Abstract
Several single nucleotide polymorphisms have been identified in cyclooxygenase-2 (COX-2) genes (e.g., −765 G>C (rs20417), −1195G>A (rs689466), and 8473 C>T (rs5275)). The association of these SNPs with the risk of different cancer types is still controversial. This study aims to evaluate the correlation between these SNPs and breast cancer risk in different ethnic groups. We have searched PubMed, Web of Knowledge, and Embase for relevant studies. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to estimate the strength of the associations. A total of 13 studies (15,330 cases and 19,260 controls) were eligible for meta-analysis. This meta-analysis showed that COX-2 rs20417 polymorphism was correlated with an increased risk of breast cancer in Caucasians, while rs689466 was associated with a decreased risk of breast cancer in Caucasians. The rs5275 polymorphism had no association with breast cancer risk.
1. Introduction
Breast cancer is the most common cancer in women worldwide [1]. It is a multifactorial disease caused by complex genetic and environmental factors [2]. Allele variants in oncogenes are candidate genetic risk factors that may alter breast cancer onset and outcome. Previous researches have suggested that the risk of breast cancer is affected by multiple environmental factors as well as genetic alterations, such as genetic polymorphisms [3, 4].
Cyclooxygenase (COX), also known as prostaglandin endoperoxide synthetase (PTGS), plays an important role in the inflammatory process by converting arachidonic acid to prostaglandins (PG) [5]. There are two COX isoforms: COX-1 and COX-2. COX-1 is present in many tissues and is involved in PG synthesis. By contrast, COX-2 is not detected in most normal tissues but is often overexpressed in many tumor types [6]. COX-2 can be rapidly induced by a variety of mitogenic and inflammatory stimuli and elevate the production of prostaglandins, which contribute to tumor occurrence and progression by modulating cell proliferation, apoptosis, and angiogenesis [6–8]. In breast cancer, several studies have suggested that moderate to high COX-2 expression is related to the genesis of mammary tumors and the expression level is associated with the aggressiveness of breast cancer, including large tumor size, positive axillary lymph node metastases, and HER2-positive tumor status [9–11]. Targeted inhibition of COX-2 blocked the proliferation of breast cancer cell lines in vitro and prevented the occurrence of rat breast cancer chemically induced by DMBA [12].
Genetic polymorphisms in COX-2 have been shown to alter its expression and influence the susceptibility to various carcinomas [13, 14], including breast cancer [15]. The human COX-2 gene (also known as PTGS2) is located on chromosome 1q25.2-q25.3 and consists of 10 exons spanning 8.3 kb [16]. Several single-nucleotide polymorphisms (SNPs) in COX-2 have been identified, of which three functional SNPs, −765 G>C (rs20417), −1195G>A (rs689466) in the promoter region, and the 8473 C>T (rs5275) in the 3′UTR of COX-2, have been widely investigated [13–15].
Previous functional studies have suggested that the rs20417 polymorphism may eliminate a Sp1-binding site but create an E2F binding site and result in altered COX-2 expression [13]. The rs5275 polymorphism was shown to be associated with the alteration of mRNA level of the gene as sequences within the 3′UTR are important for message stability and translational efficiency [17]. There are many studies that have investigated the association between COX-2 polymorphisms and breast cancer risk. However, the results are inconsistent. For example, Fawzy et al. reported that rs5275 polymorphism was associated with the BC in Egyptian women. The individuals with rs5275 CC genotypes showed significant increase in plasma PGE2 levels [18]. However, Brasky et al. demonstrated that rs5275 had no association with breast cancer risk in Caucasians [19]. In our previous study, variant genotypes of COX-2 rs20417 G>C (GC/CC) were associated with increased breast cancer risk. Furthermore, the increased risk was more prominent among the younger subjects (OR = 1.61, 95% CI = 1.00–2.61). The variant genotypes were also associated with tumor size (OR = 3.01, 95% CI = 1.47–6.12) [20].
To clarify the role of COX-2 polymorphisms in breast cancer risk, Yu et al. conducted a meta-analysis on the associations between several COX-2 polymorphisms and breast cancer risk. The results suggested borderline increased risk of breast cancer with rs5277 but no significant associations with the rs20417 and rs5275 polymorphisms [15]. However, of the studies included in their meta-analysis, only two studies were carried out in Asians [23, 28] and the rs689466 polymorphism was not involved. To make a more precise estimation, we conduct the present meta-analysis on all eligible case-control studies to evaluate the association between the three common SNPs (rs20417, rs689466, and rs5275) and breast cancer susceptibility.
2. Materials and Methods
2.1. Publication Search
We searched the electronic databases of PubMed, Web of Knowledge, and Embase to collect articles with case-control studies related to the association of COX-2 polymorphisms with breast cancer risk. The keywords were as follows: breast cancer/breast carcinoma, Cyclooxygenase-2/COX-2/PTGS, and polymorphism/genotype/SNP. All qualified studies prior to February 28, 2014, were included. The eligible literature must be published in English. Furthermore, reference lists of main reports and review articles were also reviewed manually to identify additional relevant publications.
2.2. Selection Criteria
The following criteria were used to select studies for further meta-analysis: (1) case-control studies; (2) studies that evaluated the associations between COX-2 polymorphisms and breast cancer risk; (3) studies that contained at least two comparison groups (cancer versus control); (4) studies that included detailed genotyping data.
2.3. Data Extraction and Synthesis
Articles were reviewed independently by two reviewers and data with discrepancies were discussed by all authors. For each included study, the following information was collected: first author, year of publication, country of origin, ethnicity, source of control, total number of cases and controls, and genotyping methods as well as number of cases and controls with the different genotypes. Different ethnic groups were categorized as Caucasian, Asian, African, and “mixed.” All the case and control groups were well controlled. The noncancer controls had no history of gynecologic disease, and there was no present evidence of any malignant disease. The histories of chronic inflammatory condition or other malignancies of the patients were not considered in this study.
2.4. Statistical Analysis
The associations between COX-2 polymorphisms and breast cancer risk were measured by odds ratio (OR) with 95% confidence interval (CI). The significance of the pooled OR was determined by the Z test.
The meta-analysis assessed association by using 4 different genetic models: (1) dominant genetic model—the comparison groups were the wild-type homozygous genotype versus the variant allele-positive genotypes (AA + Aa versus aa); (2) recessive genetic model—the comparison groups were the variant homozygous genotype versus the rest (AA versus aa + Aa); (3) homozygous genetic model—comparison was between the 2 homozygous genotypes (AA versus aa); and (4) allele contrast genetic model—the comparison was between the heterozygous and the homozygous wild-type genotype groups (Aa versus aa (where “a” is the wild-type allele and “A” is the variant allele)).
Statistical heterogeneity among studies was assessed with the Q and I 2 statistics. If the P value of heterogeneity test was more than 0.1 (P ≥ 0.1), the pooled OR estimate of the study was calculated by the fixed-effects model. Otherwise, the random-effects model was used [11]. The value of the I index is used to assess the degree of heterogeneity (I 2 < 25%: no heterogeneity; 25% < I 2 < 50%: moderate heterogeneity; 50% < I 2 < 75%: high heterogeneity; I 2 > 75%: extreme high heterogeneity). Publication bias was evaluated by the funnel plot and further assessed by the method of Egger's linear regression test. All statistical analyses were carried out with the review manager version 5.1 (Revman; The Cochrane Collaboration, Oxford, UK).
3. Results
3.1. Characteristics of Studies
As shown in Figure 1, a total of 378 potential publications were initially extracted. After reading the abstracts, we excluded 176 irrelevant studies, 113 studies with insufficient data, and 53 duplicated ones. In-depth reading of the remaining articles led to further exclusion of 12 articles with no detailed genotyping data, 6 studies with no case-control, 3 laboratory studies, and 4 systematic review articles. Finally, 13 studies from 11 articles were collected for this meta-analysis.
Figure 1.
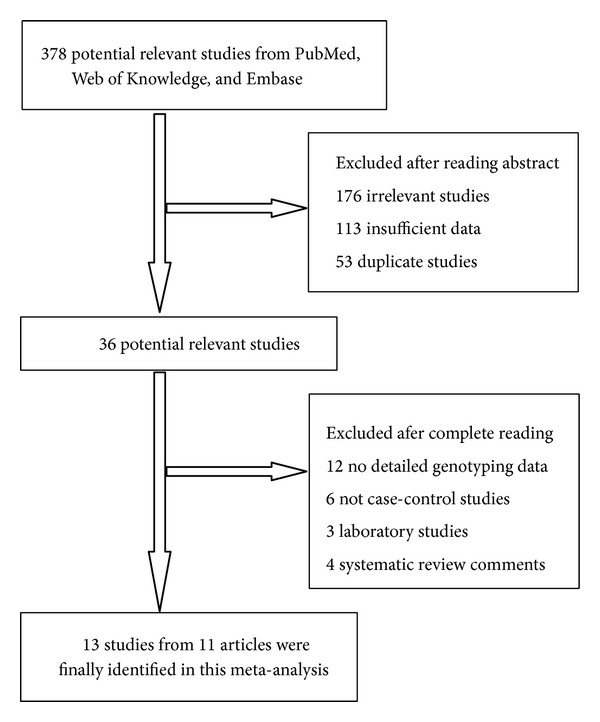
Flow chart of studies selection in this meta-analysis.
Overall, 13 studies on COX-2 polymorphisms and breast cancer risk were identified [16, 18–27], including a total of 15,330 cases and 19,260 case-free controls. The characteristics of the included studies are listed in Table 1. Among the eligible 13 studies, nine studies were carried out in Caucasians from USA, Austria, Denmark, Brazil, and nine European countries. Two were based on Asian background and were carried out in China. Only one study carried out in Egypt was based on African background. One study was on mixed ethnic groups. All studies were case-controlled. All breast cancers were confirmed by histology or pathology. Moreover, controls were mainly matched by age. Five studies were hospital-based and eight were population-based.
Table 1.
Characteristics of the studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Study design | Genotyping method | Source of control | Total sample size (case/control) | SNP number |
|---|---|---|---|---|---|---|---|---|
| Gao [20] | 2014 | China | Asian | CC | TaqMan | Hospital | 465/799 | 1, 3 |
| Fawzy [18] | 2013 | Egypt | African | CC | PCR-RFLP | Hospital | 160/150 | 3 |
| Brasky [19] | 2011 | USA | Caucasian | CC | TaqMan | Population | 1077/1910 | 2, 3 |
| Piranda [16] | 2010 | Brazil | Caucasian | CC | TaqMan | Population | 318/273 | 1, 2, 3 |
| Dossus [21] | 2010 | USA, Europe | Caucasian | CC | Illumina | Population | 6292/8135 | 1, 2, 3 |
| Abraham [22] | 2009 | EPIC | Caucasian | CC | TaqMan | Population | 2200/2280 | 3 |
| Gao [23] | 2007 | China | Asian | CC | PCR-RFLP | Hospital | 601/643 | 1, 2, 3 |
| Cox 1 [24] | 2007 | USA | Caucasian | CC | TaqMan | Population | 1270/1762 | 1, 3 |
| Cox 2 [24] | 2007 | USA | Caucasian | CC | TaqMan | Population | 317/634 | 3 |
| Cox 3 [24] | 2007 | USA | Caucasian | CC | TaqMan | Population | 702/703 | 3 |
| Vogel [25] | 2007 | Denmark | Caucasian | CC | TaqMan | Hospital | 361/361 | 3 |
| Langsenlehner [26] | 2006 | Austria | Caucasian | CC | TaqMan | Hospital | 500/500 | 3 |
| Shen [27] | 2006 | USA | Mixed | CC | PCR-RFLP | Population | 1067/1110 | 1, 3 |
CC: case-control; PCR: polymerase chain reaction; RFLP: restriction fragment length polymorphism. EPIC: European Prospective Investigation of Cancer (a prospective study of diet and cancer being carried out in nine European countries). SNP: single-nucleotide polymorphisms; SNP number 1: −765G>C (rs20417); 2: −1195G>A (rs689466); 3: 8473T>C (rs5275).
3.2. Meta-Analysis Results
As shown in Table 2, the frequencies of the minor allele varied widely across the eligible studies, ranging from 0.06 to 0.28 (rs20417), 0.12 to 0.54 (rs689466), and 0.18 to 0.45 (rs5275). The average frequencies of the minor allele in the three polymorphisms were 0.17, 0.22, and 0.33, respectively.
Table 2.
COX-2 polymorphisms genotype distribution and allele frequency in cases and controls.
| First author | Genotype (N) | Allele frequency (N) | MAF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||||||
| Total | AA | AB | BB | Total | AA | AB | BB | A | B | A | B | ||
| rs20417 | |||||||||||||
| Gao 2014 [20] | 465 | 394 | 67 | 4 | 799 | 719 | 76 | 4 | 855 | 75 | 1514 | 84 | 0.08 |
| Piranda 2010 [16] | 308 | 157 | 127 | 24 | 264 | 129 | 117 | 18 | 441 | 175 | 375 | 153 | 0.28 |
| Dossus 2010 [21] | 6254 | 4394 | 1646 | 214 | 8092 | 5694 | 2166 | 232 | 10434 | 2074 | 13554 | 2630 | 0.17 |
| Gao 2007 [23] | 601 | 526 | 73 | 2 | 643 | 582 | 59 | 2 | 1125 | 77 | 1223 | 63 | 0.06 |
| Cox 1 2007 [24] | 1243 | 865 | 336 | 42 | 1715 | 1185 | 485 | 45 | 2066 | 420 | 2855 | 575 | 0.17 |
| Shen 2006 [27] | 1067 | 670 | 387 | 1105 | 691 | 414 | — | — | — | — | — | ||
| rs689466 | |||||||||||||
| Brasky 2011 [19] | 1077 | 660 | 271 | 34 | 1910 | 1199 | 471 | 54 | 1591 | 339 | 2869 | 579 | 0.18 |
| Piranda 2010 [16] | 289 | 224 | 62 | 3 | 245 | 190 | 51 | 3 | 510 | 68 | 431 | 57 | 0.12 |
| Dossus 2010 [21] | 6247 | 4020 | 1928 | 299 | 8115 | 5143 | 2562 | 410 | 9968 | 2526 | 12848 | 3382 | 0.20 |
| Gao 2007 [23] | 601 | 121 | 305 | 175 | 643 | 150 | 327 | 166 | 547 | 655 | 627 | 659 | 0.54 |
| rs5275 | |||||||||||||
| Gao 2014 [20] | 465 | 299 | 132 | 34 | 799 | 515 | 244 | 40 | 730 | 200 | 1274 | 324 | 0.22 |
| Fawzy 2013 [18] | 160 | 53 | 71 | 36 | 150 | 69 | 67 | 14 | 177 | 143 | 205 | 95 | 0.45 |
| Brasky 2011 [19] | 1077 | 432 | 447 | 108 | 1910 | 732 | 782 | 226 | 1311 | 663 | 2246 | 1234 | 0.31 |
| Piranda 2010 [16] | 294 | 125 | 149 | 20 | 244 | 120 | 99 | 25 | 399 | 189 | 339 | 149 | 0.32 |
| Dossus 2010 [21] | 6133 | 2697 | 2664 | 772 | 7946 | 3512 | 3501 | 933 | 8058 | 4208 | 10525 | 5367 | 0.34 |
| Abraham 2009 [22] | 2172 | 927 | 985 | 260 | 2265 | 996 | 1010 | 259 | 2839 | 1505 | 3002 | 1528 | 0.35 |
| Gao 2007 [23] | 601 | 404 | 179 | 18 | 643 | 429 | 194 | 20 | 987 | 215 | 1052 | 234 | 0.18 |
| Cox 1 2007 [24] | 1249 | 541 | 567 | 141 | 1720 | 699 | 808 | 213 | 1649 | 849 | 2206 | 1234 | 0.34 |
| Cox 2 2007 [24] | 301 | 140 | 131 | 30 | 610 | 270 | 259 | 81 | 411 | 191 | 799 | 421 | 0.32 |
| Cox 3 2007 [24] | 644 | 281 | 296 | 67 | 651 | 278 | 294 | 79 | 858 | 430 | 850 | 452 | 0.33 |
| Vogel 2007 [25] | 361 | 167 | 150 | 44 | 361 | 155 | 165 | 41 | 484 | 238 | 475 | 247 | 0.33 |
| Langsenlehner 2006 [26] | 500 | 214 | 224 | 62 | 500 | 234 | 232 | 33 | 652 | 348 | 700 | 298 | 0.35 |
| Shen 2006 [27] | 1060 | 475 | 585 | 1102 | 467 | 635 | — | — | — | — | — | ||
A represents the major allele; B represents the minor allele. MAF: minor allele frequencies.
The main results of this meta-analysis were listed in Table 3. There were 6 studies with 9,938 cases and 12,618 controls for rs20417. As shown in Table 3 and Figure 2, rs20417 polymorphism has association with breast cancer risk in the overall population based on homozygote comparison (CC versus GG: OR = 1.21, 95% CI = 1.02–1.42, P = 0.03) and the recessive model (CC versus GG + GC: OR = 1.22, 95% CI = 1.03–1.43, P = 0.02). However, there are no significant associations in other genetic models (C versus G: OR = 1.04, 95% CI = 0.98–1.10, P = 0.17; heterozygote comparison (GC versus GG): OR = 0.97, 95% CI = 0.91–1.03, P = 0.35; dominant model (GC + CC versus GG): OR = 1.01, 95% CI = 0.96–1.08, P = 0.64). In the stratified analysis by ethnicity, the effects remained in Caucasians (homozygote comparison: OR = 1.20, 95% CI = 1.02–1.42, P = 0.03; recessive model: OR = 1.21, 95% CI = 1.03–1.43, P = 0.02), but not in Asians (Table 3).
Table 3.
Meta-analysis of the association between COX-2 polymorphisms and breast cancer risk.
| Comparisons | OR | 95% CI | P value | Heterogeneity | Effects model | |
|---|---|---|---|---|---|---|
| I 2 | P value | |||||
| B versus A | ||||||
| rs20417 | 1.04 | 0.98–1.10 | 0.17 | 56% | 0.06 | R |
| Caucasian | 1.02 | 0.96–1.08 | 0.50 | 0% | 0.92 | F |
| Asian | 1.45 | 1.15–1.84 | 0.002 | 0% | 0.47 | F |
| rs689466 | 0.99 | 0.94–1.04 | 0.69 | 33% | 0.21 | F |
| Caucasian | 0.97 | 0.92–1.03 | 0.34 | 0% | 0.58 | F |
| Asian | 1.14 | 0.97–1.33 | — | — | — | — |
| rs5275 | 1.01 | 0.98–1.05 | 0.50 | 56% | 0.01 | R |
| Caucasian | 1.00 | 0.97–1.04 | 0.80 | 41% | 0.09 | R |
| Asian | 1.03 | 0.89–1.19 | 0.70 | 0% | 0.51 | F |
| BB versus AA | ||||||
| rs20417 | 1.21 | 1.02–1.42 | 0.03 | 0% | 0.97 | F |
| Caucasian | 1.20 | 1.02–1.42 | 0.03 | 0% | 0.92 | F |
| Asian | 1.54 | 0.49–4.78 | 0.46 | 0% | 0.68 | F |
| rs689466 | 1.01 | 0.88–1.15 | 0.93 | 22% | 0.28 | F |
| Caucasian | 0.95 | 0.82–1.10 | 0.52 | 0% | 0.69 | F |
| Asian | 1.31 | 0.95–1.80 | — | — | — | — |
| rs5275 | 1.04 | 0.96–1.12 | 0.34 | 66% | 0.0008 | R |
| Caucasian | 1.01 | 0.94–1.10 | 0.72 | 58% | 0.01 | R |
| Asian | 1.26 | 0.85–1.85 | 0.25 | 7% | 0.30 | F |
| AB versus AA | ||||||
| rs20417 | 0.97 | 0.91–1.03 | 0.35 | 93% | <0.00001 | R |
| Caucasian | 0.94 | 0.88–1.01 | 0.07 | 95% | <0.00001 | R |
| Asian | 1.49 | 1.15–1.91 | 0.002 | 0% | 0.53 | F |
| rs689466 | 0.98 | 0.92–1.05 | 0.59 | 0% | 0.59 | F |
| Caucasian | 0.97 | 0.91–1.04 | 0.44 | 0% | 0.74 | F |
| Asian | 1.16 | 0.87–1.54 | — | — | — | — |
| rs5275 | 0.99 | 0.95–1.04 | 0.81 | 0% | 0.56 | F |
| Caucasian | 0.99 | 0.95–1.04 | 0.81 | 0% | 0.47 | F |
| Asian | 0.96 | 0.80–1.14 | 0.96 | 0% | 0.78 | F |
| AB + BB versus AA | ||||||
| rs20417 | 1.01 | 0.96–1.08 | 0.64 | 54% | 0.05 | R |
| Caucasian | 1.00 | 0.93–1.06 | 0.93 | 0% | 0.84 | F |
| Asian | 1.49 | 1.16–1.91 | 0.002 | 0% | 0.49 | F |
| rs689466 | 0.90 | 0.87–0.95 | 0.0005 | 94% | <0.00001 | R |
| Caucasian | 0.88 | 0.83–0.94 | <0.0001 | 96% | <0.00001 | R |
| Asian | 1.21 | 0.92–1.58 | — | — | — | — |
| rs5275 | 1.02 | 0.98–1.07 | 0.33 | 62% | 0.002 | R |
| Caucasian | 1.02 | 0.97–1.07 | 0.42 | 66% | 0.002 | R |
| Asian | 0.99 | 0.84–1.17 | 0.92 | 0% | 0.86 | F |
| BB versus AA + AB | ||||||
| rs20417 | 1.22 | 1.03–1.43 | 0.02 | 0% | 0.98 | F |
| Caucasian | 1.21 | 1.03–1.43 | 0.02 | 0% | 0.94 | F |
| Asian | 146 | 0.47–4.56 | 0.51 | 0% | 0.70 | F |
| rs689466 | 1.01 | 0.89–1.15 | 0.85 | 0% | 0.48 | F |
| Caucasian | 0.96 | 0.83–1.11 | 0.59 | 0% | 0.76 | F |
| Asian | 1.18 | 0.92–1.51 | — | — | — | — |
| rs5275 | 1.04 | 0.97–1.12 | 0.27 | 65% | 0.0009 | R |
| Caucasian | 1.02 | 0.95–1.10 | 0.60 | 60% | 0.01 | R |
| Asian | 1.28 | 0.87–1.87 | 0.21 | 15% | 0.28 | F |
A: represents the major allele; B: represents the minor allele; F: fixed-effects model; R: random-effects model.
Figure 2.
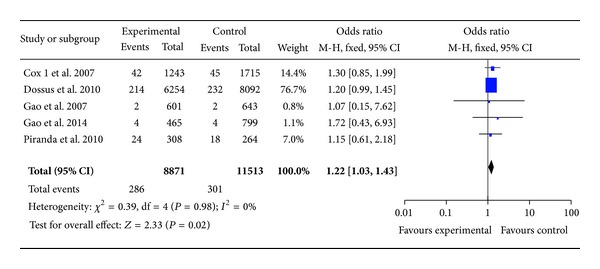
. Forest plots of COX-2 rs20417 polymorphism and breast cancer risk in the overall population (CC versus GG + GC). The squares and horizontal lines correspond to the study specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
There were 4 studies with 8,214 cases and 10,202 controls for assessing the relationship between COX-2 rs689466 polymorphism and breast cancer susceptibility. As shown in Table 3 and Figure 3, there was no association in these four genotypes (A versus G: OR = 0.99, 95% CI = 0.94–1.04, P = 0.69; homozygote comparison (AA versus GG): OR = 1.01, 95% CI = 0.88–1.15, P = 0.93; heterozygote comparison (AG versus GG): OR = 0.98, 95% CI = 0.92–1.05, P = 0.59; recessive model (AA versus GG + AG): OR = 1.01, 95% CI = 0.89–1.15, P = 0.85). However, rs689466 polymorphism has association with breast cancer risk based on the recessive model (AA + AG versus GG: OR = 0.90, 95% CI = 0.87–0.95, P = 0.002). In the stratified analysis, when analyzed by the dominant model, the OR was 0.88 (95% CI = 0.83–0.94) (P < 0.0001) among Caucasians. These results suggested that the individuals with AA or AG alleles have a 12% decreased risk of breast cancer compared with those with GG allele in Caucasians.
Figure 3.
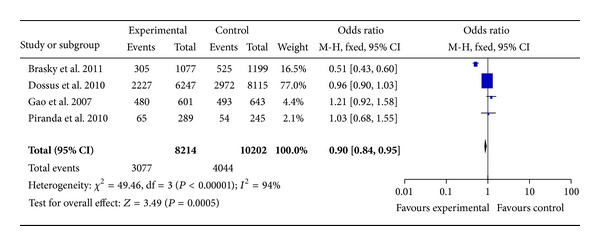
Forest plots of COX-2 rs689466 polymorphism and breast cancer risk in the overall population (AA + AG versus GG). The squares and horizontal lines correspond to the study specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
13 studies with 15,017 cases and 18,901 controls were used to evaluate the relationship between COX-2 rs5275 polymorphism with breast cancer risk. As shown in Table 3 and Figure 4, there was no significant association in rs5275 polymorphism (homozygote comparison: OR = 1.04, 95% CI = 0.96–1.12, P = 0.34; heterozygote comparison: OR = 0.99, 95% CI = 0.95–1.04, P = 0.81; dominant model: OR = 1.02, 95% CI = 0.98–1.07, P = 0.33, and recessive model: OR = 1.04, 95% CI = 0.97–1.12, P = 0.27). When stratified by ethnicity, there was also no association between rs5275 and breast cancer risk in both Caucasians and Asians (Table 3).
Figure 4.
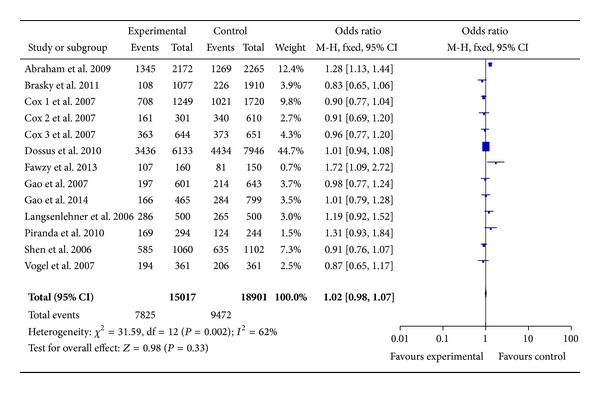
Forest plots of COX-2 rs5275 polymorphism and breast cancer risk in the overall population (TT + TC versus CC). The squares and horizontal lines correspond to the study specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
3.3. Publication Bias
In this meta-analysis, we performed funnel plot and Egger's test to access the publication bias. As shown in Figure 5, the funnel plots failed to detect any obvious asymmetry in all genotypes in overall population, and the results of Egger's test revealed no publication bias (P > 0.05). Therefore, no significant publication bias was found in this meta-analysis.
Figure 5.
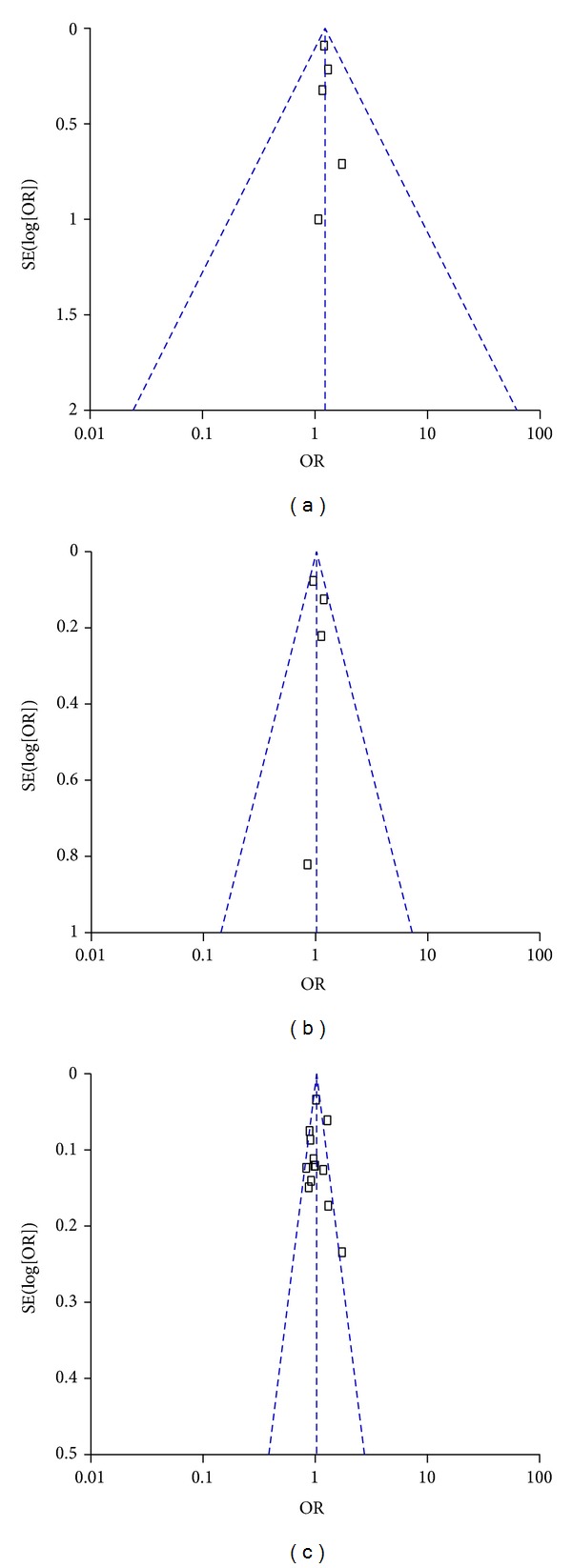
Funnel plot assessing evidence of publication bias from the eligible studies. (a) −765 G>C (rs20417); (b) −1195G>A (rs689466); and (c) 8473 C>T (rs5275).
4. Discussion
The present meta-analysis, including 15,330 cases and 19,260 controls from 13 case-control studies, was conducted to evaluate the association between the three common SNPs [−765 G>C (rs20417), −1195 G>A (rs689466), and 8473 C>T (rs5275)] in the COX-2 gene and breast cancer risk.
A previous study by Yu et al. [15] failed to detect an association between rs20417 and breast cancer risk. There were only three studies with 2,901 cases and 3,463 controls for rs20417 in Yu's meta-analysis [15]. In this study, there were six studies with 9,938 cases and 12,618 controls included to evaluate the relationship between rs20417 polymorphism and breast cancer risk. The results showed that rs20417 polymorphism was associated with breast cancer risk in the overall population based on homozygote comparison (CC versus GG: OR = 1.21, 95% CI = 1.02–1.42, P = 0.03). Moreover, in a stratified analysis by ethnicity using the recessive model, we found that the association remained in Caucasians (homozygote comparison: OR = 1.20, 95% CI = 1.02–1.42, P = 0.03; recessive model: OR = 1.21, 95% CI = 1.03–1.43, P = 0.02), but not in Asians. These results suggest ethnic differences in genetic backgrounds and the environment in which they live play a possible role in breast carcinogenesis [29].
In Zhu et al.'s meta-analysis [30], they showed that individuals with the rs20417 were associated with higher cancer risk than those with the −765GG genotype. Stratified analysis further revealed that this effect was maintained in colorectal carcinoma and esophageal cancer in Asian descents. However, the rs5275 polymorphism was not associated with cancer risk although in breast and lung cancer this allele was correlated with decreased risk.
In the present meta-analysis, 13 studies with 15,017 cases and 18,901 controls concerning the rs5275 polymorphism were included. We found no significant association of rs5275 polymorphism with breast cancer risk (homozygote comparison: OR = 1.04, 95% CI = 0.96–1.12, P = 0.34; heterozygote comparison: OR = 0.99, 95% CI = 0.95–1.04, P = 0.81; dominant model: OR = 1.02, 95% CI = 0.98–1.07, P = 0.33, and recessive model: OR = 1.04, 95% CI = 0.97–1.12, P = 0.27). When stratified by ethnicity, similar results were observed in both Caucasians and Asians.
In the previous meta-analysis by Tang et al. [14], there was an association of the rs689466 polymorphism with cancer risk in Asian populations but not in Europeans. Our results indicate that rs689466 polymorphism has association with breast cancer risk based on the recessive model (AA + AG versus GG: OR = 0.90, 95% CI = 0.87–0.95, P = 0.002). In the stratified analysis, when analyzed by the dominant model, the OR was 0.88 (95% CI = 0.83–0.94) (P < 0.0001) among Caucasians.
Some limitations of this meta-analysis should be noted. Firstly, this meta-analysis was based on pooled data and no individual data was available; thus, we could not assess the risk of cancer according to stratification by age, environment factors, and other risk factors of breast cancer. Secondly, most of the included studies did not investigate the chronic inflammatory condition and the history of taking nonsteroidal anti-inflammatory drugs. Thirdly, the included studies are mainly based on Caucasian background. There were only two studies based on Asian background and one based on African background. Larger scale multicenter studies based on Asians or Africans are warranted to further validate the association between COX-2 polymorphisms and breast cancer risk.
5. Conclusion
In summary, this meta-analysis points to the COX-2 rs20417 C allele as a risk factor for breast cancer among Caucasian subjects. On the contrary, the rs689466 allele has a decreased risk of breast cancer in Caucasians. The rs5275 C status does not seem capable of predicting breast cancer risk in both Caucasians and Asians.
Acknowledgments
This study was supported by the International Cooperative Project of Shaanxi province, China (no. 2013KW-32-01); the Fundamental Research Funds for the Central Universities, China; and Specialized Research Fund of the Second Affiliated Hospital of Xi'an Jiaotong University, China (RC (GG) 201203).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Yong-Ping Shao and Xiao-Bin Ma contributed equally to this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA: A Cancer Journal for Clinicians. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer: analyses of cohorts of twins from Sweden, Denmark, and Finland. The New England Journal of Medicine. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 3.Wolff MS, Weston A. Breast cancer risk and environmental exposures. Environmental Health Perspectives. 1997;105(4):891–896. doi: 10.1289/ehp.97105s4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulka BS, Moorman PG. Breast cancer: hormones and other risk factors. Maturitas. 2008;61(1-2):203–213. doi: 10.1016/j.maturitas.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Vane JR, Bakhle YS, Botting RM. Cyclooxygenases 1 and 2. Annual Review of Pharmacology and Toxicology. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 6.Greenhough A, Smartt HJM, Moore AE, et al. The COX-2/PGE2 pathway: key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30(3):377–386. doi: 10.1093/carcin/bgp014. [DOI] [PubMed] [Google Scholar]
- 7.Diederich M, Sobolewski C, Cerella C, Dicato M, Ghibelli L. The role of cyclooxygenase-2 in cell proliferation and cell death in human malignancies. International Journal of Cell Biology. 2010;2010:21 pages. doi: 10.1155/2010/215158.215158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalinski P. Regulation of immune responses by prostaglandin E2. Journal of Immunology. 2012;188(1):21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Half E, Tang XM, Gwyn K, Sahin A, Wathen K, Sinicrope FA. Cyclooxygenase-2 expression in human breast cancers and adjacent ductal carcinoma in situ. Cancer Research. 2002;62(6):1676–1681. [PubMed] [Google Scholar]
- 10.Subbaramaiah K, Norton L, Gerald W, Dannenberg AJ. Cyclooxygenase-2 is overexpressed in HER-2/neu-positive breast cancer: evidence for involvement of AP-1 and PEA3. The Journal of Biological Chemistry. 2002;277(21):18649–18657. doi: 10.1074/jbc.M111415200. [DOI] [PubMed] [Google Scholar]
- 11.Spizzo G, Gastl G, Wolf D, et al. Correlation of COX-2 and Ep-CAM overexpression in human invasive breast cancer and its impact on survival. British Journal of Cancer. 2003;88(4):574–578. doi: 10.1038/sj.bjc.6600741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai Z, Ma X, Kang H, et al. Antitumor activity of the selective cyclooxygenase-2 inhibitor, celecoxib, on breast cancer in Vitro and in Vivo. Cancer Cell International. 2012;12, article 53 doi: 10.1186/1475-2867-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szczeklik W, Sanak M, Szczeklik A. Functional effects and gender association of COX-2 gene polymorphism G -765C in bronchial asthma. Journal of Allergy and Clinical Immunology. 2004;114(2):248–253. doi: 10.1016/j.jaci.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 14.Tang Z, Nie Z-L, Pan Y, et al. The Cox-2 -1195 G > a polymorphism and cancer risk: a meta-analysis of 25 case-control studies. Mutagenesis. 2011;26(6):729–734. doi: 10.1093/mutage/ger040. [DOI] [PubMed] [Google Scholar]
- 15.Yu KD, Chen AX, Yang C, et al. Current evidence on the relationship between polymorphisms in the COX-2 gene and breast cancer risk: a meta-analysis. Breast Cancer Research and Treatment. 2010;122(1):251–257. doi: 10.1007/s10549-009-0688-3. [DOI] [PubMed] [Google Scholar]
- 16.Piranda DN, Festa-Vasconcellos JS, Amaral LM, Bergmann A, Vianna-Jorge R. Polymorphisms in regulatory regions of Cyclooxygenase-2 gene and breast cancer risk in Brazilians: a case-control study. BMC Cancer. 2010;10(article 613) doi: 10.1186/1471-2407-10-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cok SJ, Morrison AR. The 3′-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. The Journal of Biological Chemistry. 2001;276(25):23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- 18.Fawzy MS, Aly NMM, Shalaby SM, El-Sawy WH, Abdul-Maksoud RS. Cyclooxygenase-2 169C>G and 8473T>C gene polymorphisms and prostaglandin E2 level in breast cancer: a case-control study. Gene. 2013;527(2):601–605. doi: 10.1016/j.gene.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Brasky TM, Bonner MR, Moysich KB, et al. Genetic variants in COX-2, non-steroidal anti-inflammatory drugs, and breast cancer risk: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Research and Treatment. 2011;126(1):157–165. doi: 10.1007/s10549-010-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Kang HF, Ma XB, et al. Functional promoter −765 G>C variant in COX-2 gene is associated with the susceptibility of breast cancer in Chinese Han women. Cancer Cell International. 2014;14:p. 38. doi: 10.1186/1475-2867-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dossus L, Kaaks R, Canzian F, et al. PTGS2 and IL6 genetic variation and risk of breast and prostate cancer: results from the Breast and Prostate Cancer Cohort Consortium (BPC3) Carcinogenesis. 2010;31(3):455–461. doi: 10.1093/carcin/bgp307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham JE, Harrington P, Driver KE, et al. Common polymorphisms in the prostaglandin pathway genes and their association with breast cancer susceptibility and survival. Clinical Cancer Research. 2009;15(6):2181–2191. doi: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Ke Q, Ma H, et al. Functional polymorphisms in the cyclooxygenase 2 (COX-2) gene and risk of breast cancer in a Chinese population. Journal of Toxicology and Environmental Health A. 2007;70(11):908–915. doi: 10.1080/15287390701289966. [DOI] [PubMed] [Google Scholar]
- 24.Cox DG, Buring J, Hankinson SE, Hunter DJ. A polymorphism in the 3′ untranslated region of the gene encoding prostaglandin endoperoxide synthase 2 is not associated with an increase in breast cancer risk: a nested case-control study. Breast Cancer Research. 2007;9(1, article R3) doi: 10.1186/bcr1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel U, Christensen J, Nexø BA, Wallin H, Friis S, Tjønneland A. Peroxisome proliferator-activated receptor-γ2 Pro12Ala, interaction with alcohol intake and NSAID use, in relation to risk of breast cancer in a prospective study of Danes. Carcinogenesis. 2007;28(2):427–434. doi: 10.1093/carcin/bgl170. [DOI] [PubMed] [Google Scholar]
- 26.Langsenlehner U, Yazdani-Biuki B, Eder T, et al. The cyclooxygenase-2 (PTGS2) 8473T>C polymorphism is associated with breast cancer risk. Clinical Cancer Research. 2006;12(4):1392–1394. doi: 10.1158/1078-0432.CCR-05-2055. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Gammon MD, Terry MB, Teitelbaum SL, Neugut AI, Santella RM. Genetic polymorphisms in the cyclooxygenase-2 gene, use of nonsteroidal anti-inflammatory drugs, and breast cancer risk. Breast Cancer Research. 2006;8(6) article R71 doi: 10.1186/bcr1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F, Ren GS, Li HY, Wang XY, Chen L, Li J. A novel single nucleotide polymorphism of the cyclooxygenase-2 gene associated with breast cancer. Clinical Oncology. 2009;21(4):302–305. doi: 10.1016/j.clon.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genetics in Medicine. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Zhu W, Wei B, Shan X, Liu P. -765G>C and 8473T>C polymorphisms of COX-2 and cancer risk: a meta-analysis based on 33 case-control studies. Molecular Biology Reports. 2010;37(1):277–288. doi: 10.1007/s11033-009-9685-1. [DOI] [PubMed] [Google Scholar]


