Significance
Molecular analysis of surgically resected glioblastomas (GBM) samples has uncovered phenotypically and clinically distinct tumor subtypes. However, little is known about the molecular features of the glioma margins that are left behind after surgery. To address this key issue, we performed RNA-sequencing (RNA-seq) and histological analysis on MRI-guided biopsies from the contrast-enhancing core and nonenhancing margins of GBM. Computational deconvolution of the RNA-seq data revealed that cellular composition, including nonneoplastic cells, is a major determinant of the expression patterns at the margins of GBM. The different GBM subtypes show distinct expression patterns that relate the contrast enhancing centers to the nonenhancing margins of tumors. Understanding these patterns may provide a means to infer the molecular and cellular features of residual disease.
Keywords: glioma, tumor heterogeneity, microenvironment
Abstract
Glioblastomas (GBMs) diffusely infiltrate the brain, making complete removal by surgical resection impossible. The mixture of neoplastic and nonneoplastic cells that remain after surgery form the biological context for adjuvant therapeutic intervention and recurrence. We performed RNA-sequencing (RNA-seq) and histological analysis on radiographically guided biopsies taken from different regions of GBM and showed that the tissue contained within the contrast-enhancing (CE) core of tumors have different cellular and molecular compositions compared with tissue from the nonenhancing (NE) margins of tumors. Comparisons with the The Cancer Genome Atlas dataset showed that the samples from CE regions resembled the proneural, classical, or mesenchymal subtypes of GBM, whereas the samples from the NE regions predominantly resembled the neural subtype. Computational deconvolution of the RNA-seq data revealed that contributions from nonneoplastic brain cells significantly influence the expression pattern in the NE samples. Gene ontology analysis showed that the cell type-specific expression patterns were functionally distinct and highly enriched in genes associated with the corresponding cell phenotypes. Comparing the RNA-seq data from the GBM samples to that of nonneoplastic brain revealed that the differentially expressed genes are distributed across multiple cell types. Notably, the patterns of cell type-specific alterations varied between the different GBM subtypes: the NE regions of proneural tumors were enriched in oligodendrocyte progenitor genes, whereas the NE regions of mesenchymal GBM were enriched in astrocytic and microglial genes. These subtype-specific patterns provide new insights into molecular and cellular composition of the infiltrative margins of GBM.
Glioma cells diffusely infiltrate the brain and intermingle with neural cells in the surrounding brain tissue, resulting in a complex mixture that includes variable proportions of glioma cells, neurons, and various lineages of reactive or recruited glia. At the infiltrative margins of glioblastoma (GBM), the nonneoplastic brain cells can far outnumber the glioma cells and, therefore, will have a significant effect on the molecular features of the tissue. Expression profiling and whole genome sequencing from hundreds of GBM specimens by The Cancer Genome Atlas (TCGA) has revealed a broad spectrum of genetic alterations and discrete expression signatures or subtypes that stratify the majority of patients (1, 2). These studies analyzed tumor samples that were removed during surgery, but were not radiographically localized and, therefore, do not address the question of how the molecular signature may vary across different regions of a tumor. Recent studies have sampled multiple regions within a GBM and shown that more than one molecular subtype can coexist within a single tumor (3). However, the effect of varying cellular composition on GBM subtype, particularly the contribution of nonneoplastic cells, has not been addressed.
GBM typically appears as a contrast-enhancing mass, which represents the highly cellular core of the tumor with vascular proliferation and blood–brain barrier breakdown. This contrast-enhancing (CE) region is typically surrounded by a diffuse, nonenhancing (NE) region of abnormal T2/FLAIR signal, which represents edematous brain tissue with varying numbers of infiltrating glioma cells. The primary treatment of GBM is surgical resection, during which the surgeon removes as much of the CE mass as possible. Thus, molecular and genetic profiling of GBM, including the TCGA effort, has predominantly used samples from the CE regions of tumor. However, it is the NE regions of glioma that are left behind after surgery, which neurooncologists must treat and which inevitably give rise to recurrence. Thus, there is immense prognostic and therapeutic significance to understanding the cellular and molecular features of the NE regions of tumor, yet often these areas are not resected and, therefore, have not been directly studied.
There are two major obstacles to this goal. The first is the surgical challenge of radiographically localized sampling of the NE tumor margins. The second is the issue of the complex cellular composition that characterizes these regions of diffuse infiltration. In this study, we have addressed both challenges, and associated distinct molecular and cellular features of the NE regions of GBM with the molecular subtype, as defined by the resected CE regions of the tumor.
Results
Image-Guided Biopsies Reveal Distinct Molecular and Cellular Composition in the CE and FLAIR+ NE Regions of GBM.
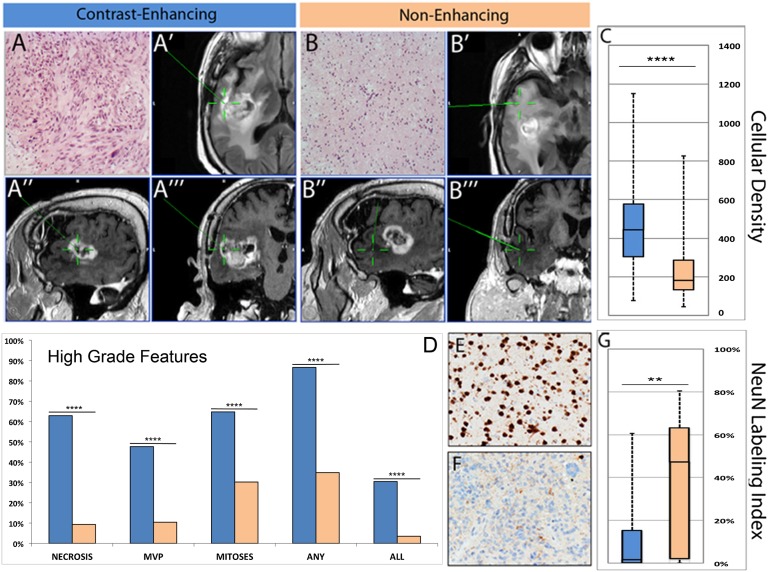
We collected radiographically localized biopsies from the CE and FLAIR+ NE regions of GBM from 69 patients. Histological and immunohistochemical analysis of the samples showed significant differences in the cellular density and cellular composition between the CE and NE samples (Fig. 1). The CE samples had significantly higher cellularity (P < 0.00001) than NE samples and were significantly more likely to contain the histological hallmarks of GBM, including glomeruloid-type vascular proliferation and necrosis (P < 0.00001 for each feature). Conversely, the NE regions showed the histological features of diffusely infiltrating glioma with neoplastic glial cells intermingled with nonneoplastic and reactive cells (SI Appendix, Fig. S1). The cellular composition of the NE samples was variable but showed significantly higher numbers of NeuN+ neurons compared with CE samples (Fig. 1). In addition to demonstrating that cellular composition varies across different radiographically localized regions of the tumor, these results show that nonneoplastic cells are a major component of the NE regions of GBM and highlight the importance of considering this complex cellular composition when analyzing the expression pattern of diffusely infiltrating gliomas.
Fig. 1.
MRI screen captures show the radiographically localized sampling of CE (A′–A′′′) and NE (B′–B′′′) regions of GBM, and micrographs of the corresponding biopsies show the histological features of the highly cellular core (A) and the infiltrative margin (B) of the tumor (stained with hematoxylin and eosin). A′ and B′ show the axial FLAIR, A′′ and B′′ show the sagittal T1 with contrast, and A′′′ and B′′′ show the coronal T1 with contrast. The green crosshairs mark the biopsy sites. Quantitative analysis of CE (blue bars) and NE (tan bars) samples shows significant differences in cellular density (C) and the presence of histopathological hallmarks of high-grade glioma (D). Immunohistochemical analysis for NeuN shows numerous positive neurons in samples from NE regions (E) and only rare entrapped neurons in samples from the CE regions (F). Quantitative analysis of theses stains shows significant differences in the fractional abundance of NeuN+ neurons in NE vs. CE samples (G). In C, D, and G, **P < 0.001; ***P < 0.0001; ****P < 0.00001.
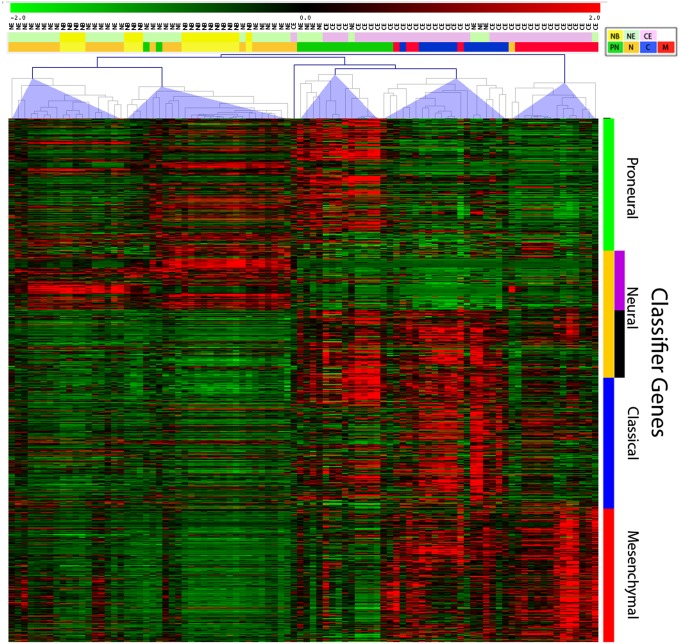
We performed RNA-seq analysis on 75 glioma samples from 27 different glioma patients including 39 samples from the CE regions and 36 samples from the NE regions (SI Appendix, Table S1). We also performed RNA-seq on 17 nonneoplastic/normal brain (NB) samples acquired from 11 patients with no oncological history undergoing ventriculoperitoneal shunt placement or resection for seizures. For each CE and NE tumor sample in the RNA-seq dataset, we determined the GBM subtype as reported (4). Briefly, we calculated the Spearman correlation between each RNA-seq profile and the subtyped TCGA profiles from Verhaak et al. (2). We used the median value of the correlation between an RNA-seq profile and the TCGA microarray data for a given subtype as a similarity score for that subtype (Fig. 2 and SI Appendix, Table S1). The majority of CE samples showed the highest correlation with the proneural, classical, or mesenchymal subtypes. In contrast, the majority of samples from NE regions showed the highest correlation with the neural subtype. A heatmap shows the expression level of the GBM subtype classifier genes across all samples (including the 17 NB samples), with the majority of CE samples clustered into three groups showing enrichment in proneural, classical, or mesenchymal genes. In contrast, most NE samples were clustered with the NB samples and were classified as neural or proneural (Fig. 2 and SI Appendix, Table S1). Notably, the NE and NB samples tended to show higher expression in only a subset of the neural classifier genes (SI Appendix, Fig. S2). This subset of neural genes is selectively up-regulated in “neural GBM” and contains genes that are normally expressed by mature neurons and oligodendrocytes (2). Together with the histological and immunohistochemical data showing that the NE samples represented infiltrated brain, these results suggest that the composite gene expression profile of the NE samples contains a significant contribution from nonneoplastic brain cells such as neurons and oligodendrocytes.
Fig. 2.
RNA-seq–based expression profiling showing the expression of the Verhaak classifier genes across 92 samples (39 CE, 36 NE, and 17 NB). The samples were clustered by using Spearman correlation into five major clusters. Two clusters are predominantly composed of NE and NB samples, and the other three clusters are predominantly composed of CE samples, with samples correlating with proneural, classical, and mesenchymal subtypes. The NE clusters contain the majority of neural samples. The colored bars above the heatmap show the sample origin (upper bar: NB, yellow; NE, green; CE, pink) and the subtype classification (lower bar: neural, brown; proneural, green; classical, blue; mesenchymal, red). The heatmap shows high levels of expression as red and low levels as green.
Computational Deconvolution of RNA-Seq Data Identifies Cell-Type Specific Expression Profiles.
As glioma cells infiltrate the brain, they encounter a uniquely complex and interconnected environment consisting of several different cell types, including astrocytes, neurons, microglia, and oligodendrocytes. Most efforts at expression profiling in glioma, including a large-scale microarray study by TCGA (1), have not addressed the complex cellular composition of the tumor. However, efforts to physically separate the various cell types by flow sorting or other methods face technical challenges and run the risk of perturbing RNA expression levels. To address these issues, we used a computational approach to deconvolve the composite RNA-seq expression data from homogenized NE samples (5–8). A report in which expression profiles of multiple neural cell types were deconvolved by microarray analysis of brain tissue is particularly relevant to our application (7). Our approach is based on the algorithm described by Kuhn et al. (7), where constrained, least-squares fitting is used to estimate average expression profiles of each cell type based on multiple homogenate expression profiles. The expression levels of marker genes are used to approximate the fractional composition of each cell type in each sample, allowing us to estimate the average expression profile of each cell type for a set of samples.
We deconvolved the NE and nonneoplastic brain RNA-seq profiles into average profiles for each of six cell types, which represent the major lineages present in brain tissue. Specifically, we seeded the algorithm with OLIG2 for oligodendrocyte progenitor-like (OPC-like) cells, CD44 for reactive astrocytes, AQP4 for unreactive gray matter astrocytes, MAL and MOG for mature oligodendrocytes, RBFOX3 (NEUN) and NEUROD6 for neurons, and AIF1 (IBA1) and CD68 for microglia. Our decision to distinguish between two different astrocyte populations (CD44+ and CD44−) was motivated by recent studies showing that astrocytes in normal and pathological brain tissue can be subdivided into CD44+ and CD44− populations, and that these cell types have very different characteristics (9). Furthermore, it has been shown that reactive astrocytes associated with a variety of pathological conditions, including GBM, up-regulate CD44 as part of their phenotypic transformation (10, 11). The other lineages that we included are also likely to contain distinct subpopulations. For example, the NE samples likely contained multiple subpopulations of neurons that were not resolved by our deconvolution algorithm, and the neuron-specific expression profile represents a composite of these different subpopulations. The microglial/monocytic lineage may contain a mixture of resident microglia and blood-derived monocytes. Notably, the NE samples also contain neoplastic glia, and their expression profiles are distributed into the six cell types in an unsupervised manner. Previous studies have shown that glioma samples contain two prominent and distinct types of tumor cells, one that expresses OLIG2 and is OPC-like, and the other that expresses CD44 and is more astrocytic (12–14). Distinct populations of reactive/nonneoplastic glial cells also express these markers. Therefore, the CD44+ populations likely contain both reactive and transformed astrocytes, and the OLIG2+ population likely contains a mixture of transformed and recruited OPCs.
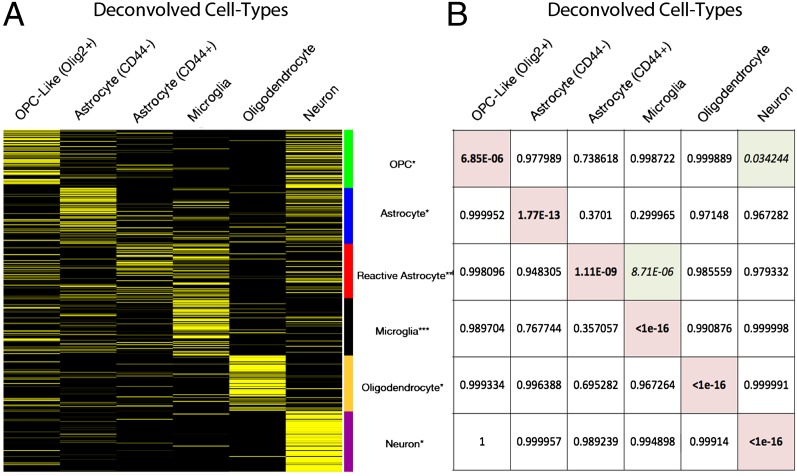
We compared the cell type-specific expression profiles derived from our computational deconvolution of the NE glioma and NB samples to expression data generated in previous studies by physical separation and purification of neural cell types from mouse brain tissue (11, 15, 16). This comparison showed that each of the deconvolved expression profiles was significantly enriched for at least one of the cell type-specific gene lists derived from these previous studies. Furthermore, for each gene list, the most significant enrichment was seen in the appropriate cell type, with P values ranging from <1 × 10−16 to 7 × 10−6 (Fig. 3 and SI Appendix, Table S3). Interestingly, two of the deconvolved cell types also showed significant enrichment for a second cell type-specific gene list: Microglia showed enrichment for genes expressed by reactive astrocytes (11), and neurons showed enrichment for genes expressed by OPCs (15). We also performed gene ontology analysis, which showed that the cell type-specific expression profiles derived from the deconvolution of the NE glioma samples were each associated with the distinct, expected cellular functions of the corresponding cell lineage (SI Appendix, Table S2). Taken together, these analyses demonstrate that the pattern of cell type-specific expression profiles predicted by computational deconvolution of the NE samples is remarkably consistent with the cell type-specific expression patterns reported in previous studies and with the expected functionality of these cell types as independently determined by gene ontology analysis. As further validation of the deconvolution analysis, immunohistochemical analysis with antibodies against three neural classifier genes that are predicted to be predominantly expressed in neurons (HPCA, CRYM, and CHN1) showed selective staining of neurons in both NB and glioma samples (SI Appendix, Fig. S3).
Fig. 3.
Computational deconvolution reveals cell type-specific expression profiles. (A) The heat map shows the expression of cell-type genes in the six cell types that were deconvolved from the NB and NE samples: High expression is yellow, and low expression is black. The lists of cell type-specific genes were derived from previous studies (11, 15, 16). (B) We performed a hypergeometric test to assess the significance of the cell type enrichment of each gene list compared with deconvolution of the whole transcriptome. Each of the six gene lists showed the most significant enrichment in the expected cell type (highlighted in red). In addition, microglia show significant enrichment for genes expressed by reactive astrocytes and neurons show significant enrichment for OPC genes (each highlighted in green). The list of cell type-specific genes and associated cell type-specific expression profiles are provided in SI Appendix, Table S3.
Differentially Expressed Genes Are Distributed in Multiple Cell Types in a GBM Subtype-Specific Pattern.
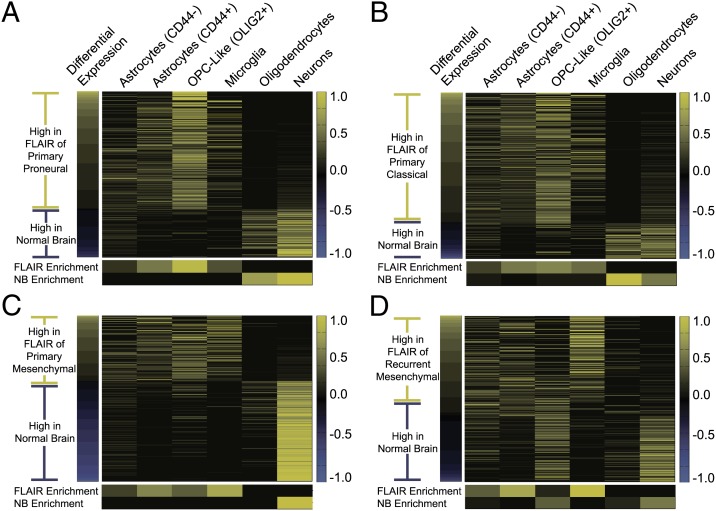
Although the above analysis demonstrated that nonneoplastic brain cells contribute significantly to the NE expression data, histological analysis showed that the NE glioma samples are distinct from normal brain. Not only do they contain infiltrating glioma cells, but they also show reactive astrocytosis and microgliosis (SI Appendix, Fig. S1). Therefore, to characterize the changes in expression patterns associated with these histological alterations, we performed differential gene expression analysis between NE glioma samples and nonneoplastic brain samples. We divided the NE glioma specimens into the different subtypes, based on the GBM subtype assignment of the CE portion of the tumor and compared the nonneoplastic brain and the NE glioma from each group. We then queried the NE deconvolution data to assess the cellular distribution of the differentially expressed genes in each sample group (Fig. 4). Although the deconvolution algorithm receives no input from the differential expression analysis, we consistently observe a sharp transition in cellular distribution between overexpressed and underexpressed genes. Genes that are highly expressed in the NE tumor tissue relative to nonneoplastic brain are expressed mainly in OPC-like cells, astrocytes, or microglia. Conversely, genes that are highly expressed in nonneoplastic brain were localized primarily to neurons and oligodendrocytes. Furthermore, the cellular distributions of differentially expressed genes in NE tissue were subtype-specific. We found that genes that were increased in NE regions of proneural tumors predominately distributed to OLIG2+ OPC-like cells, whereas the genes that were increased in the NE regions of mesenchymal tumors predominately distributed to CD44+ astrocytes and microglia (Fig. 4). We also performed differential expression analysis between nonneoplastic brain and the NE samples from recurrent mesenchymal GBM (the majority of the recurrent tumors were subtyped as mesenchymal). These results revealed a prominent expression of genes mapped to CD44+ astrocytes and microglia and a marked decrease in genes that mapped to OLIG2+ OPC-like cells (Fig. 4).
Fig. 4.
Heatmaps showing the deconvolved cellular distribution of gene expression for differentially expressed genes (P < 0.05) comparing normal brain (NB) to NE of proneural GBMs (A), NE of classical GBMs (B), NE of Primary mesenchymal GBMs (C), and NE of Recurrent mesenchymal GBMs (D). For each gene (rows), the expression level is normalized across cell types (columns) so that the value in the heat map reflects its fractional abundance in a given cell type. To obtain these cellular distributions, we deconvolved the NE and NB samples in aggregate and obtained a single average cellular distribution estimate for each gene. Although differential expression information was not provided to the deconvolution algorithm, all four heatmaps show a sharp transition in cellular composition between genes that are expressed at higher levels in the NE tumor tissue vs. genes that are expressed at higher levels in normal brain. The small heat maps that appear underneath each image represent the fraction of the total number of differentially expressed genes in each sample group are predominantly expressed in each of the six cell types.
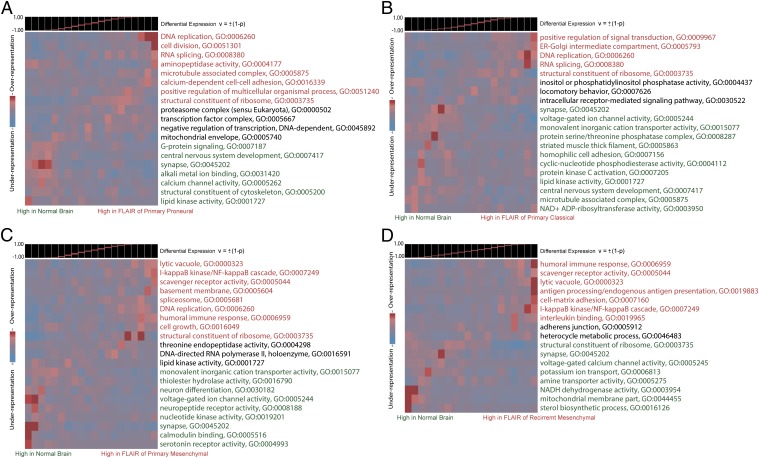
To complement our deconvolution analysis of the differentially expressed genes, we also conducted a pathway analysis for each of the four sample groups. Using iPAGE, a mutual information-based algorithm for gene ontology enrichment analysis, we determined the key pathways that are enriched among differentially expressed genes. The results, shown in Fig. 5, are remarkably consistent with our assessment of the cellular distributions for these genes. For example, iPAGE shows that cellular proliferation ontologies like DNA replication and M phase are enriched among highly expressed genes in proneural NE tissue, consistent with the predominance of proliferating OPC-like cells. However, immune response and inflammatory ontologies are preferentially enriched in the primary mesenchymal NE tissue, consistent with the microglial cellular distribution described above. In recurrent mesenchymal NE tissue, which we find to be largely devoid of OPC-like expression, we do not observe enrichment of cell cycle- and proliferation-related ontologies.
Fig. 5.
iPAGE gene ontology analysis of differentially expressed genes comparing normal brain (NB) to NE of proneural GBMs (A), NE of classical GBMs (B), NE of Primary mesenchymal GBMs (C), and NE of Recurrent mesenchymal GBMs (D). The gene ontology categories highlighted in red are associated with genes that are highly expressed in the NE tissue relative to normal brain, whereas those in green are associated with genes that are highly expressed in normal brain relative to NE tissue. Gene ontology categories in black are associated with genes that are not differentially expressed. Although cell cycle and proliferation-related pathways dominate the proneural and classical NE, an immune response/inflammatory signature dominates the mesenchymal NE. These expression signatures are consistent with the cellular distributions that we estimated for differentially expressed genes by deconvolution analysis.
Discussion
We used radiographically localized biopsies and RNA-seq to show that the molecular and cellular composition of NE regions differ significantly from those of the CE regions of a GBM. We also used a computational strategy for deconvolving composite expression profiles into cell type-specific profiles to identify GBM subtype-specific relationships between CE and NE regions. One important implication of these results is that the molecular and cellular characteristics of NE regions of a glioma, which are often left behind after surgery, may be inferred from the molecular subtype of the CE component of the tumor, which is resected during surgery. Understanding these relationships could lead to development of new therapies that target these subtype-specific alterations, which are expressed in residual tumor cells or reactive cells in the tumor microenvironment. For example, our results show that the NE regions of proneural GBM are enriched in genes expressed by OLIG2+/OPC-like cells, which most likely represent infiltrating glioma cells. Conversely, the NE regions of mesenchymal GBMs were highly enriched in genes expressed by microglia and CD44+ astrocytes. These findings are consistent with previous studies showing that proneural GBM is highly enriched in OPC genes (17, 18), whereas mesenchymal GBM harbors an inflammatory signature (13, 19, 20). However, our results provide new insight into how the expression of mesenchymal and inflammatory genes are distributed among different cell types. Notably, the deconvolution analysis shows that within the NE regions of GBM, the majority of classifier genes used to define the mesenchymal subtype are predominantly expressed by microglia, whereas a smaller subset of mesenchymal genes are expressed in CD44+ astrocytes (SI Appendix, Fig. S4 and Table S4). Similarly, differential gene expression analysis of our samples identified a set of CD44+ astrocytic and microglial genes that are up-regulated in the NE regions of mesenchymal GBM compared with nonneoplastic brain. Gene ontology analysis shows that these genes are associated with inflammatory and immune response, and include cytokines and cytokine receptors that may mediate reciprocal signaling between glioma cells and microglia in the infiltrative margins of mesenchymal GBM.
Gene ontology analysis of the deconvolved expression data shows that both OLIG2+ and CD44+ cells are highly enriched in genes involved in cellular proliferation (Fig. 5 and SI Appendix, Fig. S5), suggesting that these cell types include the neoplastic glioma cells. Previous studies have shown that CD44 and OLIG2 are markers for distinct populations of glioma cells (13, 14). Our deconvolution analysis suggests that these two cell types have different expression patterns, with the OLIG2+ cells most resembling OPCs and CD44+ cells most closely resembling reactive astrocytes (Fig. 3). In addition, the expression patterns of these two cell types differ with respect to the classifier genes used to define different GBM subtypes. Notably, OLIG2+ cells show the highest expression of proneural and classical genes and low levels of mesenchymal genes, whereas CD44+ cells show high levels of classical and mesenchymal genes and lower levels of proneural genes (SI Appendix, Fig. S4). These findings suggest that the NE regions of proneural gliomas predominantly contain OLIG2+, OPC-like glioma cells, whereas NE regions of mesenchymal gliomas contain a larger proportion of CD44+ astrocyte-like tumor cells. Notably, Sox2, a proneural classifier gene and stem cell marker that has been implicated in glioma growth and progression (21, 22) is expressed by both OLIG2+ cells and CD44+ cells. Immunohistochemical stains of NE glioma samples showed that SOX2 is expressed in a high percentage of infiltrating glioma cells in primary samples; however, recurrent gliomas showed a significantly lower SOX2 labeling index (SI Appendix, Fig. S6). These findings are consistent with our RNA-seq analysis and with previous published results showing that SOX2 is widely expressed in primary glioma, but is reduced in recurrent glioma (22).
Our results also suggest that some samples may contain a mixture of OLIG2+ and CD44+ glioma cells, particularly in the NE regions of classical GBMs where genes from both cell types are highly represented. In a recent study, Pietras et al. show that CD44 is differentially expressed across the three major GBM subtypes (23). In mesenchymal and classical GBM, high levels of CD44 were seen throughout the tumors, whereas in proneural GBM CD44 expression was predominantly localized to perivascular cells. They also show a significant correlation between CD44 expression levels and survival in patients with proneural GBM. These findings suggest that even a minor population of cells (such as CD44+ cells in proneural GBM) can potentially confer an aggressive behavior, highlighting the importance of resolving the cellular composition of residual tumor tissue.
In further support of these subtype specific differences in lineage relationships of the transformed cell populations, the deconvolution showed that EGFR, which is frequently amplified in the classical subtype of GBM, is predominantly expressed by CD44+ astrocytes, whereas PDGFRa, which is amplified or overexpressed in proneural GBM, is predominantly expressed by OPC-like cells (SI Appendix, Tables S3 and S4). These two populations are likely to have different susceptibilities to chemotherapy and radiation, and what is therapeutically effective for one population may be relatively ineffective for the other. Consistent with this idea, the deconvolution analysis also revealed a marked loss of the OLIG2+ population in GBM samples that have recurred after treatment (Fig. 4). Previous studies have shown that OPCs are highly sensitive to chemotherapy and radiation (24–26). OLIG2+ OPC-like glioma cells are more sensitive to radiation than CD44+ astrocytic tumor cells (13). It is also possible the OPC-like glioma cells undergo mesenchymal transformation to CD44+ glioma cells. In either case, such a shift in cellular composition might explain the previously reported tendency for recurrent tumors to acquire a mesenchymal phenotype (20, 27). Future studies with greater numbers of primary and recurrent tumor specimens will be needed to address this possibility.
The computational approach used in this study could be used to characterize the cell type-specific expression patterns of any sample with a heterogeneous cellular composition. However, the samples taken from the margins of glioma, which contain a variable mixture of infiltrating glioma cells and nonneoplastic brain cells, provide a particularly robust and clinically relevant dataset for this type of analysis. The deconvolution of cell type-specific expression profiles offers a means to disentangle the molecular signature of neoplastic cells, which can be a minority of the total cell population, from the nonneoplastic populations, which may have significant molecular alterations of their own. In future applications, this approach can be used to refine the analyses of intracellular signaling pathways and transcription networks, which presume expression in the same cell. Conversely, identifying signaling molecules (such as ligand-receptor partners) that are distributed in different cell types may give new insights into paracrine signaling interactions. This type of analysis can be used to identify and access new therapeutic targets that may be expressed by infiltrating glioma cells or by nonneoplastic/reactive cells that also play a role in disease progression. Finally, understanding common patterns that relate the center of the tumor, which is commonly resected during surgery, to the infiltrative margins, which are often left behind, will greatly facilitate the design of clinical trials that target residual disease.
Materials and Methods
This study included 69 adult patients presenting for open surgical resection of glioma. MRI-localized biopsies measuring ∼1.0 cm × 0.5 cm × 0.5 cm were obtained before surgical debulking. Sampled regions included areas within the gadolinium enhancing core of the tumors (CE) and areas of nonenhancing, FLAIR hyperintense tissue at the margins of the tumors (NE). Radiographic localization was confirmed by using intraoperative stereotaxis with T1+Gadolinium and FLAIR sequences using the Brainlab Neuronavigation interface (Brainlab). Samples from each region were divided into two pieces in the operating suite: One piece was immediately flash frozen in liquid nitrogen; the other piece was fixed in 10% (vol/vol) formalin and used for histological and immunohistochemical analysis. A total of 17 nonneoplastic brain tissue samples were collected from 11 patients, with no oncological history, who were undergoing ventriculoperitoneal shunt placement for normal pressure hydrocephalus or surgical resection for seizure control. A small biopsy was obtained at the cortical entry point before passing the ventricular catheter. These samples are referred to as nonneoplastic/normal brain (NB). A detailed description of the methods used for immunohistochemistry, microscopy, RNA extraction, sequencing, computational deconvolution of RNA-seq data, gene ontology, and differential expression analysis are available in SI Appendix, SI Materials and Methods. All RNA-seq data is available on the Gene Expression Omnibus (accession no. GSE59612).
Supplementary Material
Acknowledgments
P.A.S. is supported by National Institutes of Health (NIH)/National Institute of Biomedical Imaging and Bioengineering Grant 1K01EB016071. P.C. is supported by NIH/National Institute of Neurological Disorders and Stroke Grant 1R01NS066955. P.A.S. and P.C. are part of the Brain Tumor Ecology Collaborative supported by the James S. McDonnell Foundation. The project was also supported by the Collaborative and Multidisciplinary Pilot Research Award from the Irving Institute for Clinical and Translational Research, Columbia University Medical Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE59612).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405839111/-/DCSupplemental.
References
- 1.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonabend AM, et al. Murine cell line model of proneural glioma for evaluation of anti-tumor therapies. J Neurooncol. 2013;112(3):375–382. doi: 10.1007/s11060-013-1082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen-Orr SS, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods. 2010;7(4):287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong T, et al. Optimal deconvolution of transcriptional profiling data using quadratic programming with application to complex clinical blood samples. PLoS ONE. 2011;6(11):e27156. doi: 10.1371/journal.pone.0027156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn A, Thu D, Waldvogel HJ, Faull RLM, Luthi-Carter R. Population-specific expression analysis (PSEA) reveals molecular changes in diseased brain. Nat Methods. 2011;8(11):945–947. doi: 10.1038/nmeth.1710. [DOI] [PubMed] [Google Scholar]
- 8.Sims JS, et al. Patterns of gene-specific and total transcriptional activity during the Plasmodium falciparum intraerythrocytic developmental cycle. Eukaryot Cell. 2009;8(3):327–338. doi: 10.1128/EC.00340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sosunov AA, et al. Phenotypic heterogeneity and plasticity of isocortical and hippocampal astrocytes in the human brain. J Neurosci. 2014;34(6):2285–2298. doi: 10.1523/JNEUROSCI.4037-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz AM, et al. Astrocyte-specific expression patterns associated with the PDGF-induced glioma microenvironment. PLoS ONE. 2012;7(2):e32453. doi: 10.1371/journal.pone.0032453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci. 2012;32(18):6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motomura K, et al. Immunohistochemical analysis-based proteomic subclassification of newly diagnosed glioblastomas. Cancer Sci. 2012;103(10):1871–1879. doi: 10.1111/j.1349-7006.2012.02377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhat KP, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu IM, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Reports. 2013;4(2):385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei L, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS ONE. 2011;6(5):e20041. doi: 10.1371/journal.pone.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonabend AM, et al. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 2014;74(5):1440–1451. doi: 10.1158/0008-5472.CAN-13-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engler JR, et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PLoS ONE. 2012;7(8):e43339. doi: 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piao Y, et al. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19(16):4392–4403. doi: 10.1158/1078-0432.CCR-12-1557. [DOI] [PubMed] [Google Scholar]
- 21.Favaro R, et al. Sox2 is required to maintain cancer stem cells in a mouse model of high-grade oligodendroglioma. Cancer Res. 2014;74(6):1833–1844. doi: 10.1158/0008-5472.CAN-13-1942. [DOI] [PubMed] [Google Scholar]
- 22.Berezovsky AD, et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16(3):193–206. doi: 10.1016/j.neo.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pietras A, et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukuda A, et al. Age-dependent sensitivity of the developing brain to irradiation is correlated with the number and vulnerability of progenitor cells. J Neurochem. 2005;92(3):569–584. doi: 10.1111/j.1471-4159.2004.02894.x. [DOI] [PubMed] [Google Scholar]
- 25.Dietrich J, Han R, Yang Y, Mayer-Pröschel M, Noble M. CNS progenitor cells and oligodendrocytes are targets of chemotherapeutic agents in vitro and in vivo. J Biol. 2006;5(7):22. doi: 10.1186/jbiol50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez KA, et al. Convection-enhanced delivery of topotecan into a PDGF-driven model of glioblastoma prolongs survival and ablates both tumor-initiating cells and recruited glial progenitors. Cancer Res. 2011;71(11):3963–3971. doi: 10.1158/0008-5472.CAN-10-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.