Significance
MYC is an essential transcriptional regulator that controls cell proliferation. Elevated MYC is a driving force in most human cancers, yet MYC has been an exceedingly challenging target for small-molecule inhibitors. Here we describe a novel MYC inhibitor that interacts directly with MYC and interferes with its transcriptional and oncogenic activities.
Keywords: transcriptional control, protein–protein interactions, xenograft, combinatorial library, gene signature
Abstract
In a fluorescence polarization screen for the MYC–MAX interaction, we have identified a novel small-molecule inhibitor of MYC, KJ-Pyr-9, from a Kröhnke pyridine library. The Kd of KJ-Pyr-9 for MYC in vitro is 6.5 ± 1.0 nM, as determined by backscattering interferometry; KJ-Pyr-9 also interferes with MYC–MAX complex formation in the cell, as shown in a protein fragment complementation assay. KJ-Pyr-9 specifically inhibits MYC-induced oncogenic transformation in cell culture; it has no or only weak effects on the oncogenic activity of several unrelated oncoproteins. KJ-Pyr-9 preferentially interferes with the proliferation of MYC-overexpressing human and avian cells and specifically reduces the MYC-driven transcriptional signature. In vivo, KJ-Pyr-9 effectively blocks the growth of a xenotransplant of MYC-amplified human cancer cells.
MYC is a transcriptional regulator that occupies an apex position in the organizational hierarchy of the cell (1–3). It belongs to a family of basic helix–loop–helix leucine zipper (bHLH-LZ) proteins that dimerize with the small bHLH-LZ protein MAX to become functional (4). The MYC–MAX heterodimer preferentially binds to the palindromic DNA sequence CACGTG, referred to as the E-box motif. As a transcription factor, MYC can bind to the promoters of target genes to stimulate or repress transcriptional activity (5–7). The human genome contains three MYC genes, c-MYC, N-MYC, and L-MYC. Throughout this paper, we will use “MYC” to indicate the protein product of the c-MYC gene.
MYC is involved in almost all cancers (8, 9). It is rarely mutated, but achieves gain of function through overexpression or amplification. Because of this broad pathogenic significance, MYC is an important cancer target. However, both conceptual and practical difficulties have stood in the way of identifying potent and effective small-molecule inhibitors of MYC. The conceptual obstacles reflect concern about inhibiting a gene that controls essential cellular activities. Because MYC plays an important role in cell proliferation (10, 11), it is often argued that inhibition of this function would lead to broad and unacceptable side effects in vivo. However, studies with the dominant-negative MYC construct Omomyc have shown that inhibiting MYC has only mild and rapidly reversible effects on normal, fast-proliferating tissues (8, 12, 13). The main practical difficulty in targeting MYC is the absence of pockets or grooves that could serve as binding sites for small molecules (14).
The preferred strategy for the identification of potential MYC inhibitors has been interference with MYC–MAX dimerization (15–18). The formation of the MYC–MAX heterodimer involves the bHLH-LZ domains of the two partner molecules with a protein–protein interaction (PPI) surface of ∼3,200 Å2. This surface lacks well-defined binding sites for small molecules and therefore is widely considered as “undruggable.” However, despite the large interaction surface, a single-amino acid substitution can completely disrupt the dimerization of MYC with MAX (14). This observation provides proof of principle that a high-affinity ligand to a portion of the interaction surface would be sufficient to disrupt the interaction.
Early inhibitors of MYC–MAX dimerization were small molecules designed to target the MYC–MAX interface. The best of these were able to inhibit MYC–MAX dimerization and oncogenic cellular transformation induced by MYC (15, 16). The most widely used MYC inhibitor, 10058-F4 (16), has an effect on the transcriptome that strikingly resembles that of MYC-targeting shRNA (19). These compounds are useful as experimental tools in cell culture, but lack the potency or appropriate pharmacokinetic properties for in vivo applications.
As part of our continuing efforts to identify small molecules able to target structural “sweet spots” and disrupt PPIs, we have recently discovered a new series of small-molecule antagonists of the MYC–MAX PPI. The most potent member of this family of compounds binds to both MYC and MYC–MAX with nanomolar affinity. It also inhibits MYC-driven oncogenic transformation as well as MYC-dependent transcriptional regulation. The promising pharmacokinetic properties of this molecule allowed preliminary in vivo studies. This new inhibitor of the MYC–MAX PPI effectively interfered with the growth of a MYC-driven xenograft tumor, making it to our knowledge a first-in-class chemical probe for investigating the modulation of the MYC–MAX PPI as an anticancer strategy. In this communication, we present the chemical and biological properties of this compound.
Results
A Library of Pyridine Compounds Yields Effective Inhibitors of MYC.
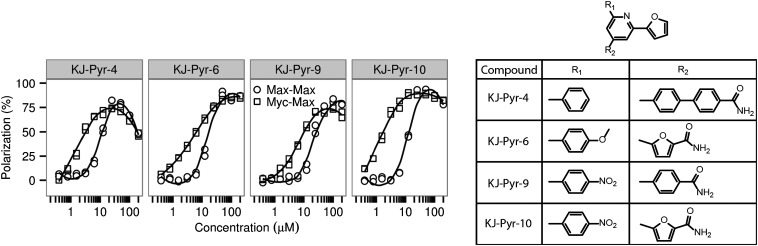
A previously described Kröhnke pyridine library (20) was screened by fluorescence polarization (21) for inhibition of MYC–MAX dimerization. The human MYC and MAX bHLH-LZ domains were expressed in Escherichia coli and combined with an E-box–containing DNA duplex labeled with Alexa Fluor 594. When these three components are mixed, MYC and MAX heterodimerize and bind to the E-box DNA. A binding event results in an increase in the fluorescence polarization, whereas compounds that inhibit the formation of this complex cause a decrease in the fluorescence polarization. Initial library screening was conducted with mixtures (Fig. S1). Those mixtures that showed the strongest inhibition were resynthesized as individual compounds and rescreened, yielding four effective molecules, shown in Fig. 1. The relative binding affinities of each of these compounds for MYC–MAX and MAX–MAX were reassessed, vide supra, and each displayed significantly higher affinity for MYC–MAX over MAX–MAX dimers (Binding of KJ-Pyr-9 to MYC).
Fig. 1.
(Left) Fluorescence polarization data of KJ-Pyr-4, KJ-Pyr-6, KJ-Pyr-9, and KJ-Pyr-10, comparing binding to MYC–MAX with the interaction with MAX–MAX. (Right) Structures of the four compounds.
Specificity of Inhibition.
An assay of MYC-induced oncogenic transformation in chicken embryo fibroblasts (CEF) was used as a secondary screen to determine inhibition of MYC in a biological setting. CEF were infected with the retroviral expression vector RCAS, mediating expression of ATG-MYC, a variant of human MYC that has the noncanonical CTG start codon replaced by an ATG start codon. The ATG start codon mediates higher expression and greater potency in oncogenic transformation, resulting in rapid formation of focal microtumors in the cell monolayer. We also used RCAS-expressing MYC from the wild-type sequence as well as a modified construct expressing only the smaller isoform of MYC (Fig. S2). The effects of the inhibitors were the same as with ATG-MYC. In these experiments, only KJ-Pyr-9 and KJ-Pyr-10 interfered with the oncogenic activity of MYC (Table 1). We surmise that failure to show a cellular effect is likely the result of poor compound solubility in culture media. Because KJ-Pyr-9 had the best aqueous solubility of the four compounds selected, all further experiments were conducted with KJ-Pyr-9.
Table 1.
Effects of inhibitor compounds on the efficiency of MYC-induced oncogenic transformation
| Compound (10 µM) | EOT* |
| KJ-Pyr-4 | 1.12 |
| KJ-Pyr-6 | 0.79 |
| KJ-Pyr-9 | 0.00083 |
| KJ-Pyr-10 | 0.00017 |
Efficiency of transformation indicates focus counts in the presence of inhibitor over focus counts of control plates. Oncogenic transformation was determined in chicken embryo fibroblasts using the RCAS vector expressing ATG-MYC in which the noncanonical initiation codon CTG had been optimized to ATG.
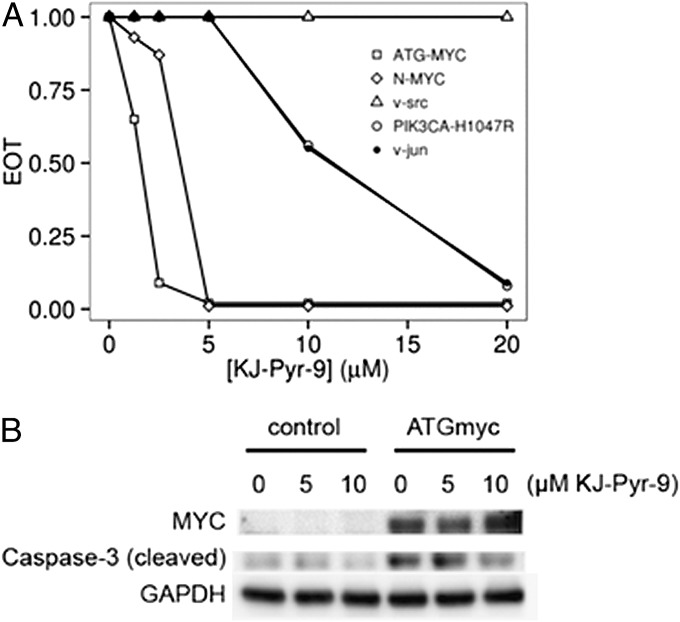
The assay of oncogene-induced cellular transformation in CEF was also used to determine selectivity of KJ-Pyr-9 for MYC. KJ-Pyr-9 was tested against ATG-MYC, N-MYC, and three unrelated oncoproteins, v-Src, v-Jun, and the H1047R mutant of phosphatidylinositol 3-kinase (PI3K) (Fig. 2A). The oncogenic activity of N-MYC and ATG-MYC was strongly inhibited by KJ-Pyr-9, whereas the unrelated oncoproteins were either unaffected (v-Src) or were inhibited at significantly higher concentrations (v-Jun, PI3K H1047R). Massive overexpression of MYC induces apoptosis in CEF. As shown in Fig. 2B, KJ-Pyr-9 effectively inhibits the apoptosis-linked emergence of cleaved caspase 3.
Fig. 2.
(A) Dose–response of KJ-Pyr-9 versus oncogenic transformation induced by MYC, N-MYC, v-Src, PIK3CA-H1047R, and v-Jun. Data from a representative experiment conducted with cells derived from a single chicken embryo. In these transformation assays, the MYC expression vector produced the larger isoform of MYC, initiated by the first initiation codon that had been mutated from CTG to ATG. Identical results were obtained by expressing MYC from the wild-type mRNA sequence and from an mRNA sequence in which the first initiation codon CTG is mutated to a nonfunctional CAG, thus forcing expression of only the smaller isoform of MYC. EOT, efficiency of transformation. (B) CEF were infected at a multiplicity of infection of 10 with the RCAS retroviral vector expressing ATG-MYC. Cells were transferred on days 3 and 5 postinfection and treated or not treated with KJ-Pyr-9, and cleaved caspase 3 was determined by Western blot on day 9.
Binding of KJ-Pyr-9 to MYC.
Despite being the most water-soluble of the series, KJ-Pyr-9 has a low solubility in water (12.5 µM). Because of this limitation, many commonly used methods such as analytical ultracentrifugation, isothermal calorimetry, and noncovalent mass spectrometry fail to provide definitive evidence for direct binding. However, we were able to demonstrate a direct interaction and determine a binding constant for KJ-Pyr-9 and MYC using backscattering interferometry (BSI) (17, 22) (Table 2). The results show that KJ-Pyr-9 directly binds to MYC (6.5 nM) as well as to the MYC–MAX heterodimer (13.4 nM), but only weakly to the MAX homodimer (>1 μM). The data suggest that KJ-Pyr-9 is capable of binding to the disordered monomeric form of MYC and that it can dissociate the intact MYC–MAX complex.
Table 2.
Dissociation constants of KJ-Pyr-9 and MYC, MAX, and the MYC–MAX dimer as measured by backscattering interferometry
| Protein | Kd |
| MYC | 6.5 ± 1.0 nM |
| MYC–MAX | 13.4 ± 3.9 nM |
| MAX | >1.0 µM |
To test the ability of KJ-Pyr-9 to enter cells and specifically interfere with the formation of a functional MYC–MAX complex, we applied a protein fragment complementation assay (PCA) based on Renilla luciferase (Rluc). Rluc-based PCA sentinels have been designed and used to study the dynamics of PPIs in vivo (23, 24). An advantage of this assay is that it reports absolute values of protein-complex formation in real time. We used an efficient and sensitive PCA biosensor that is based on the PPI involving the full-length MAX protein and the C-terminal bHLH-LZ in MYC (MYC332–439). The assay showed that KJ-Pyr-9 selectively reduced complex formation of MYC332–439 with MAX, compared with the effect on an unrelated biosensor based on the homodimer of the protein kinase A regulatory subunits (RII:RII) used here as a control (Fig. S3). KJ-Pyr-9 also interfered with MAX homodimerization, albeit to a lesser degree than MYC–MAX heterodimerization (Fig. S3). These data support the conclusion that KJ-Pyr-9 enters cells and specifically interferes with MYC–MAX complex formation.
The Effect of KJ-Pyr-9 on Cellular Proliferation.
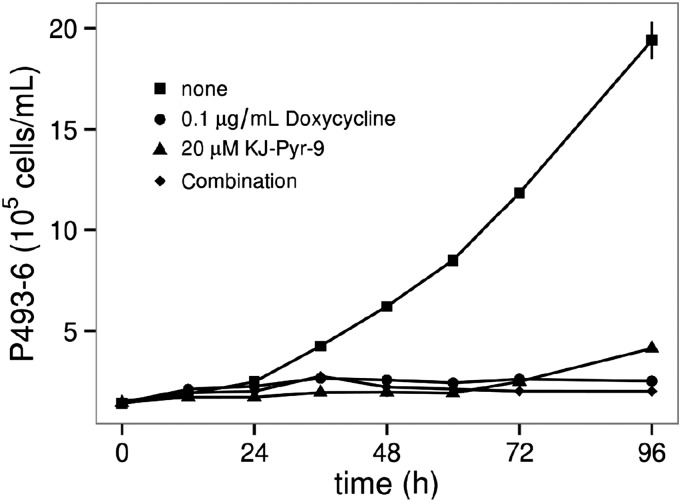
For the study of the effect of KJ-Pyr-9 on MYC-driven cellular proliferation, we used the human B-cell line P493-6. In these cells, the expression of MYC is under the control of a Tet-off promoter. In the absence of doxycycline, MYC is expressed, leading to robust cell proliferation. As an inhibitor of MYC dimerization, KJ-Pyr-9 should have no activity in the absence of the MYC protein. Both KJ-Pyr-9 and doxycycline inhibited the growth of P493-6, as shown in Fig. 3. Importantly, when KJ-Pyr-9 and doxycycline are used in combination, there is no additional inhibition beyond either compound alone. Elevated activity of MYC is also essential in the proliferation of numerous cancer cell lines. We tested KJ-Pyr-9 against three cell lines known to be dependent on increased MYC activity: NCI-H460, MDA-MB-231, and SUM-159PT. The proliferation of all cell lines tested was inhibited, with IC50 values between 5 and 10 μM (Fig. S4A). Additionally, the proliferation of Burkitt lymphoma cell lines, which show constitutively high expression of c-MYC, is more sensitive to KJ-Pyr-9 (IC50 values between 1 and 2.5 μM) (Fig. S4B). KJ-Pyr-9 was further assessed against selected proliferating avian and human cells (Fig. S5). Whereas normal quail embryo fibroblasts (QEFs) and QEFs oncogenically transformed by the v-jun oncogene or by the chemical carcinogen methylcholanthrene were only slightly impaired in their growth, cells transformed by the retroviral v-myc were significantly more affected (Fig. S5A). A similar result was obtained with human fibroblasts and distinct human cancer cell lines. Notably, the leukemia cell lines K-562, MOLT-4, and HL-60, which express high levels of MYC, were strongly inhibited in their proliferation, whereas human fibroblasts and the colon carcinoma cell line SW-480 were not affected (Fig. S5B).
Fig. 3.
Effect of KJ-Pyr-9 on the proliferation of P493-6 cells. Expression of MYC in these cells is suppressed by doxycycline, resulting in cessation of cell proliferation. Cell proliferation is also halted by KJ-Pyr-9 alone or in combination with doxycycline. Error bars represent standard error of the mean (SEM).
Interference with the Transcriptional Signature of MYC.
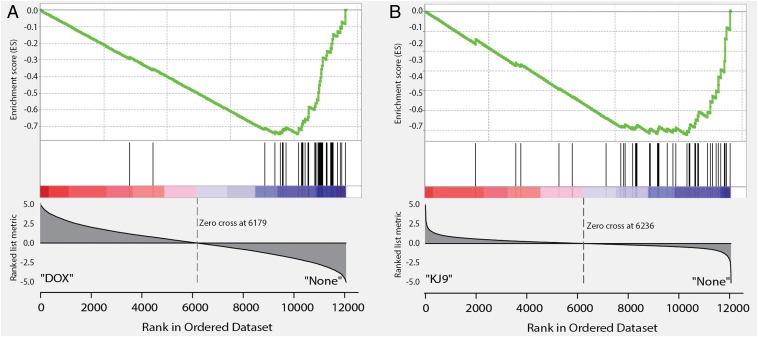
To study the effect of KJ-Pyr-9 on MYC-dependent transcription, we again chose P493-6 cells, in which MYC expression can be suppressed with doxycycline. These cells have been widely used as a model system to investigate the transcriptional targets of MYC (7, 25–27). RNA sequencing (RNAseq) was performed in triplicate on untreated P493-6 cells as well as those treated separately with KJ-Pyr-9 and doxycycline to assess the effect of KJ-Pyr-9 on the transcriptional targets of MYC. The data were analyzed by gene set enrichment analysis (GSEA) (28, 29) and are summarized in Fig. 4 and Table S1. Doxycycline-treated cells showed strong negative enrichment for previously established gene signatures of MYC (25–27) (Table S1). Treatment of P493-6 cells with KJ-Pyr-9 also resulted in this negative enrichment of essentially the same MYC-driven transcriptional signatures. We conclude that KJ-Pyr-9 specifically interferes with the MYC-dependent transcriptional program.
Fig. 4.
GSEA of RNAseq comparing the effect of doxycycline (A) with the effect of KJ-Pyr-9 (B) on the MYC gene signature published by O’Donnell et al. (26). Both drugs target the established MYC signature; the overlap between their effects is highly significant, but not 100%.
In Vivo Activity of KJ-Pyr-9.
Previous MYC inhibitors have not been effective in animal model studies. These failures resulted from insufficient affinity toward MYC or poor pharmacokinetic properties. We investigated the pharmacokinetic properties of KJ-Pyr-9 in mouse and rat and found that the concentrations of KJ-Pyr-9 achievable in the blood are sufficient to cause inhibition of MYC in vitro (Materials and Methods). We observed no signs of acute toxicity at a dose of 10 mg/kg. Rather surprisingly, KJ-Pyr-9 also crossed the blood–brain barrier and was present at higher concentrations in brain tissue than in the blood after 4 h.
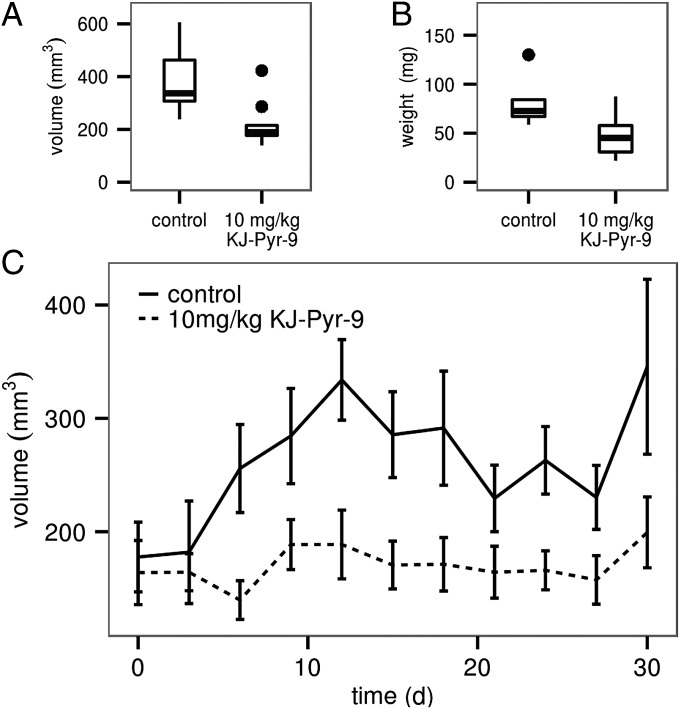
To test the in vivo effectiveness of KJ-Pyr-9, nude mice received a xenograft of MDA-MB-231 cells suspended in Matrigel and injected s.c. into the left and right flanks. When the tumors had reached an average volume of 100 mm3, mice were treated daily with 10 mg/kg KJ-Pyr-9 or vehicle control by i.p. injection for 31 d. Inhibition of tumor growth by KJ-Pyr-9 was noted after 8 d of treatment. By day 31, the tumor volume in the KJ-Pyr-9–treated animals had not increased significantly (Fig. 5). At the conclusion of the experiment the tumors were extracted and weighed. The weight measurements were in agreement with the volume determinations and confirmed the ability of KJ-Pyr-9 to halt tumor growth (Fig. 5). Treatment with KJ-Pyr-9 had no effect on the body weight of the animals.
Fig. 5.
KJ-Pyr-9 interferes with the growth of a xenograft of MDA-MB-231 cells. Mice were injected with 5 × 106 MDA-MB-231 cells s.c. into the left and right flanks. When tumors reached a volume of 100 mm3, half of the mice were given daily i.p. injections of 10 mg/kg KJ-Pyr-9 and the other half received vehicle only. Tumor growth was followed for 31 d. (A) Tumor volumes of treated and untreated animals. (B) Tumor weights of treated and untreated animals. (C) Time course of tumor volumes. Error bars indicate 95% confidence intervals.
To determine drug activity in tumor cells, protein lysates were prepared from frozen tumor samples and analyzed by Western blot. These blots showed that the expression of the MYC suppression target NDRG1 was significantly increased by treatment with KJ-Pyr-9. The degree of this enhancement varied between tumors. Immunofluorescence staining of NDRG1 and histological analysis suggested that the variability was caused by differences in the amounts of necrotic areas within the different tumors (Fig. S6). These observations suggest that KJ-Pyr-9 gained access to the tumor tissue and inhibited the transcriptional activity of MYC.
Discussion
Protein–protein interactions are difficult targets for small-molecule ligands (18, 30–38). The interacting surfaces are large and devoid of structural landmarks, and the interaction energies are typically not focused within a cleft. There is no theoretical method of identifying accessible critical residues or “hot spots” that could induce a conformational change resulting in the disruption of the PPI. These problems are augmented by the bias in available libraries that target enzymes and GPCRs, taking advantage of distinct pockets and grooves that are suitable for high-affinity interactions with small molecules. However, highly effective small-molecule inhibitors of PPI have been identified, and these successes serve as encouraging proofs of principle (18, 31–34, 38–41). New strategies for identifying weak binders have become available (42), biophysical and cell-based screening methods are being improved, and different types of libraries are being generated.
The importance of MYC in cancer has steadily risen since it was first identified as the oncogenic component in a retrovirus (43, 44) and then linked to human disease by the chromosomal translocation in Burkitt lymphoma (45). A gain of function in MYC is seen in nearly all human cancers. In addition, animal model systems have revealed unexpected roles of this oncoprotein in cancers that nominally have a non-MYC etiology. Thus, a KRAS-driven lung cancer can be eradicated by dominant-negative MYC (46). In human cancer cell lines, acquired resistance to inhibitors of PI3K can be mediated by MYC (47).
The inhibitory action of KJ-Pyr-9 on oncogenic cellular transformation is most potent against MYC and far less effective against several other oncoproteins. In this context, the data on v-Src are in apparent conflict with previous work that has shown a dependence of Src-mediated oncogenic transformation on MYC (48). However, the inhibition of MYC by KJ-Pyr-9 is not complete, and residual MYC activity could suffice to allow Src-induced cellular transformation. KJ-Pyr-9 has a strong effect on the viability and proliferative capacity of several human cancer cell lines. These effects are particularly striking in the case of leukemia cells, but they extend to cell lines derived from solid tumors as well. In the P493-6 cell model, KJ-Pyr-9–induced inhibition of proliferation is MYC-dependent. The data on the xenotransplant of a MYC-dependent human cancer cell line show a block of tumor growth in the presence of the compound. However, the effects of KJ-Pyr-9 in cell culture and in vivo are cytostatic, not cytocidal, in contrast to the effect of Omomyc overexpression in tumors (49). This may stem from incomplete inhibition of MYC, with residual activity facilitating cell survival, or it may reflect fundamental differences between Omomyc and KJ-Pyr-9 as inhibitors of MYC. Omomyc sequesters MYC into a heterodimeric complex away from MAX, whereas KJ-Pyr-9 interferes with the binding of MYC to MAX. In a cancer setting, a merely cytostatic action might appear as a therapeutic disadvantage. However, it would also reduce side effects on rapidly proliferating normal tissues, and the effect on cancer cells could be enhanced by a combination with other possibly targeted drugs.
Direct binding of KJ-Pyr-9 to MYC was demonstrated by BSI (22). The choice of this novel and highly sensitive technique was prompted by the low solubility of KJ-Pyr-9. Low solubility imposes some experimental restrictions and, for animal studies that require higher injectable concentrations of KJ-Pyr-9, we were able to increase solubility by using Tween 80 in the vehicle without any toxic effect. Low solubilities have been observed with other PPI inhibitors including Taxol (50, 51) and ABT-737 (52) and, although not ideal, the low solubility does not interfere with their clinical effectiveness. Direct, intracellular binding of KJ-Pyr-9 to MYC is also supported by our experiments using PCA constructs.
GSEA of the RNAseq data obtained with the doxycycline-treated P493-6 cells (MYC-off) shows strong negative enrichment scores that are in full concordance with the numerous previously published MYC gene signatures (25–27). The enrichment scores generated from the RNAseq data of KJ-Pyr-9–treated cells reveal a highly significant overlap with the same MYC gene signatures and with the doxycycline effect on P493-6 cells. The great majority of genes down-regulated by doxycycline are also down-regulated by KJ-Pyr-9. However, the doxycycline and KJ-Pyr-9 gene sets are not completely overlapping; an explanation of the small minority of “outliers” will require additional work.
Our work raises several questions that need to be addressed by future studies. Among these is a determination of the KJ-Pyr-9 binding mode and binding site(s) to MYC. Such data would probably also help to explain the selectivity for MYC over MAX. Additionally, in vivo observations are very limited at this time. They need to be expanded to other animal models and include an examination of normal rapidly proliferating tissues.
Materials and Methods
Fluorescence Polarization Assay.
His-tagged bHLH-LZ domains of MYC and MAX were expressed in E. coli and purified by HisTrap. Fluorescence polarization assays were conducted as described (21), except that 5-carboxyfluorescein was replaced with Alexa Fluor 594.
Renilla Luciferase-Based Protein Fragment Complementation Assay.
HEK293 cells were grown in DMEM supplemented with 10% (vol/vol) FBS. The indicated Rluc-PCA expression constructs were transiently overexpressed in a 24-well plate format. Twenty-four or 48 h posttransfection, confluent cells were treated with 20 µM KJ-Pyr-9. Following treatment, the growth medium was exchanged and cells were resuspended in PBS. Cell suspensions were transferred to white-walled 96-well plates and subjected to bioluminescence analysis using the LMax II 384 luminometer (Molecular Devices). Rluc bioluminescence signals were integrated for 10 s following addition of the Rluc substrate benzylcoelenterazine to intact cells (5 µM; NanoLight). For immunoblot analyses, anti-Renilla luciferase antibodies to detect either F[1]-fused (Millipore; MAB4410) or F[2]-fused (Millipore; MAB4400) hybrid proteins were used (23, 24).
Assay for Oncogenic Transformation in Cell Culture.
Oncogenic transformation was determined in cultures of chicken embryo fibroblasts as previously described (53, 54). CEF were infected with a series of 10-fold dilutions of the indicated virus. Compounds were added to the nutrient agar overlay. Additional compound-containing overlay was added every 3 d until experimental end point. ATG-MYC was created by site-directed mutagenesis changing the CTG start codon of human c-MYC to ATG. It was then inserted into the retroviral expression vector RCAS(A).sfi (55).
Backscattering Interferometry.
Data were collected on an instrument built by Molecular Sensing. N-terminal His-tagged maltose-binding protein fusions of the MYC and MAX bHLH-LZ domains were cloned and expressed in E. coli. These fusion proteins were purified by Ni-NTA column chromatography and buffer-exchanged to 60 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 9 mM MgCl2, 3 mM EDTA by dialysis in Slide-a-Lyzer cassettes (Thermo Scientific). Constant amounts of MYC, MAX, or MYC–MAX heterodimer were mixed with increasing amounts of KJ-Pyr-9 and analyzed by backscattering interferometry. The resulting changes in the index of refraction were plotted and fitted to a logistic curve to determine Kd.
Cell Lines.
P493-6 cells were kindly provided by Chi Van Dang (University of Pennsylvania, Philadelphia). P493-6 cells were grown in RPMI-1640 (Life Technologies) supplemented with 10% tetracycline-free FBS (Gemini) and 1× penicillin, streptomycin, l-glutamine (Sigma). Cells were incubated at 37 °C in a 5% CO2 atmosphere. The human cancer cell lines were NCI-H460 (ATCC) (large-cell lung cancer), MDA-MB-231 (National Cancer Institute) (adenocarcinoma of the breast), SUM-159PT (Asterand) (estrogen-independent breast cancer), and SW-480 (ATCC) (colorectal carcinoma). K-562, MOLT-4, and HL-60 were derived from chronic myeloid leukemia in blast crisis, acute lymphoblastic leukemia, and acute myeloid leukemia, respectively. Human immortalized fibroblasts were provided by J. Troppmair (Medical University Innsbruck, Innsbruck, Austria). Cell culture of quail (Coturnix japonica) embryo fibroblasts and of the established quail cell lines Q8, QEF/MC29, VJ, and QT6 was performed as described (56).
Cell-Proliferation Assays.
Assays used staining with the redox dye resazurin (Sigma) to measure cell viability (57). Cells were seeded at 103 per 100-µL well in 96-well plates and grown in the presence of 2.5% FBS. MDA-MB-231 cells were cultured in DMEM; SUM-159PT cells were cultured in HAM’s F12; and NCI-H460 cells were cultured in RPMI-1640. MDA-MB-231 cells were exposed to KJ-Pyr-9 for 216 h with fresh compound-containing medium supplied at 120 and 192 h; SUM-159PT cells were exposed to the compound for 120 h and fresh medium with the appropriate compound concentrations was supplied at 48 h; and NCI-H460 cells were grown with compound for 72 h. Triplicate cultures of P493-6 cells were grown in six-well plates from a starting density of 1 × 105 cells per mL in 4 mL culture medium per well. Compounds were added immediately following cell seeding. Following compound addition, cells were distributed by vortexing the plate at 400 rpm for 10 s. One hundred-microliter samples were taken after vortexing and counted using a Beckman Coulter Z1 counter at 0, 12, 24, 36, 48, 60, 72, and 96 h of incubation.
Pharmacokinetics.
Three mice were injected with 10 mg/kg KJ-Pyr-9 dissolved in 10:10:80 Tween 80:DMSO:5% dextrose in water intraperitoneally. At 4 h, concentrations of KJ-Pyr-9 in the plasma and in the brain were 3.5 and 12.4 µM, respectively. Rats were dosed with 1 mg/kg i.v.; elimination half-lives in plasma were ∼1.84 h (rats).
Xenograft Experiments.
Ten 8-wk-old female nude mice (HSD:athymic nude-Foxn1nu) were injected with 5 × 106 MDA-MB-231 cells s.c. into the left and right flanks. Cells were suspended in high-concentration Matrigel (BD Biosciences) before injection. Xenograft tumors were allowed to grow until the average volume of the tumors reached 100 mm3, as measured by external calipers. At this point, the mice were divided into two groups. One received 10 mg/kg KJ-Pyr-9 and the other received vehicle only, dosed daily by i.p. injection. Tumor volume and mouse weight were measured daily. Vehicle used in all cases was 10:10:80 Tween 80:DMSO:5% dextrose in water. The mice were treated for a period of 31 d. At the end of the experiment, the mice were euthanized and tumors were excised. Tumors were weighed. Samples of each tumor were fixed in formalin for histology and frozen for Western blotting. All vertebrate experiments were conducted with the approval of The Scripps Research Institute (TSRI) Institutional Animal Care and Use Committee.
Statistical Methods.
Tumor volumes were calculated using (length) × (width)2. Tumor volumes were evaluated using Efron’s bootstrap (58) procedures as implemented in R (59–62). Mean volumes and 95% confidence intervals were determined using bootstrap resampling with 106 randomizations and graphed using the ggplot2 package (63). P values were determined using the resampling permutation method with 106 randomizations.
Supplementary Material
Acknowledgments
We are indebted to Peter A. Wells (Pfizer, San Diego) for sharing data on the fluorescence polarization assay involving MYC–MAX and pyridine libraries. The authors also gratefully acknowledge the kind help of M. G. Finn in the backscattering interferometry measurements. Hugh Rosen, Steven Brown, and Michael Cameron of TSRI Translational Research Institute–Drug Metabolism and Pharmacokinetics carried out the pharmacokinetic determinations. RNAseq was performed at TSRI Next Generation Sequencing Core with Steven Head and Lana Schaffer. The initial stages of this investigation received support from Pfizer under SPF-1892. This work was supported by the National Cancer Institute under Award R01 CA078230 (to P.K.V.), The Skaggs Institute for Chemical Biology (K.D.J.), and Austrian Science Fund Grants P23652 (to K.B.) and P22608 (to E.S.). This is manuscript 25089 of The Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE58168).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319488111/-/DCSupplemental.
References
- 1.Lee LA, Dang CV. Myc target transcriptomes. Curr Top Microbiol Immunol. 2006;302:145–167. doi: 10.1007/3-540-32952-8_6. [DOI] [PubMed] [Google Scholar]
- 2.Eisenman RN. The Max network: Coordinated transcriptional regulation of cell growth and proliferation. Harvey Lect. 2000–2001;96:1–32. [PubMed] [Google Scholar]
- 3.Eisenman RN. Deconstructing Myc. Genes Dev. 2001;15(16):2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- 4.Blackwood EM, Eisenman RN. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251(4998):1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 5.Ji H, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE. 2011;6(10):e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Z, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151(1):68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsson LG, Henriksson MA. The Yin and Yang functions of the Myc oncoprotein in cancer development and as targets for therapy. Exp Cell Res. 2010;316(8):1429–1437. doi: 10.1016/j.yexcr.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 11.Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: The relationship between c-Myc and the cell cycle. Oncogene. 1999;18(19):2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 12.Lawlor ER, et al. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006;66(9):4591–4601. doi: 10.1158/0008-5472.CAN-05-3826. [DOI] [PubMed] [Google Scholar]
- 13.Sodir NM, et al. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 2011;25(9):907–916. doi: 10.1101/gad.2038411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112(2):193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 15.Berg T, et al. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci USA. 2002;99(6):3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22(40):6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 17.Prochownik EV, Vogt PK. Therapeutic targeting of Myc. Genes Cancer. 2010;1(6):650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg T. Small-molecule modulators of c-Myc/Max and Max/Max interactions. Curr Top Microbiol Immunol. 2011;348:139–149. doi: 10.1007/82_2010_90. [DOI] [PubMed] [Google Scholar]
- 19.Zirath H, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci USA. 2013;110(25):10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimori T, Wirsching P, Janda KD. Preparation of a Kröhnke pyridine combinatorial library suitable for solution-phase biological screening. J Comb Chem. 2003;5(5):625–631. [Google Scholar]
- 21.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem Biol. 2006;13(7):745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Baksh MM, Kussrow AK, Mileni M, Finn MG, Bornhop DJ. Label-free quantification of membrane-ligand interactions using backscattering interferometry. Nat Biotechnol. 2011;29(4):357–360. doi: 10.1038/nbt.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachmann VA, et al. Reciprocal regulation of PKA and Rac signaling. Proc Natl Acad Sci USA. 2013;110(21):8531–8536. doi: 10.1073/pnas.1215902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stefan E, et al. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc Natl Acad Sci USA. 2007;104(43):16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuhmacher M, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29(2):397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell KA, et al. Activation of transferrin receptor 1 by c-Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol. 2006;26(6):2373–2386. doi: 10.1128/MCB.26.6.2373-2386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 2003;4(10):R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 30.Raj M, Bullock BN, Arora PS. Plucking the high hanging fruit: A systematic approach for targeting protein-protein interactions. Bioorg Med Chem. 2013;21(14):4051–4057. doi: 10.1016/j.bmc.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson CG, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: Lessons learned and applied. Curr Top Microbiol Immunol. 2011;348:25–59. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- 32.Adler MJ, Jamieson AG, Hamilton AD. Hydrogen-bonded synthetic mimics of protein secondary structure as disruptors of protein-protein interactions. Curr Top Microbiol Immunol. 2011;348:1–23. doi: 10.1007/82_2010_91. [DOI] [PubMed] [Google Scholar]
- 33.Vu BT, Vassilev L. Small-molecule inhibitors of the p53-MDM2 interaction. Curr Top Microbiol Immunol. 2011;348:151–172. doi: 10.1007/82_2010_110. [DOI] [PubMed] [Google Scholar]
- 34.Zeitlin BD, Nör JE. Small-molecule inhibitors reveal a new function for Bcl-2 as a proangiogenic signaling molecule. Curr Top Microbiol Immunol. 2011;348:115–137. doi: 10.1007/82_2010_109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arkin MR, Whitty A. The road less traveled: Modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13(3):284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 36.Arkin M. Protein-protein interactions and cancer: Small molecules going in for the kill. Curr Opin Chem Biol. 2005;9(3):317–324. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: Progressing towards the dream. Nat Rev Drug Discov. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 38.Garner AL, Janda KD. Protein-protein interactions and cancer: Targeting the central dogma. Curr Top Med Chem. 2011;11(3):258–280. doi: 10.2174/156802611794072614. [DOI] [PubMed] [Google Scholar]
- 39.Haggarty SJ, Schreiber SL. 2007. Forward chemical genetics. Chemical Biology: From Small Molecules to Systems Biology and Drug Design, eds Schreiber SL, Kapoor TM, Wess G (Wiley-VCH, Weinheim, Germany), Vol 1, pp 299–354.
- 40.Gough JD, Crews CM. 2007. Using natural products to unravel cell biology. Chemical Biology: From Small Molecules to Systems Biology and Drug Design, eds Schreiber SL, Kapoor TM, Wess G (Wiley-VCH, Weinheim, Germany), Vol 1, pp 95–114.
- 41.Lampson MA, Kapoor TM. 2007. Using small molecules to unravel biological mechanisms. Chemical Biology: From Small Molecules to Systems Biology and Drug Design, eds Schreiber SL, Kapoor TM, Wess G (Wiley-VCH, Weinheim, Germany), Vol 1, pp 71–94.
- 42.Erlanson DA, Wells JA, Braisted AC. Tethering: Fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 43.Duesberg PH, Bister K, Vogt PK. The RNA of avian acute leukemia virus MC29. Proc Natl Acad Sci USA. 1977;74(10):4320–4324. doi: 10.1073/pnas.74.10.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bister K. MC29 avian myelocytomatosis virus. In: Maloy S, Hughes K, editors. Brenner’s Encyclopedia of Genetics. 2nd Ed. Vol 4. San Diego: Academic Press; 2013. pp. 330–332. [Google Scholar]
- 45.Dalla-Favera R, et al. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79(24):7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soucek L, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27(5):504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu P, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17(9):1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bowman T, et al. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci USA. 2001;98(13):7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savino M, et al. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS ONE. 2011;6(7):e22284. doi: 10.1371/journal.pone.0022284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scripture CD, Figg WD, Sparreboom A. Paclitaxel chemotherapy: From empiricism to a mechanism-based formulation strategy. Ther Clin Risk Manag. 2005;1(2):107–114. doi: 10.2147/tcrm.1.2.107.62910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adams JD, et al. Taxol: A history of pharmaceutical development and current pharmaceutical concerns. J Natl Cancer Inst Monogr. 1993;(15):141–147. [PubMed] [Google Scholar]
- 52.Petros AM, et al. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J Med Chem. 2006;49(2):656–663. doi: 10.1021/jm0507532. [DOI] [PubMed] [Google Scholar]
- 53.Duff RG, Vogt PK. Characteristics of two new avian tumor virus subgroups. Virology. 1969;39(1):18–30. doi: 10.1016/0042-6822(69)90344-4. [DOI] [PubMed] [Google Scholar]
- 54.Bos TJ, et al. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4(10):1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 55.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The Akt kinase: Molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95(25):14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartl M, Nist A, Khan MI, Valovka T, Bister K. Inhibition of Myc-induced cell transformation by brain acid-soluble protein 1 (BASP1) Proc Natl Acad Sci USA. 2009;106(14):5604–5609. doi: 10.1073/pnas.0812101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahmed SA, Gogal RM, Jr, Walsh JE. A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: An alternative to [3H]thymidine incorporation assay. J Immunol Methods. 1994;170(2):211–224. doi: 10.1016/0022-1759(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 58.Efron B, Tibshirani R. An Introduction to the Bootstrap. Boca Raton, FL: Chapman & Hall/CRC; 1994. [Google Scholar]
- 59.Davison A, Hinkley D. Bootstrap Methods and Their Application. New York: Cambridge Univ Press; 1997. [Google Scholar]
- 60.Canty A, Ripley B. 2014. boot: Bootstrap R (S-Plus) Functions, R package Version 1.3-11 (Oxford, UK)
- 61.The R Development Core Team 2014. R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna)
- 62.Wu J, Houghton PJ. Assessing cytotoxic treatment effects in preclinical tumor xenograft models. J Biopharm Stat. 2009;19(5):755–762. doi: 10.1080/10543400903105158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickham H. 2009. ggplot2: Elegant Graphics for Data Analysis (Springer, New York)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.