, cAMP and cGMP are synthesized by adenylyl cyclase (AC) and guanylate cyclase (GC), respectively, and both cytosolic and membrane-bound forms of these two enzymes have been identified [18, 52, 53]. After synthesis, these cyclic nucleotides either degrade to their corresponding monophosphates by the action of different isoforms of phosphodiesterases (PDEs) or efflux out of the cell by the action of endogenous membrane transporters, such as MRP4/5 [2, 11, 13]. Among the eleven isoforms of PDEs, PDE 4, 7 and 8 are cAMP specific and PDE 5, 6 and 9 are cGMP specific, whereas other isoforms exhibit dual specificity [13, 54]. The distinct localizations and specificities of the enzymes maintain the discrete microdomains of cyclic nucleotides in response to different intrinsic and extrinsic signals. The spatial organization of different effector molecules by the scaffolding proteins also assists the establishment of the specific compartments where MRP4/5, together with several isoforms of PDEs, play pivotal roles in maintaining the localized restricted diffusion of these second messengers, instead of a rapid and global modulation, to induce a more-specific response [2].
In biology, the most-commonly used fluorophore, green fluorescence protein (GFP), has inherent limitations, such as slow rotation and spatially-restricted behavior, that make it unsuitable for use as a FRET indicator in bioimaging [55, 56]. However, spectral mutants of GFP, cyan (CFP) and yellow (YFP), form the best pair of FRET fluorophores [29, 57]. To visualize the spatiotemporal dynamics of cAMP, fluorophores have been attached to the cAMP-dependent protein kinase A (PKA) subunit. Unlike the bimolecular Ca++ indicator ‘Cameleon’ that consists of CFP-calmodulin and calmodulin-binding peptide M13-YFP and gives a larger FRET signal upon Ca++ binding [49, 58, 59], binding of cAMP to the PKA-regulatory subunit induces the release of catalytic subunits and therefore disrupts the FRET signal, followed by an increase in the donor-to-FRET ratio [51, 60]. Using the PKA-based FRET indicator, it has been determined that migrating cells have polarized accumulation of cAMP at the leading edge [61], and recently we have shown that inhibition of MRP4 augments the polarized cAMP concentration at the leading edge of a migrating fibroblast and thus facilitates cell migration [41]. To overcome the variability in expression of different subunits and endogenous unlabeled-molecules-mediated error in FRET detection, the need for a unimolecular fluorescent indicator arises. The cAMP-dependent conformational change in Exchange protein directly activated by cAMP (EPAC) makes it a readily targetable FRET-based sensor for cAMP by fusing the N terminus of EPAC with CFP and the C terminus with the YFP analogue [62]. Upon cAMP binding, EPAC undergoes a conformational change to liberate the catalytic domain, which moves CFP and YFP apart from each other to cause a reduced FRET signal [26, 27, 41]. Considering the relatively larger conformational change induced by cGMP and an almost100-fold higher selectivity for cGMP compared to cAMP, cGMP-dependent protein kinase (PKG) has been modified and flanked by CFP and an improved pH-insensitive YFP variant. Thus, an efficient genetically encoded FRET-based cGMP sensor called Cygnet 2.1 has been developed to visualize cGMP regulation in real time. Cygnet 2.1 undergoes a 1.4- to 1.5-fold increase in cyan-to-yellow emission upon saturation, and this dynamic range is comparable to the cAMP sensor [50, 63]. The mechanism of FRET sensors specific for cAMP and cGMP is illustrated in Figure 1. These fluorescent sensors not only resolve the precise augmentation of intracellular cAMP and cGMP dynamics, but also provide useful tools to determine the functional efficacy of various PDE and MRP4/5 inhibitors through the real-time monitoring of the competitive inhibition of cyclic nucleotides catabolism and efflux, respectively.
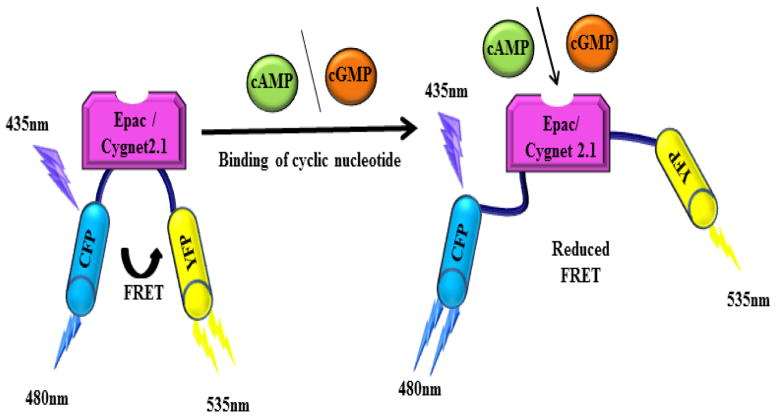
Figure 1.
Mechanism of FRET based sensors specific for cyclic nucleotides. Binding of cAMP and cGMP to the Epac and Cygnet 2.1 sensor respectively induces a conformation change to the corresponding sensors. The change in conformation takes the donor and acceptor fluorophores apart from each other which leads to a reduced FRET signal.
Our laboratory has successfully determined the spatiotemporal alteration of cyclic nucleotides at or near the plasma membrane by the effect of cilostazol (PDE3 inhibitor), zaprinast (PDE5 inhibitor) and MK571 (MRP4 inhibitor) in various cell types [26, 41, 43]. Unlike conventional cell population-based assays, the fluorescent single-cell imaging method also is capable of potentially discriminating cell-to-cell heterogeneity in particular signaling processes [64, 65].
Signaling involving membrane proteins is expected to be more profound at or near the membrane. A further modification of the FRET sensors by inserting a myristoylation-palmitoylation site into the N-terminus of the backbone sequence, anchored the protein in the plasma membrane [66]. These membrane-bound sensors can allow the resolution between global and membrane-restricted alterations of signaling messengers and provide a better understanding of the spatiotemporal regulation of various signaling cascades. Similarly, the insertions of other targeting motifs (e.g., specific for mitochondria or the nucleus) into the FRET sensor will allow a direct sensing of cyclic nucleotide dynamics at different subcellular regions inside the cell and the differential regulation of cellular targets [66–68].
4. FRET in protein-protein interactions
Recent advancements in fluorescent-protein biology, imaging methods and analyzing technologies enable the visualization of not only protein concentration, localization and trafficking, but also the mobility and interactions crucial for the function and regulation of a particular protein [69–73]. Interactions between proteins have been well studied and found to be responsible for a defined yet segregated functional and structural regulation of the interacting partners in a highly compartmentalized fashion. Regular biochemical approaches such as co-immunoprecipitation and yeast-two-hybrid screens are unable to map the specific association of various proteins in real time in living cells. FRET has added breadth to this by detecting the proximity of donor and acceptor fluorophore-fused interacting partners [72, 74–76]. Using this instantaneous and reversible method [77], both association and dissociation of the interacting proteins can be monitored under physiological conditions, which is impossible to obtain by the conventional techniques. FRET microscopy does not require the protein partners to touch each other and contributes no attraction or repulsion by itself, but typically measures the emission of both donor and acceptor after excitation of only the donor molecule [36]. For example, the most popular donor-acceptor pair, CFP/YFP, is only able to produce FRET when located closer to each other than 100Å, although they cannot come closer than 30Å to each other after being buried 15Å into the target proteins [30]. Figure 2 illustrates the induction of FRET signals by interaction of two donor and acceptor fluorophore-conjugated proteins. Recent development of DsRed protein, with increased tissue penetration and better spectral separation from cellular auto-fluorescence [78], can provide another useful FRET pair with GFP [79, 80]. In fixed cells, FRET also can be measured by immunofluorescence techniques using fluorescently tagged proteins or by microinjection of dye. FRET cannot be measured by ratio of donor and acceptor emission when donor and acceptor are two separate molecules, due to the difference in expression levels and involvement of mixed complexes between fluorescent and endogenous proteins [35]. The mathematical processing of three images with necessary correction for Direct/Sensitized FRET is required for visualization and quantification of energy transfer. In order to perform direct FRET, specific filters for either three channels (donor, acceptor and FRET) or two channels (donor or acceptor and FRET) should be configured with respective excitation and emission. Subsequent corrections are necessary to minimize the effect of background and crosstalk between channels [43, 44].
Figure 2.
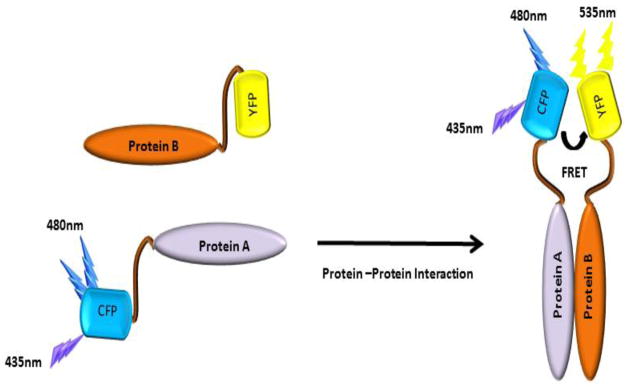
Induction of FRET signal by protein-protein interaction. Two proteins conjugated with donor and acceptor fluorophores respectively, can induce FRET signal when they interact with each other and are located closer than 100Å.
FRET allows the visualization of receptor dimerization and signaling protein interactions associated with various signaling cascades in real time [69, 70]. In our laboratory, we have detected significant FRET signal using YFP-CFTR as acceptor and CFP-PDE3A as donor molecules with a high spatiotemporal resolution, which allowed direct visualization of CFTR-containing macromolecular complexes at or near the plasma membrane and modulation of interaction sensitivity by the cyclic nucleotides levels in the vicinity [43]. In addition, the membrane surface density of the membrane-localized proteins, depending on their interactive properties and state of oligomerization in membrane bilayer surface, also can be resolved by FRET technology [81]. Both inter- and intra-molecular interactions that mediate stabilization of the multi-domain signaling protein PKCα also were studied using FRET to understand the auto-inhibition and activation of the kinase and subsequent nodal regulation of cell signaling by PKCα [16]. Not only protein-protein interactions, but also protein-nucleotide interactions can be visualized by FRET imaging. Altered interaction between RNA-binding proteins and RNA has been implicated in a number of disorders including myotonic dystrophy. Recently, acceptor photo-bleaching FRET techniques revealed important RNA motifs and RNA-binding domains that are involved in RNA toxicity [82]. Similarly, interactions between proteins and DNA are important for regulation of gene expression. The interaction between DNA and transcriptional activator or repressor proteins can be detected by FRET and thus, epigenetic regulation and other transcription modulations can be explored in real time and space [83]. With the development of drug delivery, efforts have been made to develop protein-conjugated formulations for targeted drug therapy. The efficacy of drug-protein conjugation, as well as proper intracellular targeting of drug molecules, can be evaluated by measuring the FRET signal between the drug core and the protein shell or the delivered formulation and targeted receptors in live cells [84].
However, false-negative and false-positive detection are sometimes associated with this intermolecular FRET system. Although the weak affinity of GFP variants mainly for membrane-anchored proteins is responsible for false-positive FRET to some extent, the development of new mutated monomerized GFPs eliminates this defect [35, 77]. Improper orientation of the florescent donor and acceptor molecules sometimes distances them from each other and thus abrogates the FRET signal, even though the fusion partners are interacting. Therefore, obtaining higher spatiotemporal resolution of an already known interaction may be a better use of the FRET technique than screening the proteomes of unknown interacting partners.
5. Signaling effectors and FRET
In addition to cyclic nucleotides, myriads of effector molecules play important roles in compartmentalized signaling. Various FRET sensors have been used extensively to study signaling effectors in real time and space. Unimolecular FRET-based biosensor for Ca2+ has been developed for real-time monitoring of highly localized Ca2+ signals in live cells. This improved Cameleon indicator consists of a tandem fusion of CFP, calmodulin, calmodulin-binding peptide M13 and YFP. Upon Ca2+ binding, calmodulin muffles with M13 and gives a higher FRET signal [49]. Further modifications of the acceptor and donor fluorophores improved the pH sensitivity of the sensor [48]. With the development of Troponin C (TnC)-based FRET that possesses higher sensitivity, better ion selectivity and complete inertness toward the host-cell biochemistry, the in vivo Ca 2+ imaging gained improved response kinetics, signal size, stability, and temporal resolution [85]. Using specifically targeted unimolecular FRET sensors comprised of the ε subunit of bacterial F0F1-ATP synthase, Imamura, et al. monitored real-time dynamics of ATP, the major energy currency of cells, at different subcellular compartments and found a direct correlation between glycolysis and ATP generation specifically in the mitochondria [86]. Protease activities also have been monitored in real time using various protease substrates containing FRET sensors that get cleaved by specific proteases causing a disruption in FRET signal [87]. In cells, cyclic nucleotides execute their function primarily via the activation of their effector kinase (e.g., PKA and PKG), and FRET can be used as a powerful tool for visualization of the kinase activities [88, 89] by using the specific phosphorylation substrate peptide flanked by CFP and YFP as sensors [90–93]. The improved, membrane-targeted, forkhead-associated domain 1 (FHA1) and the PKA substrate sequence LRRATLVD containing sensor (pmAKAR3) has been used for monitoring the polarized PKA activation in the leading edge, which is an essential early hallmark step for directional cell migration [61, 94]. The use of FRET-based kinase sensors may significantly contribute to the identification of downstream effector kinases that mediate regulation of various signaling processes and detection of the correlation between cyclic nucleotide dynamics and kinase activity in real time and space. Both PKA and PKG have been shown to dichotomously regulate various complex and integrated biological processes in a Rac GTPase-dependent manner and also via involvement of other signaling molecules [14, 17, 18, 95, 96]. Rac activation in membrane ruffling at the leading edge of a migrating cell has been observed by FRET technology using p21-activated kinase (PAK1)-based sensors that specifically bind to activated Rac [21]. Similarly, growth factor-induced activation of Ras at the plasma membrane was unraveled using a unimolecular FRET sensor consisting of H-Ras and the Ras-binding domain of Raf (called Raichu-Ras), whereas a simultaneous activation of Rap1 in the perinuclear region was detected using an identical Raichu-Rap sensor [20, 28].
6. Future prospects
Considering the complexity of different endogenous signaling molecules, currently available and yet-to-be-developed specific FRET-based sensors can be used as a direct indicator of protein function with high spatiotemporal resolution in live cells. For the successful development of new sensors, overlap of the excitation and emission spectra for donor and acceptor need to be minimized rather than maximizing the overlap between the donor emission and acceptor excitation spectra [40]. With advanced FRET technology, it is no more impossible to visualize whether, together with the uneven expression levels, the differential protein activities at different subcellular localization and in various macromolecular environments in the vicinity are responsible for the precise regulation of specific cellular events. Using FRET-based methods, the contribution of protein-protein interactions in restricted cellular compartments can be monitored in real time, and the heterogeneity of signaling cascades at different locations under the influence of a unique cue can be revealed and interpreted. To some extent, our group and others have tried to use FRET as a novel approach to identify the spatial regulation of various protein interactomes and define the repertoire of signaling events. Thus, beyond the extensively studied whole-cell signaling mediated by effector molecules, an emphasis has been continually imparted on the spatiotemporal regulation of various physiological and pathological signaling phenomena. However, the scope of FRET technology with perpetual improvements is yet to be explored for resolving the complexity of biological processes. With the commercial availability of high-content microscopy [97, 98], we await a new era with FRET indicators available for rapid and instantaneous optical imaging of biochemical and physiological functions as well as for high-throughput screening of inhibitor, potentiator and interacting partners of specific protein molecules. In addition, nucleotide-interacting and -modulating proteins can be identified using FRET-based methods that can provide attractive strategies for development of genetic and epigenetic therapies. Even for the evaluation of targeted drug delivery, FRET technology has been implicated to be important. Development of fluorescent reporters with more sensitivity for the subtle conformational changes associated with some interesting intracellular events will provide visualization of activation and localization of endogenous biomolecules involved in diverse signaling cascades. Advanced single-molecule spectroscopy holds great promise in revealing the lateral mobility, trafficking and interaction patterns of a single protein molecule in live cells [43, 99]. Altogether, taking the advantages from live cell fluorescent markers in conjunction with advanced FRET-based imaging technology, researchers will continue to assess and understand the functional outcomes associated with the heterogeneous and multilayered cell signaling processes.
High Lights.
FRET is the transfer of energy from an excited fluorophore to a nearby chromophore.
Compartmentalized signaling regulates the preciseness of cell behaviors.
Spatio-temporal regulation of cellular events can be monitored by FRET in live cells.
We explored application of FRET in understanding the compartmentalized cell signaling.
Acknowledgments
We thank J. Denise Wetzel, CCHMC Medical Writer, for critical review and editing of our manuscript. This work was supported by grants RO1- DK080834 and RO1-DK093045 to A. P. Naren.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora K, Sinha C, Zhang W, Ren A, Moon CS, Yarlagadda S, Naren AP. Compartmentalization of cyclic nucleotide signaling: a question of when, where, and why? Pflugers Arch. 2013;465:1397–1407. doi: 10.1007/s00424-013-1280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheepala S, Hulot JS, Morgan JA, Sassi Y, Zhang W, Naren AP, Schuetz JD. Cyclic nucleotide compartmentalization: contributions of phosphodiesterases and ATP-binding cassette transporters. Annu Rev Pharmacol Toxicol. 2013;53:231–253. doi: 10.1146/annurev-pharmtox-010611-134609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elferink JG, de Koster BM. The effect of cyclic GMP and cyclic AMP on migration by electroporated human neutrophils. Eur J Pharmacol. 1993;246:157–161. doi: 10.1016/0922-4106(93)90093-o. [DOI] [PubMed] [Google Scholar]
- 5.Elferink JG, VanUffelen BE. The role of cyclic nucleotides in neutrophil migration. Gen Pharmacol. 1996;27:387–393. doi: 10.1016/0306-3623(95)00070-4. [DOI] [PubMed] [Google Scholar]
- 6.Schuetz JD, Connelly MC, Sun D, Paibir SG, Flynn PM, Srinivas RV, Kumar A, Fridland A. MRP4: A previously unidentified factor in resistance to nucleoside-based antiviral drugs. Nat Med. 1999;5:1048–1051. doi: 10.1038/12487. [DOI] [PubMed] [Google Scholar]
- 7.Hara Y, Sassi Y, Guibert C, Gambaryan N, Dorfmuller P, Eddahibi S, Lompre AM, Humbert M, Hulot JS. Inhibition of MRP4 prevents and reverses pulmonary hypertension in mice. J Clin Invest. 2011;121:2888–2897. doi: 10.1172/JCI45023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van de Ven R, Scheffer GL, Reurs AW, Lindenberg JJ, Oerlemans R, Jansen G, Gillet JP, Glasgow JN, Pereboev A, Curiel DT, Scheper RJ, de Gruijl TD. A role for multidrug resistance protein 4 (MRP4; ABCC4) in human dendritic cell migration. Blood. 2008;112:2353–2359. doi: 10.1182/blood-2008-03-147850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Copsel S, Garcia C, Diez F, Vermeulem M, Baldi A, Bianciotti LG, Russel FG, Shayo C, Davio C. Multidrug resistance protein 4 (MRP4/ABCC4) regulates cAMP cellular levels and controls human leukemia cell proliferation and differentiation. J Biol Chem. 2011;286:6979–6988. doi: 10.1074/jbc.M110.166868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tagami M, Kusuhara S, Imai H, Uemura A, Honda S, Tsukahara Y, Negi A. MRP4 knockdown enhances migration, suppresses apoptosis, and produces aggregated morphology in human retinal vascular endothelial cells. Biochem Biophys Res Commun. 2010;400:593–598. doi: 10.1016/j.bbrc.2010.08.109. [DOI] [PubMed] [Google Scholar]
- 11.Russel FG, Koenderink JB, Masereeuw R. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol Sci. 2008;29:200–207. doi: 10.1016/j.tips.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Schinkel AH, Jonker JW. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev. 2003;55:3–29. doi: 10.1016/s0169-409x(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 13.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 14.Howe AK, Baldor LC, Hogan BP. Spatial regulation of the cAMP-dependent protein kinase during chemotactic cell migration. Proc Natl Acad Sci U S A. 2005;102:14320–14325. doi: 10.1073/pnas.0507072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulucci-Holthauzen AA, Vergara LA, Bellot LJ, Canton D, Scott JD, O’Connor KL. Spatial distribution of protein kinase A activity during cell migration is mediated by A-kinase anchoring protein AKAP Lbc. J Biol Chem. 2009;284:5956–5967. doi: 10.1074/jbc.M805606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swanson CJ, Ritt M, Wang W, Lang M, Narayan A, Tesmer J, Westfall M, Sivaramakrishnan S. Conserved Modular Domains Team Up to Latch-Open Active PKCalpha. J Biol Chem. 2014 doi: 10.1074/jbc.M113.534750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negash S, Narasimhan SR, Zhou W, Liu J, Wei FL, Tian J, Raj JU. Role of cGMP-dependent protein kinase in regulation of pulmonary vascular smooth muscle cell adhesion and migration: effect of hypoxia. Am J Physiol Heart Circ Physiol. 2009;297:H304–312. doi: 10.1152/ajpheart.00077.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo D, Tan YC, Wang D, Madhusoodanan KS, Zheng Y, Maack T, Zhang JJ, Huang XY. A Rac-cGMP signaling pathway. Cell. 2007;128:341–355. doi: 10.1016/j.cell.2006.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 20.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 21.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 22.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 23.Milligan G. G protein-coupled receptor dimerization: function and ligand pharmacology. Mol Pharmacol. 2004;66:1–7. doi: 10.1124/mol.104.000497.. [DOI] [PubMed] [Google Scholar]
- 24.Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Naren AP. CFTR chloride channel in the apical compartments: spatiotemporal coupling to its interacting partners. Integr Biol (Camb) 2010;2:161–177. doi: 10.1039/b924455g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Krishnamurthy PC, Penmatsa H, Marrs KL, Wang XQ, Zaccolo M, Jalink K, Li M, Nelson DJ, Schuetz JD, Naren AP. Spatiotemporal coupling of cAMP transporter to CFTR chloride channel function in the gut epithelia. Cell. 2007;131:940–951. doi: 10.1016/j.cell.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sellers ZM, Naren AP, Xiang Y, Best PM. MRP4 and CFTR in the regulation of cAMP and beta-adrenergic contraction in cardiac myocytes. Eur J Pharmacol. 2012;681:80–87. doi: 10.1016/j.ejphar.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Roessel P, Brand AH. Imaging into the future: visualizing gene expression and protein interactions with fluorescent proteins. Nat Cell Biol. 2002;4:E15–20. doi: 10.1038/ncb0102-e15. [DOI] [PubMed] [Google Scholar]
- 29.Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 31.Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- 32.Tsien RY. Fluorescent probes of cell signaling. Annu Rev Neurosci. 1989;12:227–253. doi: 10.1146/annurev.ne.12.030189.001303. [DOI] [PubMed] [Google Scholar]
- 33.Tsien RY. Fluorescence ratio imaging of dynamic intracellular signals. Acta Physiol Scand Suppl. 1989;582:6. doi: 10.1117/12.962706. [DOI] [PubMed] [Google Scholar]
- 34.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Annalen der Physik. 1948;437:55–75. [Google Scholar]
- 35.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 36.Forster T. Energy migration and fluorescence. 1946. J Biomed Opt. 2012;17:011002. doi: 10.1117/1.JBO.17.1.011002. [DOI] [PubMed] [Google Scholar]
- 37.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 38.Gordon GW, Berry G, Liang XH, Levine B, Herman B. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J. 1998;74:2702–2713. doi: 10.1016/S0006-3495(98)77976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Z, Liu Y. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys J. 2001;81:2395–2402. doi: 10.1016/S0006-3495(01)75886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsien RY. Indicators based on fluorescence resonance energy transfer (FRET) Cold Spring Harb Protoc. 2009 doi: 10.1101/pdb.top57. pdb top57. [DOI] [PubMed] [Google Scholar]
- 41.Sinha C, Ren A, Arora K, Moon CS, Yarlagadda S, Zhang W, Cheepala SB, Schuetz JD, Naren AP. Multi-drug resistance protein 4 (MRP4)-mediated regulation of fibroblast cell migration reflects a dichotomous role of intracellular cyclic nucleotides. J Biol Chem. 2013;288:3786–3794. doi: 10.1074/jbc.M112.435925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Penmatsa H, Ren A, Punchihewa C, Lemoff A, Yan B, Fujii N, Naren AP. Functional regulation of cystic fibrosis transmembrane conductance regulator-containing macromolecular complexes: a small-molecule inhibitor approach. Biochem J. 2011;435:451–462. doi: 10.1042/BJ20101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penmatsa H, Zhang W, Yarlagadda S, Li C, Conoley VG, Yue J, Bahouth SW, Buddington RK, Zhang G, Nelson DJ, Sonecha MD, Manganiello V, Wine JJ, Naren AP. Compartmentalized cyclic adenosine 3′,5′-monophosphate at the plasma membrane clusters PDE3A and cystic fibrosis transmembrane conductance regulator into microdomains. Mol Biol Cell. 2010;21:1097–1110. doi: 10.1091/mbc.E09-08-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galperin E, Sorkin A. Visualization of Rab5 activity in living cells by FRET microscopy and influence of plasma-membrane-targeted Rab5 on clathrin-dependent endocytosis. J Cell Sci. 2003;116:4799–4810. doi: 10.1242/jcs.00801. [DOI] [PubMed] [Google Scholar]
- 45.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol. 2000;10:1395–1398. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 46.Bastiaens PI, Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/s0962-8924(98)01410-x. [DOI] [PubMed] [Google Scholar]
- 47.van Munster EB, Gadella TW. Fluorescence lifetime imaging microscopy (FLIM) Adv Biochem Eng Biotechnol. 2005;95:143–175. doi: 10.1007/b102213. [DOI] [PubMed] [Google Scholar]
- 48.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 50.Honda A, Adams SR, Sawyer CL, Lev-Ram V, Tsien RY, Dostmann WR. Spatiotemporal dynamics of guanosine 3′,5′-cyclic monophosphate revealed by a genetically encoded, fluorescent indicator. Proc Natl Acad Sci U S A. 2001;98:2437–2442. doi: 10.1073/pnas.051631298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DiPilato LM, Cheng X, Zhang J. Fluorescent indicators of cAMP and Epac activation reveal differential dynamics of cAMP signaling within discrete subcellular compartments. Proc Natl Acad Sci U S A. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Denninger JW, Marletta MA. Guanylate cyclase and the .NO/cGMP signaling pathway. Biochim Biophys Acta. 1999;1411:334–350. doi: 10.1016/s0005-2728(99)00024-9. [DOI] [PubMed] [Google Scholar]
- 53.Hanoune J, Defer N. Regulation and role of adenylyl cyclase isoforms. Annu Rev Pharmacol Toxicol. 2001;41:145–174. doi: 10.1146/annurev.pharmtox.41.1.145. [DOI] [PubMed] [Google Scholar]
- 54.Houslay MD, Milligan G. Tailoring cAMP-signalling responses through isoform multiplicity. Trends Biochem Sci. 1997;22:217–224. doi: 10.1016/s0968-0004(97)01050-5. [DOI] [PubMed] [Google Scholar]
- 55.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 56.Tsien RY, Bacskai BJ, Adams SR. FRET for studying intracellular signalling. Trends Cell Biol. 1993;3:242–245. doi: 10.1016/0962-8924(93)90124-j. [DOI] [PubMed] [Google Scholar]
- 57.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 58.Emmanouilidou E, Teschemacher AG, Pouli AE, Nicholls LI, Seward EP, Rutter GA. Imaging Ca2+ concentration changes at the secretory vesicle surface with a recombinant targeted cameleon. Curr Biol. 1999;9:915–918. doi: 10.1016/s0960-9822(99)80398-4. [DOI] [PubMed] [Google Scholar]
- 59.Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem. 1997;272:13270–13274. doi: 10.1074/jbc.272.20.13270. [DOI] [PubMed] [Google Scholar]
- 60.Zaccolo M, De Giorgi F, Cho CY, Feng L, Knapp T, Negulescu PA, Taylor SS, Tsien RY, Pozzan T. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat Cell Biol. 2000;2:25–29. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- 61.Lim CJ, Kain KH, Tkachenko E, Goldfinger LE, Gutierrez E, Allen MD, Groisman A, Zhang J, Ginsberg MH. Integrin-mediated protein kinase A activation at the leading edge of migrating cells. Mol Biol Cell. 2008;19:4930–4941. doi: 10.1091/mbc.E08-06-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ponsioen B, Zhao J, Riedl J, Zwartkruis F, van der Krogt G, Zaccolo M, Moolenaar WH, Bos JL, Jalink K. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 2004;5:1176–1180. doi: 10.1038/sj.embor.7400290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato M, Hida N, Ozawa T, Umezawa Y. Fluorescent indicators for cyclic GMP based on cyclic GMP-dependent protein kinase Ialpha and green fluorescent proteins. Anal Chem. 2000;72:5918–5924. doi: 10.1021/ac0006167. [DOI] [PubMed] [Google Scholar]
- 64.Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–1715. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- 65.Adams SR, Harootunian AT, Buechler YJ, Taylor SS, Tsien RY. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–697. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- 66.Terrin A, Di Benedetto G, Pertegato V, Cheung YF, Baillie G, Lynch MJ, Elvassore N, Prinz A, Herberg FW, Houslay MD, Zaccolo M. PGE(1) stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. J Cell Biol. 2006;175:441–451. doi: 10.1083/jcb.200605050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliveira RF, Terrin A, Di Benedetto G, Cannon RC, Koh W, Kim M, Zaccolo M, Blackwell KT. The role of type 4 phosphodiesterases in generating microdomains of cAMP: large scale stochastic simulations. PloS one. 2010;5:e11725. doi: 10.1371/journal.pone.0011725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stangherlin A, Koschinski A, Terrin A, Zoccarato A, Jiang H, Fields LA, Zaccolo M. Analysis of compartmentalized cAMP: a method to compare signals from differently targeted FRET reporters. Methods Mol Biol. 2014;1071:59–71. doi: 10.1007/978-1-62703-622-1_5. [DOI] [PubMed] [Google Scholar]
- 69.Li HY, Ng EK, Lee SM, Kotaka M, Tsui SK, Lee CY, Fung KP, Waye MM. Protein-protein interaction of FHL3 with FHL2 and visualization of their interaction by green fluorescent proteins (GFP) two-fusion fluorescence resonance energy transfer (FRET) J Cell Biochem. 2001;80:293–303. [PubMed] [Google Scholar]
- 70.Tertoolen LG, Blanchetot C, Jiang G, Overvoorde J, Gadella TW, Jr, Hunter T, den Hertog J. Dimerization of receptor protein-tyrosine phosphatase alpha in living cells. BMC Cell Biol. 2001;2:8. doi: 10.1186/1471-2121-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siegel RM, Frederiksen JK, Zacharias DA, Chan FK, Johnson M, Lynch D, Tsien RY, Lenardo MJ. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 72.Ruehr ML, Zakhary DR, Damron DS, Bond M. Cyclic AMP-dependent protein kinase binding to A-kinase anchoring proteins in living cells by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. J Biol Chem. 1999;274:33092–33096. doi: 10.1074/jbc.274.46.33092. [DOI] [PubMed] [Google Scholar]
- 73.Janetopoulos C, Jin T, Devreotes P. Receptor-mediated activation of heterotrimeric G-proteins in living cells. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 74.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol. 2000;2:168–172. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 75.Day RN. Visualization of Pit-1 transcription factor interactions in the living cell nucleus by fluorescence resonance energy transfer microscopy. Mol Endocrinol. 1998;12:1410–1419. doi: 10.1210/mend.12.9.0168. [DOI] [PubMed] [Google Scholar]
- 76.Prufer K, Racz A, Lin GC, Barsony J. Dimerization with retinoid X receptors promotes nuclear localization and subnuclear targeting of vitamin D receptors. J Biol Chem. 2000;275:41114–41123. doi: 10.1074/jbc.M003791200. [DOI] [PubMed] [Google Scholar]
- 77.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 78.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 79.Tsien RY. Rosy dawn for fluorescent proteins. Nat Biotechnol. 1999;17:956–957. doi: 10.1038/13648. [DOI] [PubMed] [Google Scholar]
- 80.Zacharias DA. Sticky caveats in an otherwise glowing report: oligomerizing fluorescent proteins and their use in cell biology. Sci STKE. 2002;2002:pe23. doi: 10.1126/stke.2002.131.pe23. [DOI] [PubMed] [Google Scholar]
- 81.Melo AM, Fedorov A, Prieto M, Coutinho A. Exploring homo-FRET to quantify the oligomer stoichiometry of membrane-bound proteins involved in a cooperative partition equilibrium. Phys Chem Chem Phys: PCCP. 2014 doi: 10.1039/c4cp00060a. [DOI] [PubMed] [Google Scholar]
- 82.Rehman S, Gladman JT, Periasamy A, Sun Y, Mahadevan MS. Development of an AP-FRET Based Analysis for Characterizing RNA-Protein Interactions in Myotonic Dystrophy (DM1) PloS one. 2014;9:e95957. doi: 10.1371/journal.pone.0095957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wyhs N, Walker D, Giovinazzo H, Yegnasubramanian S, Nelson WG. Time-Resolved Fluorescence Resonance Energy Transfer Assay for Discovery of Small-Molecule Inhibitors of Methyl-CpG Binding Domain Protein 2. J Biomol Screen. 2014 doi: 10.1177/1087057114526433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banerjee V, Das KP. Structure and Functional Properties of a Multimeric Protein alphaA-Crystallin Adsorbed on Silver Nanoparticle Surface. Langmuir. 2014;30:4775–4783. doi: 10.1021/la5007007. [DOI] [PubMed] [Google Scholar]
- 85.Qian T, Lu S, Ma H, Fang J, Zhong W, Wang Y. FRET imaging of calcium signaling in live cells in the microenvironment. Integr Biol (Camb) 2013;5:431–438. doi: 10.1039/c2ib20264f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim GB, Kim YP. Analysis of protease activity using quantum dots and resonance energy transfer. Theranostics. 2012;2:127–138. doi: 10.7150/thno.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wouters FS, Bastiaens PI. Fluorescence lifetime imaging of receptor tyrosine kinase activity in cells. Curr Biol. 1999;9:1127–1130. doi: 10.1016/s0960-9822(99)80484-9. [DOI] [PubMed] [Google Scholar]
- 89.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, Hanby A, Harris W, Barnes D, Schmidt S, Mellor H, Bastiaens PI, Parker PJ. Imaging protein kinase Calpha activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 90.Kurokawa K, Mochizuki N, Ohba Y, Mizuno H, Miyawaki A, Matsuda M. A pair of fluorescent resonance energy transfer-based probes for tyrosine phosphorylation of the CrkII adaptor protein in vivo. J Biol Chem. 2001;276:31305–31310. doi: 10.1074/jbc.M104341200. [DOI] [PubMed] [Google Scholar]
- 91.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato M, Ozawa T, Inukai K, Asano T, Umezawa Y. Fluorescent indicators for imaging protein phosphorylation in single living cells. Nat Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- 93.Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sachs BD, Baillie GS, McCall JR, Passino MA, Schachtrup C, Wallace DA, Dunlop AJ, MacKenzie KF, Klussmann E, Lynch MJ, Sikorski SL, Nuriel T, Tsigelny I, Zhang J, Houslay MD, Chao MV, Akassoglou K. p75 neurotrophin receptor regulates tissue fibrosis through inhibition of plasminogen activation via a PDE4/cAMP/PKA pathway. J Cell Biol. 2007;177:1119–1132. doi: 10.1083/jcb.200701040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen L, Zhang JJ, Huang XY. cAMP inhibits cell migration by interfering with Rac-induced lamellipodium formation. J Biol Chem. 2008;283:13799–13805. doi: 10.1074/jbc.M800555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 97.Abraham VC, Taylor DL, Haskins JR. High content screening applied to large-scale cell biology. Trends Biotechnol. 2004;22:15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 98.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat Rev Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 99.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]