Abstract
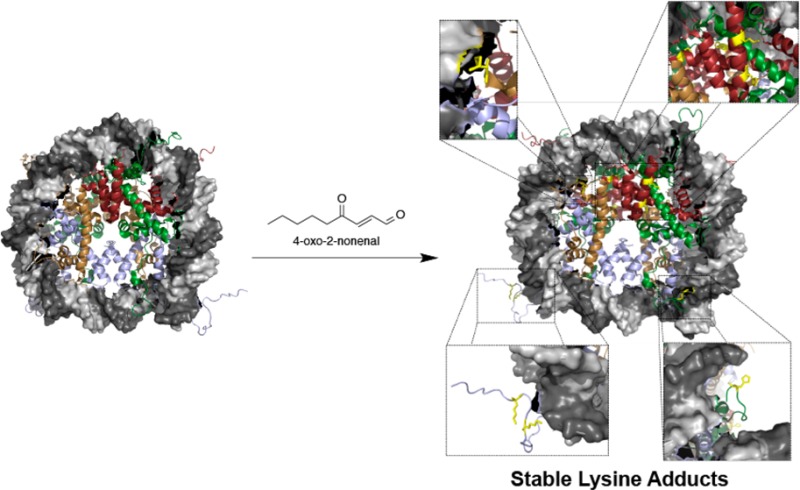
Lipid electrophiles modify cellular targets, altering their function. Here, we describe histones as major targets for modification by 4-oxo-2-nonenal, resulting in a stable Lys modification structurally analogous to other histone Lys acylations. Seven adducts were identified in chromatin isolated from intact cells: four 4-ketoamides to Lys and three Michael adducts to His. A 4-ketoamide adduct residing at H3K27 was identified in stimulated macrophages. Modification of histones H3 and H4 prevented nucleosome assembly.
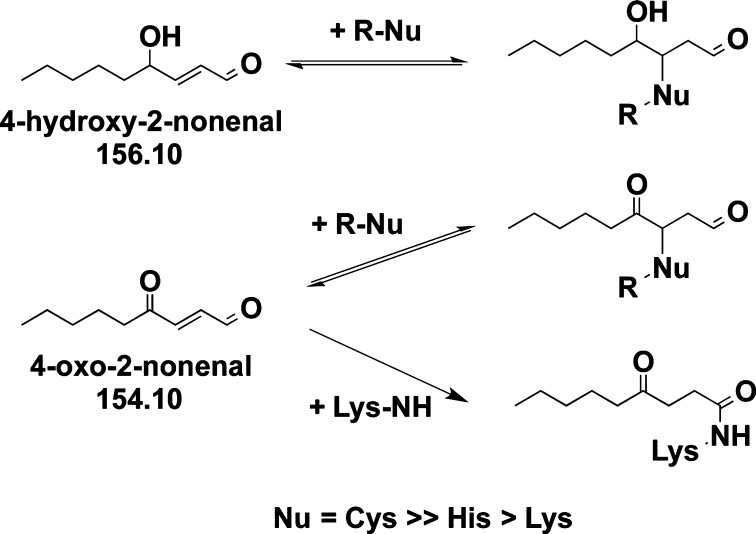
Reactive oxygen species have been implicated in the pathogenesis of numerous diseases.1−3 The oxidation of cellular lipids results in the generation of α,β-unsaturated alkenals, including 4-hydroxy-2-nonenal (4-HNE) and 4-oxo-2-nonenal (4-ONE).4 These electrophilic molecules adduct both DNA and protein, resulting in altered structure and function.2,5−7 Although similar in structure, 4-HNE and 4-ONE differ in reactivity in that 4-ONE can form stable ketoamide adducts via 1,2 addition to Lys (Scheme 1 and Supporting Information, Figure S1).8,9 ONE-derived 4-ketoamide adducts have been observed in atherosclerotic lesions, suggesting a role in inflammation-related disease pathogenesis.10
Scheme 1.
Histones are lysine-rich, cysteine-deficient proteins that regulate chromatin structure and gene expression.11−13 They are interesting targets for 4-ketoamide adduction and may offer a novel link between oxidative stress and gene expression.12 To test this hypothesis, RKO cells were treated with either alkynyl-4-HNE or alkynyl-4-ONE. Chromatin was isolated and click chemistry performed to attach biotin moieties to the alkyne groups. Adducted proteins were visualized on gels with fluorescently labeled streptavidin. Alkynyl-4-ONE demonstrated high selectivity for histone modification; all four histones were preferentially adducted in a concentration- (Figure 1A) and time-dependent manner (Figure 1B). H2B appeared to be the most heavily adducted species. In contrast, alkynyl-4-HNE reacted with higher molecular weight chromatin-associated proteins, likely because of the higher cysteine and histidine content of these non-histone proteins.
Figure 1.
(A) Alkynyl-4-ONE preferentially modifies histone proteins in chromatin isolated from RKO cells. (B) Time-course of histone adduction with alkynyl-4-ONE.
Histones have been identified as targets for modification by reactive species in vitro; however, sites of modification have yet to determined in intact cells.14−16 Analysis of post-translational modification of histones by mass spectrometry is a significant challenge because the abundance of Lys and Arg residues makes sequence coverage very difficult.17 To identify the sites of modification by 4-ONE, we adapted a series of chemical derivatizations and enzymatic digests of histones isolated from chromatin in RKO cells to achieve 100% sequence coverage on all four proteins (Figure S4, Table S1). These methods were used to establish a standard protocol to efficiently analyze histones in complex samples. Table S1 offers a map for sequence coverage resulting from each method, demonstrating the large diversity in peptide yield and post-translational modifications. Utilizing these methods, seven site-specific histone adducts were identified in RKO cells treated with 4-ONE: four 4-ketoamide adducts to Lys and three Michael adducts to His (Tables 1 and S2; Figures S5–S11).
Table 1. 4-ONE Adducts Identified on Chromatin Isolated from RKO Cellsa.
| protein | peptide | residue adducted | species |
|---|---|---|---|
| H3 | KprQLATK*AAR | K23 | ketoamide (9) |
| K*SAPATGGVK | K27 | ketoamide (9) | |
| H2B | LAH*YNKR | H82 | Michael adduct (3) |
| H*AVSEGTK | H109 | Michael adduct (3) | |
| HAVSEGTK*AVTK | K116 | ketoamide (9) | |
| H2A | KTESH*HK | H123 | Michael adduct (3) |
| H4 | K*TVTAMDVVYALKR | K79 | ketoamide (9) |
Sites of modification are indicated with an asterisk. Numbers correspond to adducts shown in Figure S1.
The modified amino acids were mapped to the crystal structure of the nucleosome core particle. As shown in Figure 2A, the adducted residues are located on solvent-exposed side chains (denoted in yellow). H4K79 (Figure 2B) is located at the DNA–protein interface, and the positive charge presented by this lateral surface is critical to maintaining heterochromatin.12,18 Modifications identified on H2B (Figure 2C) reside on the lateral surface of the nucleosome, suggesting a minimal role in regulating protein–DNA electrostatics. H2AH123 (Figure 2E) resides on the C-terminal tail of H2A and interacts with the phosphodiester backbone of the DNA. Perhaps the most intriguing adducts are the ketoamides at H3K23 and H3K27 (Figure 2D). Previous studies have demonstrated that acetylation or methylation of these residues plays a role in numerous pathologies, including cancer.19−21
Figure 2.
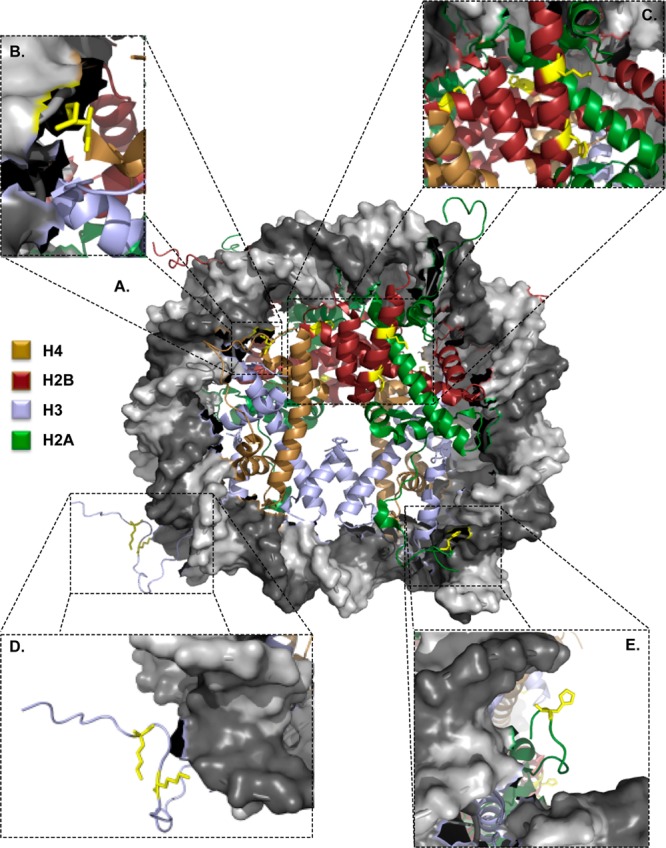
4-ONE adducts are located on surface-exposed amino acids. (A) Crystal structure of the nucleosome. Adducts are located at (B) H4K79, (C) H2BH82, H109, and K116, (D) H3K23 and K27, and (E) H2AH123. Structure obtained from PDB 1AOI.
These structural insights led us to investigate the impact of histone adduction on DNA binding. Utilizing recombinant histones and a 203 bp oligomer, nucleosomes were reconstituted in vitro (Figure S12), resulting in a shift from ∼200 to ∼900 bp (Figure 3, lanes 1 and 2, respectively). Treatment of assembled nucleosomes with increasing concentrations of alkynyl-4-ONE resulted in only minor alterations in nucleosome structure (lanes 3–5). Likewise, pretreatment of H2A/H2B dimers with alkynyl-4-ONE did not disrupt nucleosome assembly (lanes 6–8), presumably due to the sites of modification residing on the lateral surface of the nucleosome. Pretreatment of H3/H4 tetramers, however, resulted in inhibition of nucleosome assembly (lanes 9–11), similar to lysine acetylation (Figure S13). These data support our cellular and in silico data, which suggest that although H2B is the most heavily adducted histone, the effects on nucleosome structure are minimal, whereas adduction of free H3 or H4 appears to alter histone dynamics and compromise canonical nucleosome formation.
Figure 3.
Adduction of H3/H4 results in disrupted nucleosome formation.
To evaluate the presence of histone modifications in a more physiologically relevant model, fatty acid-deficient RAW264.7 macrophages were enriched with arachidonic acid.22 Following stimulation with a chemically defined lipopolysaccharide, KLA, chromatin was harvested and subjected to MS. This resulted in the positive identification of a 4-ketoamide adduct at H3K27, which was absent in untreated cells (Figure 4). These data reveal the presence of a novel histone modification linked to oxidative stress generated in a well-characterized cellular model for inflammation.
Figure 4.
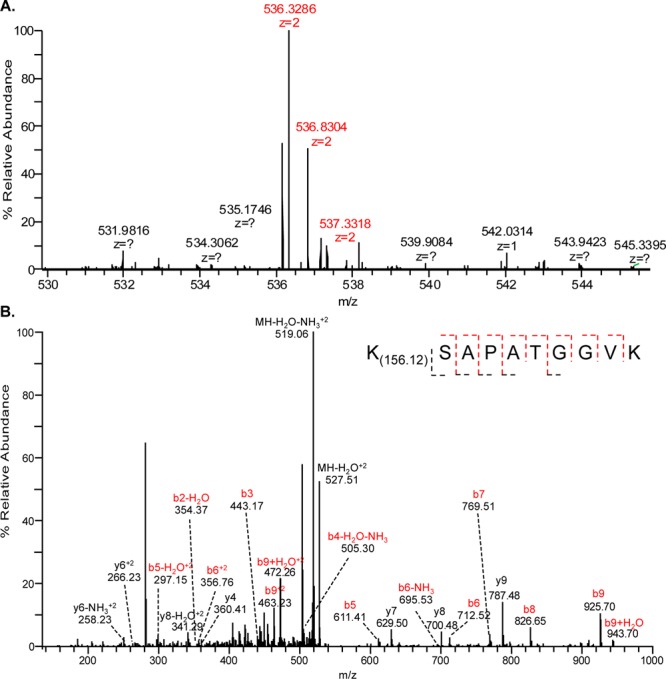
KLA stimulation results in stable 4-ketoamide adduction of H3K27 in RAW264.7 macrophages. (A) MS of the product ion (Δ8.57 ppm). (B) MS/MS reveals H3K27 as a site for modification by 4-ONE. Fragment ions confirming the adducted peptide are indicated in red.
We have identified four novel sites of lysine modification in chromatin isolated from intact cells treated with the lipid peroxidation end product, 4-ONE. The 4-ketoamide adduct at H3K27 was also identified in macrophages stimulated with the lipopolysaccharide mimetic, KLA. Adduction of H3 and H4 may disrupt chromatin dynamics and alter histone turnover. Additionally, the presence of these adducts on well-characterized acetylation and methylation sites, such as H3K23 and H3K27, may impact epigenetic patterning. This offers a potential feed-forward mechanism whereby low-level oxidative acylation of histones leads to alterations in gene expression.
Acknowledgments
This work was supported by research grants from the NIH [R37 CA087819 (L.J.M.), RO1 GM067728 (W.P.T.), and T32 ES007028 (J.J.G.)]. The LTQ Orbitrap Velos mass spectrometer used in these studies was purchased with funds from the NIH S10 grant RR027714 awarded to the Vanderbilt proteomics shared resource.
Supporting Information Available
Procedures for all experiments; Figures S1–S13, showing products of lysine adduction by 4-ONE and their masses, expansion of Figure 1 with loading controls, and mass spectra of identified adducts; expansion of Table 1 with m/z and Δppm (Tables S1 and S2, Excel spreadsheet). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Andersen J. K. Nat. Med. 2004, 10Suppl.S18. [DOI] [PubMed] [Google Scholar]
- Esterbauer H.; Schaur R. J.; Zollner H. Free Radical Biol. Med. 1991, 11, 81. [DOI] [PubMed] [Google Scholar]
- Van Gaal L. F.; Mertens I. L.; De Block C. E. Nature 2006, 444, 875. [DOI] [PubMed] [Google Scholar]
- Skrzydlewska E.; Sulkowski S.; Koda M.; Zalewski B.; Kanczuga-Koda L.; Sulkowska M. World J. Gastroenterol.: WJG 2005, 11, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G.; Schaur R. J.; Siems W. G.; Leonarduzzi G. Med. Res. Rev. 2008, 28, 569. [DOI] [PubMed] [Google Scholar]
- West J. D.; Marnett L. J. Chem. Res. Toxicol. 2005, 18, 1642. [DOI] [PubMed] [Google Scholar]
- Wang C.; Weerapana E.; Blewett M. M.; Cravatt B. F. Nat. Methods 2014, 11, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.; Sayre L. M. Chem. Res. Toxicol. 2007, 20, 165. [DOI] [PubMed] [Google Scholar]
- Zhu X.; Sayre L. M. Redox Rep.: Commun. Free Radical Res. 2007, 12, 45. [DOI] [PubMed] [Google Scholar]
- Shibata T.; Shimozu Y.; Wakita C.; Shibata N.; Kobayashi M.; Machida S.; Kato R.; Itabe H.; Zhu X.; Sayre L. M.; Uchida K. J. Biol. Chem. 2011, 286, 19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Nature 1997, 389, 349. [DOI] [PubMed] [Google Scholar]
- Luger K.; Mader A. W.; Richmond R. K.; Sargent D. F.; Richmond T. J. Nature 1997, 389, 251. [DOI] [PubMed] [Google Scholar]
- Strahl B. D.; Allis C. D. Nature 2000, 403, 41. [DOI] [PubMed] [Google Scholar]
- Chen D.; Fang L.; Li H.; Tang M. S.; Jin C. J. Biol. Chem. 2013, 288, 21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oe T.; Arora J. S.; Lee S. H.; Blair I. A. J. Biol. Chem. 2003, 278, 42098. [DOI] [PubMed] [Google Scholar]
- Carrier E. J.; Zagol-Ikapitte I.; Amarnath V.; Boutaud O.; Oates J. A. Biochemistry 2014, 53, 2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia B. A.; Mollah S.; Ueberheide B. M.; Busby S. A.; Muratore T. L.; Shabanowitz J.; Hunt D. F. Nat. Protoc. 2007, 2, 933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland E. M.; Cosgrove M. S.; Molina H.; Wang D.; Pandey A.; Cottee R. J.; Boeke J. D. Mol. Cell. Biol. 2005, 25, 10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe M. T.; Ott H. M.; Ganji G.; Korenchuk S.; Thompson C.; Van Aller G. S.; Liu Y.; Graves A. P.; Della Pietra A. III; Diaz E.; LaFrance L. V.; Mellinger M.; Duquenne C.; Tian X.; Kruger R. G.; McHugh C. F.; Brandt M.; Miller W. H.; Dhanak D.; Verma S. K.; Tummino P. J.; Creasy C. L. Nature 2012, 492, 108. [DOI] [PubMed] [Google Scholar]
- Roche J.; Nasarre P.; Gemmill R.; Baldys A.; Pontis J.; Korch C.; Guilhot J.; Ait-Si-Ali S.; Drabkin H. Cancers 2013, 5, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S.; Black K. L.; Kovermann S. K.; Brock H. W.; Mazo A. Nat. Commun. 2013, 4, 2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer C. A.; Ivanova P. T.; Byrne M. O.; Milne S. B.; Marnett L. J.; Brown H. A. Biochemistry 2006, 45, 14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.