Significance
Enzymes that use the same active site to catalyze two native, sequential reactions are extraordinary. Structural studies of phosphohexose mutases are particularly informative, permitting direct comparison of the organization of catalysis of phosphoryl transfer involving two different substrates. The present study of β-phosphoglucomutase (βPGM) deploys chemical synthesis of substrate analogs to enable detailed NMR and X-ray structural analysis of both steps of its catalytic activity. It reveals how βPGM conserves fidelity of transition state organization while maintaining substrate recognition for its two steps by prioritizing positioning of both phosphates over direct hexose recognition for the second step. It identifies the structural basis for the strong discrimination by βPGM between two, diastereoisomeric α-fluoromethylenephosphonate analogs of β-d-glucose 1-phosphate.
Keywords: phosphonate analogs, phosphoryl transfer mechanism, 19F NMR, X-ray crystallography, water-mediated substrate recognition
Abstract
β-Phosphoglucomutase (βPGM) catalyzes isomerization of β-d-glucose 1-phosphate (βG1P) into d-glucose 6-phosphate (G6P) via sequential phosphoryl transfer steps using a β-d-glucose 1,6-bisphosphate (βG16BP) intermediate. Synthetic fluoromethylenephosphonate and methylenephosphonate analogs of βG1P deliver novel step 1 transition state analog (TSA) complexes for βPGM, incorporating trifluoromagnesate and tetrafluoroaluminate surrogates of the phosphoryl group. Within an invariant protein conformation, the β-d-glucopyranose ring in the βG1P TSA complexes (step 1) is flipped over and shifted relative to the G6P TSA complexes (step 2). Its equatorial hydroxyl groups are hydrogen-bonded directly to the enzyme rather than indirectly via water molecules as in step 2. The (C)O–P bond orientation for binding the phosphate in the inert phosphate site differs by ∼30° between steps 1 and 2. By contrast, the orientations for the axial O–Mg–O alignment for the TSA of the phosphoryl group in the catalytic site differ by only ∼5°, and the atoms representing the five phosphorus-bonded oxygens in the two transition states (TSs) are virtually superimposable. The conformation of βG16BP in step 1 does not fit into the same invariant active site for step 2 by simple positional interchange of the phosphates: the TS alignment is achieved by conformational change of the hexose rather than the protein.
Efficient enzyme catalysis of the manipulation of phosphates is one of the great achievements of evolution (1). Enzymes that operate on phosphate monoesters and anhydrides transfer the phosphoryl moiety, PO3−, with rate accelerations approaching 1021 for monoesters, placing them among the most proficient of all enzymes (1). Phosphomutases, including α-phosphoglucomutase (αPGM) (2, 3) and β-phosphoglucomutase (βPGM) (4–6), phosphoglycerate mutase (7), α-phosphomannomutase (αPMM/PGM) (8), and N-acetylglucosamine-phosphate mutase (9), merit special attention because these enzymes have to be effective in donating a phosphoryl group to either of two hydroxyl groups that have intrinsically different reactivity. Only when both half-reactions of a phosphomutase are accessible to mechanistic analysis can the problem of how an enzyme accommodates two distinct chemistries within a single active site be resolved. Hexose 1-phosphate mutases, including enzymes central to glycolysis and other metabolic pathways, are well characterized (10, 11). They are generally activated by phosphorylation to form a covalent phosphoenzyme, which then donates its PO3− group to either of its substrates to deliver a common, transient, hexose 1,6-bisphosphate intermediate species. However, structural studies on phosphomutases are complicated by the rapid and often imbalanced equilibrium position between the substrates, and kinetic studies are problematic because of competitive, parallel pathways of enzyme activation and substrate inhibition (12, 13). As a result, transition states (TSs) for both half-reactions have not hitherto been accessible for mechanistic analysis.
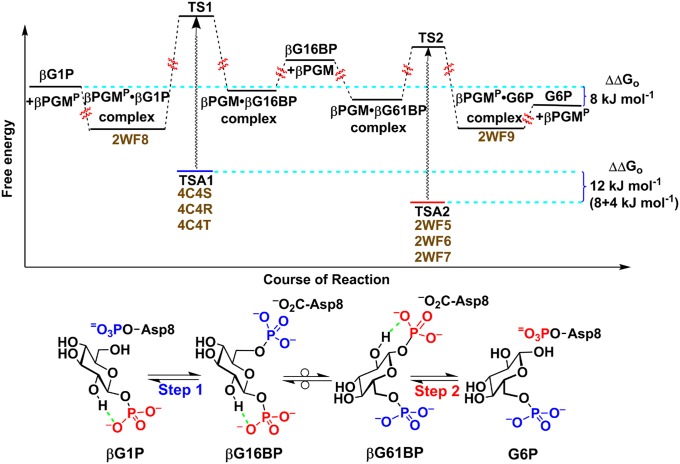
βPGM is the best-characterized hexose 1-phosphate mutase and is a member of the haloacid dehalogenase (HAD) superfamily (14), which has 58 HAD homologs in Homo sapiens (11). The key cellular role for βPGM is to support growth on maltose (14), which demands isomerization of β-d-glucose 1-phosphate (βG1P) via β-d-glucose 1,6-bisphosphate (βG16BP) into d-glucose 6-phosphate (G6P), a universal source of cellular energy. This interconversion is achieved via a transient, covalent phosphoenzyme intermediate involving an essential aspartic acid, Asp8, to conserve the phosphoryl group that migrates intermolecularly (Fig. 1). Mechanistically, this pathway demands the architecture of the catalytic site to be effective in promoting phosphoryl transfer from phospho-Asp8 to the 6-OH group of βG1P (step 1), followed by reverse phosphoryl transfer from 1β-OH of βG16BP to Asp8 (step 2).
Fig. 1.
Reaction scheme and free energy profile for the conversion of βG1P into G6P via βG16BP catalyzed by βPGM. The phosphoryl transfer reaction between βG1P and the phosphoenzyme (βPGMP) is step 1 (transferring phosphate is shown in blue), and the equivalent reaction between G6P and the phosphoenzyme is step 2 (transferring phosphate is shown in red). The two intermediate complexes are labeled βG16BP and βG61BP to indicate the two orientations of bound β-bisphosphoglucose. Intramolecular hydrogen bonds within the glucose phosphates are indicated in green. The PDB ID codes (shown in brown) for the structures of metal fluoride ground state analog (GSA) and TSA complexes are listed next to the corresponding steps. G6P is ca. 8 kJ⋅mol−1 lower in free energy than βG1P at equilibrium (12). βG1P binds fivefold less tightly than G6P in an AlF4− TSA complex, corresponding to a binding energy difference of ca. 4 kJ⋅mol−1. This places the TSA for step 1 (blue) ca. 12 kJ⋅mol−1 (4 kJ⋅mol−1 + 8 kJ⋅mol−1) higher in free energy than the TSA for step 2 (red). The free energy levels of TS1 and TS2 are placed only approximately, using the assumption that the free energy difference (wavy arrows) between the TSA complex and the true TS is similar for both step 1 and step 2. The approximate relative free energy levels for the intermediate enzyme-bound states denoted with βG16BP and βG61BP are based on published data (13).
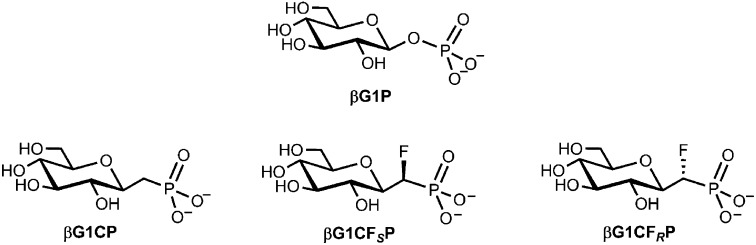
Step 2 has been studied intensively, with analyses focused on structural studies of trifluoromagnesate (MgF3−) and tetrafluoroaluminate (AlF4−) transition state analogs (TSAs) and trifluoroberyllate ground state analogs for G6P complexes (4–6, 15). 19F NMR resonances for these complexes additionally have provided in situ probes for the electronic and protonic environment of the phosphate moiety in the active site (4–6, 15, 16). Such studies have confirmed a trigonal bipyramidal (tbp) TS associated with inline stereochemistry and general acid–base catalysis, following the rearrangement of near-attack conformers (6). By contrast, step 1, involving phosphorylation of the 6-OH group of βG1P, is not well understood. The corresponding TSA complexes hitherto have proved inaccessible; attempted crystallization of the mutase using βG1P with magnesium and fluoride provides the same MgF3− TSA complex as is formed directly with G6P because residual enzyme activity catalyzes mutation of βG1P into G6P at a rate competitive with crystallization of the complex (17). Similarly, although 19F NMR studies have identified a transient TSA complex for an AlF4− complex of βG1P, it readily isomerizes into the corresponding TSA complex of G6P (SI Appendix, Fig. S1). This impasse is resolved here by the synthesis and use of stable analogs of βG1P that resist mutase-catalyzed isomerization. Because it has been established that α-fluorination of 6-phosphonomethyl-6-deoxy-glucose (G6CP) can enhance or impair analog binding to glucose 6-phosphate dehydrogenase, depending on the stereochemistry of the α-fluorine substituent (18), we have synthesized both diastereoisomeric α-monofluoromethylenephosphonate analogs of βG1P, its methylenephosphonate analog, and the three corresponding phosphonate 1α-hydroxyl analogs. We have identified the two best-binding analogs by 19F NMR and measured their affinities with βPGM in TSA complexes using fluorescence titration. We have thereby obtained three high-resolution crystal structures of TSA complexes for step 1 of the mutase catalytic reaction. Their comparison with TSA complexes for step 2 establishes the substantially different binding modes for βG1P and G6P in their respective reactions.
Results
Synthesis of Stable Analogs of βG1P.
The stable analogs of βG1P, with the bridging oxygen replaced with a methylene or chirally defined fluoromethylene group, are shown (Fig. 2). Their syntheses used chirally defined precursor benzyl ethers having an additional hydroxyl group in the 1α-position (19). This hydroxyl was removed by Barton–McCombie deoxygenation (20–22), followed by hydrogenolysis to remove benzyl protecting groups to give three analogs of βG1P: the ammonium salts of 1-β-phosphonomethylene-1-deoxy-d-glucopyranose (βG1CP) (19), (S)-1-β-phosphonofluoromethylene-1-deoxy-d-glucopyranose (βG1CFSP), and (R)-1-β-phosphonofluoromethylene-1-deoxy-d-glucopyranose (βG1CFRP) (Fig. 2), as described in SI Appendix. The three benzyl intermediates were also hydrogenolyzed to give three further corresponding analogs of βG1P, with each having the additional hydroxyl group in the 1α-position (SI Appendix, Fig. S2).
Fig. 2.
Dianion structures of βG1P and its nonhydrolyzable analogs: βG1CP, βG1CFSP, and βG1CFRP.
Structural Analysis of TSA Complexes for Steps 1 and 2.
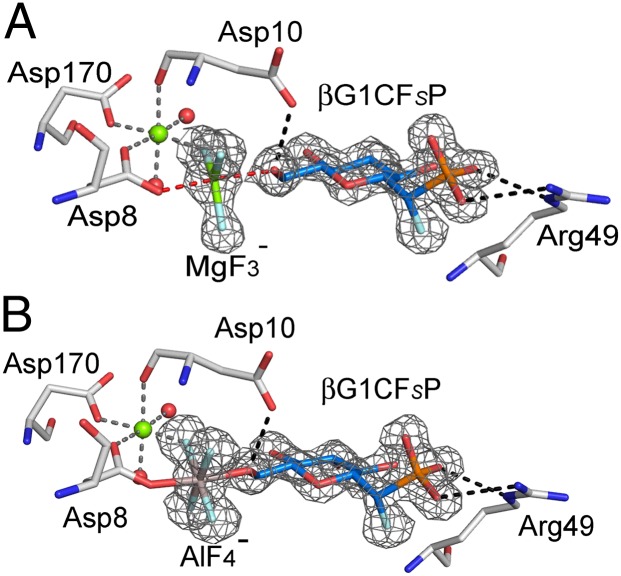
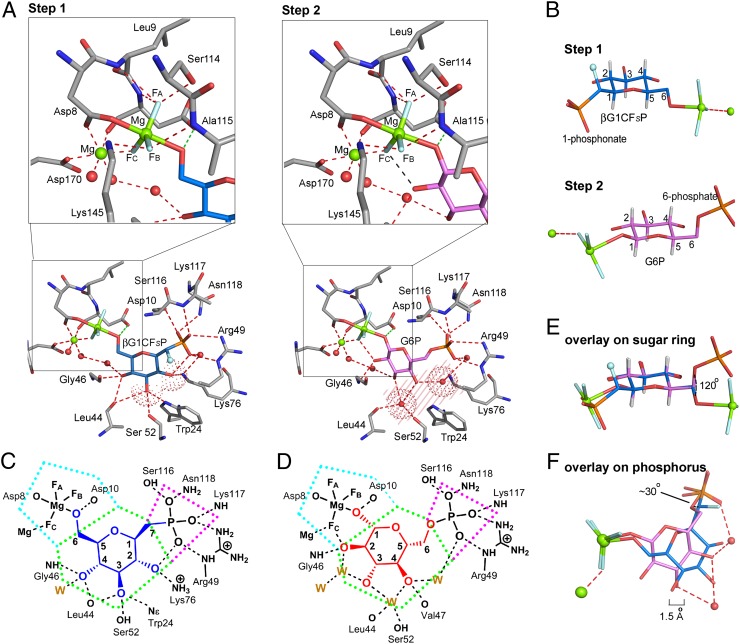
According to 19F NMR measurements, only analogs βG1CFSP and βG1CP formed metal fluoride TSA complexes, and these were screened in crystallization trials. Crystals of TSA complexes for step 1 were obtained in three cases, βPGM-MgF3−-βG1CFSP [Protein Data Bank (PDB) ID code 4C4S], βPGM-AlF4−-βG1CFSP (PDB ID code 4C4T), and βPGM-MgF3−-βG1CP (PDB ID code 4C4R), and their structures were solved at resolutions between 1.1 Å and 1.5 Å (SI Appendix, Fig. S3 A–C and Table S1). The structure of the βPGM-MgF3−-βG1CFSP TSA complex confirms the (S)-stereochemistry for the C7-fluorine (Fig. 3A). Its tbp MgF3− core mimics the TS phosphoryl group with equatorial Mg–F bonds of 1.8 Å (4), axial ligands to glucose-O6 (2.2 Å) and Asp8 Oδ (2.0 Å), and an Oax–Mg–Oax angle of 176°. The three equatorial fluorine atoms are coordinated: FA to Leu9 NH (3.0 Å), Asp10 NH (2.8 Å), and Ser114 OHδ (2.6 Å); FB to Ala115 NH (2.9 Å) and Lys145 NHζ (2.7 Å); and FC to the catalytic magnesium (2.0 Å) (SI Appendix, Fig. S3E). The catalytic magnesium is octahedral, liganded by FC (2.0 Å), by Asp8 Oδ (2.1 Å), by the backbone carbonyl of Asp10 (2.1 Å), by Asp170 Oδ (2.1 Å), and by two water molecules (Fig. 4A, Left), whereas the nucleophilic glucose-O6 is hydrogen-bonded to the carboxylate group of the general base catalyst, Asp10 (2.6 Å). The phosphonate moiety in the inert phosphate binding site coordinates to Arg49 NHε (2.9 Å) and NHη (3.1 Å), Ser116 OHδ (2.6 Å), Lys117 NH (3.0 Å), and Asn118 NHδ (3.1 Å), forming five intermolecular hydrogen bonds and having a sixth intramolecular hydrogen bond to the 2-OH group (2.8 Å). The C7-fluorine does not make any hydrogen bond and is 2.8 Å from C2 and 3.0 Å from O5, thereby reducing the eclipsed interaction through C1–C7 bond rotation (SI Appendix, Fig. S4B). The glucopyranose ring has an undistorted 4C1 chair form (Fig. 4B). Its three equatorial hydroxyls form six hydrogen bonds directly to amino acid residues: 2-OH to Lys76 Nε (3.0 Å) and a phosphonate oxygen (2.8 Å); 3-OH to Trp24 Nε (3.0 Å), Leu44 CO (3.4 Å), and Ser52 OHδ (3.0 Å); and 4-OH to Leu44 CO (2.8 Å), Gly46 NH (3.2 Å), and a water molecule (2.8 Å) (Fig. 4A). The βPGM-AlF4−-βG1CFSP TSA complex aligns very closely with the βPGM-MgF3−-βG1CFSP TSA complex (average pairwise rmsd for Cα atoms is 0.075 Å) (Fig. 3B). In the βPGM-MgF3−-βG1CP TSA complex, the C7 methylene group is shifted 0.3 Å relative to its position in βG1CFSP (SI Appendix, Fig. S4A), and it aligns well with the βPGM-MgF3−-βG1CFSP TSA complex in all other respects (average pairwise rmsd for Cα atoms is 0.086 Å). For both complexes, C7 is in a staggered conformation with the phosphonate oxygen atoms (SI Appendix, Fig. S4C), whereas the P–C7 bond eclipses H1 on the glucopyranose ring (dihedral angle of −10° for βG1CP; SI Appendix, Fig. S4D). There are no hydrogen bonds to O5, which is in van der Waals contact with Ala115 and Ser116, which flank the glucose ring on one edge. The hexose ring α-face is capped by the imidazole ring of His20, whereas its β-face is capped primarily by five amino acid residues, located in the Leu44-to-Ser52 loop between helices 2 and 3 in the cap domain (14) (SI Appendix, Fig. S5A).
Fig. 3.
Difference density (Fo − Fc) for βG1CFSP in the MgF3− (A) and AlF4− (B) TSA complexes. The difference electron density is shown as a gray mesh contoured at 4σ. The octahedrally coordinated catalytic magnesium is shown in green, fluorine is shown in pale blue, and waters are shown as red spheres. In A, the tbp magnesium is shown in green, whereas in B, the octahedral aluminum is shown in gray. The carbon atoms of the sugar are shown in midblue.
Fig. 4.
Comparison of the active site detail for step 1 and step 2 TSA complexes. (A, Left) For step 1, the coordination of βG1CFSP (midblue) in the βPGM-MgF3−-βG1CFSP TSA complex shows direct interaction between the 2-OH and 3-OH groups (red dotted surfaces) and protein residues (gray). The α-fluorine is shown as a pale blue sphere. (A, Right) For step 2, the coordination of G6P (magenta) in the βPGM-MgF3−-G6P TSA complex shows a space between the substrate and protein (pink shaded area), which is occupied by two conserved waters (red dotted surfaces). (Upper) Magnified views highlight the coordination of the MgF3− moieties. The identities of the fluorines are those assigned using 19F NMR. (B) Hexose rings of βG1CFSP (midblue, for step 1) and G6P (magenta, for step 2) from their respective βPGM-MgF3− TSA complexes show indistinguishable 4C1 chair conformations. All sugar carbon atoms are numbered. Substrate coordination for βG1CP (C, shown in blue) and G6P (D, shown in red) in the βPGM-MgF3− TSA complexes. The inert phosphate site is outlined in magenta, the hexose ring site is outlined in green, and the phosphoryl transfer site is outlined in cyan. Hydrogen bonds (black dashes) to the protein and to water molecules (W) are shown. (E) Overlay of the different conformations of the sugar substrates illustrating the ∼120° rotation of the C5–C6 exocyclic bond between step 1 and step 2. (F) Overlay of the different conformations of the sugar substrates illustrating the extent to which the sugar ring moves closer to the phosphoryl transfer site in step 2 (magenta) compared with step 1 (midblue). The 1.5-Å scale bar is aligned with the center of the sugar rings. The ∼30° angular difference between the orientation of the (C)O–P inert bonds for steps 1 and 2 is marked. In B, E, and F, magnesiums are shown as green spheres, fluorines are shown in pale blue, phosphorus is shown in orange, hydrogens are shown in white, and waters are shown as red spheres.
To monitor the substitution of phosphonate for phosphate in step 2, we determined the βPGM-AlF4−-G6CP TSA complex structure for the phosphonate analog (4) of G6P (PDB ID code 2WF7) at a resolution of 1.05 Å (SI Appendix, Fig. S3D and Table S1). The average pairwise rmsd for Cα atoms is 0.093 Å, compared with the βPGM-AlF4−-G6P TSA complex (PDB ID code 2WF6). The 6-phosphonate is bound in the inert phosphate site and is superimposable on the 6-phosphate. It has the same rotamer conformation as seen (4, 5) in the βPGM-MgF3−-G6P (PDB ID code 2WF5) and βPGM-AlF4−-G6P (PDB ID code 2WF6) TSA complexes, with two conserved water molecules bridging 3-OH and 4-OH, and 4-OH and the 6-phosphonate oxygen to the protein (SI Appendix, Fig. S6). All of these step 2 complexes show G6P (or G6CP) having a conformation with the C5–C6 bond rotated, making O6 (or C7) anti to H5 (−176 ± 2° dihedral angle). By contrast, the step 1 complexes with the βG1P analogs βG1CFSP and βG1CP bind with the C5–C6 bond staggered with O6 anti to C4 (177 ± 1°) (Fig. 4B).
Polarization of the Fluorine Coordinating the Catalytic Magnesium in TSA Complexes.
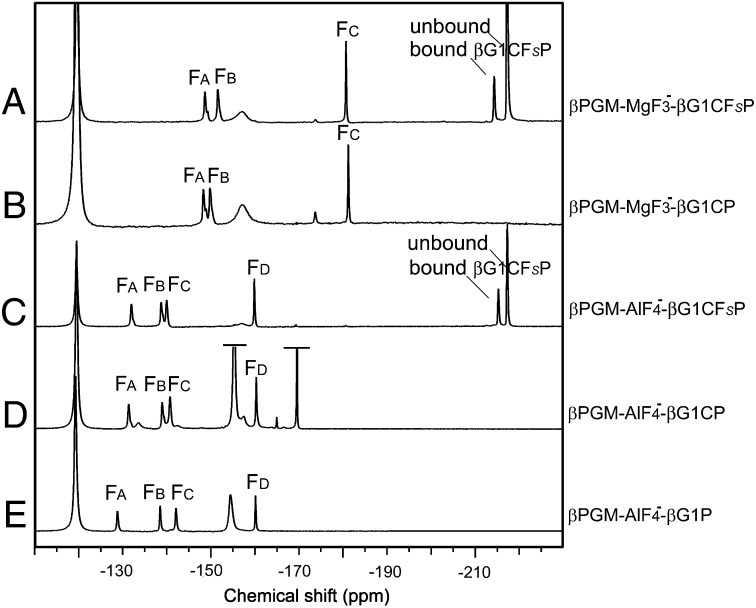
The 19F NMR spectra corresponding to the βG1CFSP and βG1CP TSA complexes with MgF3− and with AlF4− ligands are shown in Fig. 5. 19F resonances were assigned by chemical shift and solvent-induced isotope shift (SIIS), and these data provide in situ probes of the electronic and protonic environment in the chemical transfer step (4, 5, 15) (SI Appendix, Figs. S3 E and F and S7 and Table S2). Although a direct comparison of the MgF3− TSA complex of βG1CFSP or βG1CP with that of βG1P is not experimentally possible, the four 19F NMR resonances in the AlF4− complexes with βG1CFSP and βG1CP closely match those for the corresponding βG1P complex in solution (Fig. 5 C–E and SI Appendix, Table S2). This shows that both βG1CFSP and βG1CP are good mimics of the natural substrate βG1P. The α-fluorine resonance of βG1CFSP (−214 ppm) has the same peak integral as those of the three MgF3− and four AlF4− resonances, establishing that βG1CFSP is bound to βPGM exclusively as a TSA complex.
Fig. 5.
19F NMR spectra of βPGM TSA complexes for step 1. βPGM-MgF3−-βG1CFSP TSA complex (A) and βPGM-MgF3−-βG1CP TSA complex (B), recorded for 1 mM βPGM, 5 mM MgCl2, 10 mM NH4F, and 5 mM βG1CFSP or βG1CP at pH 7.2. (C) βPGM-AlF4−-βG1CFSP TSA complex after addition of 1 mM AlCl3 to A. (D) βPGM-AlF4−-βG1CP TSA complex after addition of 1 mM AlCl3 to B. (E) βPGM-AlF4−-βG1P TSA complex under conditions equivalent to those in C and D. Free and bound βG1CFSP in A and C show peaks at −217 ppm and −214 ppm, respectively. Resonances at −119, −155, and −170 ppm are from free F−, free MgF+ plus AlFx (truncated for clarity in D), and unbound sugar-AlFx species (truncated for clarity in D), respectively.
For the MgF3− TSA complexes of βG1CFSP and βG1CP, the chemical shifts of FA (−148 ppm) and FB (−150 ppm) are close to those for G6P and G6CP complexes (SI Appendix, Table S2). However, FC (coordinated to the catalytic magnesium) has a resonance 22 ppm up-field compared with FC in the complexes for step 2. Likewise, the resonance for FD (coordinated to the catalytic magnesium) in the AlF4− complexes (5) for βG1CFSP and βG1CP is shifted up-field by 16 ppm. This significant up-field shift indicates that the electron density associated with this fluoride is substantially increased in the step 1 TSA complexes (4). The SIIS values (SI Appendix, Fig. S7) are near zero for this same fluoride in the step 1 TSA complexes and are substantially smaller than those for the step 2 TSA complexes (SI Appendix, Table S2). They identify greatly reduced hydrogen bonding from any donor to the equivalent oxygen in step 1. This has a clear structural basis: in the MgF3− TSA complex with G6P for step 2, FC is directly hydrogen-bonded to the 2-OH group at 2.7 Å (Fig. 4A, Right). No such hydrogen bond donor occurs in step 1, where it is obviated by the 6-CH2 group in the TS for phosphorylation of the 6-OH group and places the nearest water 4.0 Å from FC (Fig. 4A, Left). In addition, 19F NMR analysis established that the three 1α-hydroxy-1β-methylenephosphonates do not form metal fluoride TSA complexes with βPGM (compounds 4a, 5a, and 6a in SI Appendix, Fig. S2). By modeling, this can be attributed to the steric opposition of the 1α-OH group functionality with His20 (SI Appendix, Fig. S8 A and B).
Binding Affinity in Metal Fluoride TSA Complexes.
Apparent dissociation constants of the complexes were measured from fluorescence changes on TSA formation (SI Appendix, Fig. S9 A and B and Table S3). For MgF3− TSA complexes, the apparent Kd for the (S)-isomer βG1CFSP is 0.66 ± 0.07 mM, whereas the binding of the (R)-isomer βG1CFRP is too weak for determination (Kd > 50 mM). The Kd for βG1CP is 1.30 ± 0.09 mM, which is 4.5-fold weaker than that of G6CP [Kd = 0.3 ± 0.1 mM (4)], and equates to a difference in binding energy of ∼4 kJ⋅mol−1. G6P binds in the MgF3− TSA complex 300-fold more strongly than G6CP. Although the measured apparent Kd values are relatively high, the very low formation constant for MgF3− in water (estimated at less than 1 μM) would mean that the true affinity of βPGM for the TSA complexes with these analogs is likely to be in the nanomolar range (12, 23, 24).
It was not possible to determine the Kd for βG1P in the MgF3− TSA complex because enzymatic isomerization gives a mixture of hexose phosphates. However, the Kd values for the step 1 ligands were obtained for the AlF4− TSA complexes: βG1P (Kd = 46 ± 4 μM) binds 26-fold stronger than the phosphonate analog βG1CP (Kd = 1.2 ± 0.1 mM). This behavior compares favorably with the performance of α-d-glucose 1-phosphate (αG1P; pKa2 = 6.15), which binds to αPGM 800-fold more strongly than the methylenephosphonate analog of αG1P (25). Furthermore, βG1P binds fivefold less tightly than G6P (Kd = 9.0 ± 1.0 μM), corresponding to a binding energy difference of ∼4 kJ⋅mol−1. This fivefold difference in the Kd was confirmed by a direct 19F NMR competition experiment between βG1P and G6P in the same sample at saturating conditions. It showed that a 1:1 ratio of βG1P-AlF4− to G6P-AlF4− TSA complexes with βPGM (1 mM) for step 1 and step 2 is achieved with a fivefold higher concentration of βG1P (25 mM) than of G6P (5 mM) at pH 7.2 (SI Appendix, Fig. S9 C–E). Because G6P is ca. 8 kJ⋅mol−1 lower in free energy than βG1P at equilibrium (12) (Fig. 1), this places the TSA for step 1 at ca.12 kJ⋅mol−1 (8 kJ⋅mol−1 + 4 kJ⋅mol−1) higher in free energy than the TSA for step 2. It predicts that the free energy of TS1 is higher than that of TS2 if the free energy difference between the AlF4− TSA complex and the true TS is similar for both step 1 and step 2 (Fig. 1).
βPGM Has Very High α-Fluorophosphonate Epimer Discrimination.
The greater than 100-fold epimer discrimination in favor of the (S)-α-fluoromethylenephosphonate analog βG1CFSP over the (R)-isomer βG1CFRP is much larger than that seen for glucose 6-phosphate dehydrogenase, where the α-fluorophosphonate analogs of G6P showed only a 10-fold kcat/Km advantage for the (R)-isomer over the (S)-isomer (18), whereas with thymidylyltransferase Cps2L, there is only a fivefold greater activity for the (S)-fluoromethylenephosphonate substrate isomer over the (R)-fluoromethylenephosphonate substrate isomer (19). This discrimination has a clear structural basis. Modeling an (R)-fluorine onto C7 in the βPGM-MgF3−-βG1CP TSA complex structure shows strong eclipsing between C7–F and C1–O5 bonds (−9°), with the fluorine 2.4 Å from O5, 2.6 Å from the carbonyl oxygen of Ala115, and 1.9 Å from the α-hydrogen of Ser116 (SI Appendix, Fig. S8C). The (R)-isomer βG1CFRP is thus likely to be strongly disfavored by a combination of adverse steric and dipolar interactions. The increase in affinity resulting from α-fluorination in βG1CFSP vs. βG1CP can be attributed to their relative pKa values (18, 26): βG1CFSP is fully dianionic in solution at pH 7.2 (pKa2 = 5.43), whereas βG1CP is 35% monoanionic (pKa2 = 6.94).
Discussion
The rational design and chemical synthesis of novel phosphonate analogs of βG1P has delivered probes for structural and mechanistic analysis of exclusively the first phosphoryl transfer step of βPGM. It has thereby enabled direct comparison of organization between the two consecutive TSs of a phosphomutase. This establishes that βPGM accomplishes step 1 and step 2 of its reaction within an unchanged, closed protein conformation (SI Appendix, Fig. S5 C and D). The primary differences between the TSA complexes for steps 1 and 2 lie in substrate accommodation. In comparing these TSAs, it is convenient to apportion the substrate cavity into three zones: the inert phosphate site, the hexose ring site, and the phosphoryl transfer site (Fig. 4 C and D). These will be discussed serially.
The amino acids that coordinate the inert phosphate have virtually identical pairing interactions for both steps of the mutase reaction in all six of the TSA complex structures determined to date (PDB ID codes 4C4R, 4C4S, 4C4T, 2WF5, 2WF6, and 2WF7). This shows that the enzyme coordinates the negatively charged phosphoryl group in essentially the same way for both steps (Fig. 4 C and D), although it tolerates an angular difference between the orientation of the (C)O–P inert bonds for steps 1 and 2 of ∼30° (Fig. 4F).
By contrast, recognition in the hexose ring site differs markedly between step 1 and step 2 (Fig. 4A). In the step 1 TSA complexes, the α-face of the glucopyranose ring is adjacent to the imidazole ring of His20 (SI Appendix, Fig. S5A). In the step 2 TSA complexes, the glucopyranose ring has the same regular 4C1 chair form but is rotated to place its β-face adjacent to the imidazole ring of His20 (SI Appendix, Fig. S5B). This relocation to accommodate the positional interchange of the two phosphates results in the glucopyranose ring atoms being shifted ∼1.5 Å closer to the phosphoryl transfer site (Fig. 4F). However, to fit both inert and transferring phosphate groups into their respective sites without changing the protein conformation, the C5–C6 exocyclic bond is rotated ∼120° between two different staggered conformations in the step 1 and step 2 TSA complexes (Fig. 4E).
These differences in the two binding modes of the hexose ring impose a very different coordination of βPGM onto the three equatorial hydroxyl groups (2-OH, 3-OH, and 4-OH). In step 1, specific ligation to these three equatorial hydroxyl groups in the hexose ring site is made directly from five amino acids through six hydrogen bonds (Fig. 4A, Left), while accommodating inline presentation of O6 to accept the phosphate from phospho-Asp8 in the phosphoryl transfer site. By contrast, inline location of the 1-phosphate in the phosphoryl transfer site in step 2, combined with tight coordination of the 6-phosphate in the inert phosphate site and the invariant protein conformation, results in the 3-OH and 4-OH groups being displaced away from their hydrogen bonding partners (Fig. 4A, Right). The resulting space is occupied by two water molecules not present in step 1, and they mediate indirect hydrogen bonding between substrate and enzyme in the hexose ring site, leaving only one direct hydrogen bond to Gly46.
For both steps 1 and 2, βPGM locates the transferring tbp phosphoryl group in the phosphoryl transfer site by the same five hydrogen bond donors to coordinate two of the equatorial oxygen atoms, whereas the third equatorial oxygen is coordinated by the catalytic magnesium. In each case, Asp10 is positioned to provide general base catalysis by short hydrogen bonding to the nucleophile (Fig. 4A), as is generally observed for nucleophilic hydroxyl groups (27). Also, the nucleophilic carboxylate of Asp8, chelated by the catalytic magnesium, has an identical orientation in both steps. Both TSAs have the same inline geometry (both Oax–Mg–Oax bond angles are 176°) and short Oax–Mg–Oax distances (4.1 Å and 4.3 Å). The angle between Asp8 Oδ and the hexose nucleophilic oxygens for these two steps is close to zero (<5°). The two step 1 and step 2 TSA structures correspond to Oax–P–Oax distances that require the nonbridging, equatorial oxygens of the substrate and product phosphate monoesters to be virtually coincident within the catalytic site. The migrating phosphorus atom is thus located within a trigonal bipyramid whose apices are the five oxygens, which means that the phosphorus atom simply migrates ∼1.0 Å along the major axis to effect phosphoryl group transfer (SI Appendix, Fig. S10).
The behavior of βPGM shows significant differences from that of αPGM, which is not a HAD superfamily enzyme like βPGM but is a member of the structurally unrelated α-phosphohexomutase superfamily. αPGM catalyzes isomerization of the αG1P into G6P involving α-d-glucose 1,6-bisphosphate (αG16BP) as an intermediate. Although both mutases use a single magnesium ion as the preferred catalytic metal (28, 29), they use fundamentally different chemistry: αPGM uses a phosphoserine (30) as an enzyme intermediate, whereas βPGM uses a phosphoaspartate. Classical kinetic studies on rabbit αPGM, based on the binding contributions of various components of the glucose phosphate moiety to the enzyme, were analyzed in terms of a substrate-induced rate effect (31) that has received much support (32–35). The conservation of the interactions between βPGM and the different inert phosphates in the two TSA complexes is consistent with this analysis. From a structural perspective, however, αPGM has not yet proven to be as amenable as βPGM to analysis of TSA complexes for the two steps in its reaction. The only deposited TSA structure is of rabbit αPGM (PDB ID code 1C4G) in which a tbp vanadate complex bridges the OH group of the nucleophilic serine and the 6-OH group of the αG1P (36). This complex contained cobalt in place of the catalytic magnesium ion and only diffracted to 2.7 Å, which precluded a clear definition of recognition of the hexose, the inert phosphate, or the catalytic metal ion.
Higher resolution structures have been determined for a related prokaryotic α-phosphohexomutase, αPMM/PGM, from Pseudomonas aeruginosa (8, 37), although these are of ground state complexes in which the enzyme is deactivated by replacing the catalytic magnesium with a tetrahedral zinc ion. In a series of structures in which the nucleophilic serine is phosphorylated, four isomeric sugar phosphates representing steps 1 and 2 for this enzyme, namely, αG1P, G6P, α-mannose 1-phosphate, and mannose 6-phosphate, have indistinguishable coordination of their phosphates in the inert binding site (i.e., the angular difference of ∼30° observed for the TSA structures of steps 1 and 2 for the βPGM structures is not present). In these structures, however, the nucleophilic oxygens are significantly out of line with the breaking O–P bond, indicating that they are similar to the near-attack conformations (6) rather than the TSA structures determined for βPGM. However, these αPMM/PGM structures suggest that the interchange of the O1 and O6 atoms between step 1 and step 2 in αPMM/PGM can be accomplished simply by rotation of the hexose bisphosphate about a virtual twofold C2 axis defined by the ring oxygen and the midpoint of the C3–C4 bond (SI Appendix, Fig. S11), and without the change in conformation of the exocyclic C5–C6 bond that is a feature of βPGM. The more simple rotation of the hexose bisphosphate in αPMM/PGM interchanges the 3-OH and 4-OH groups and maintains direct recognition in both steps by hydrogen bonding to a single glutamate carboxylate and a serine hydroxyl group (8, 10). It is also noteworthy that the cavity accommodating the substrate in the TSA complexes of βPGM (730 Å3 for PDB ID code 2WF5) is far smaller than that observed in the near-attack conformation structures of αPMM/PGM [1,700 Å3 for PDB ID codes 1P5D and 1P5G (38)]. Although αPMM/PGM is capable of accommodating both α-glucose and α-mannose substrates, βPGM does not accommodate β-mannose substrates because, for instance, the axial 2-OH group would clash with the imidazole ring of His20 in the step 2 TS.
In conclusion, the synthesis and employment of α-fluorophosphonates show how βPGM prioritizes protein TS conformation over direct hexose recognition in the two consecutive steps of the mutase reaction. βPGM delivers highly proficient catalysis by achieving virtual coincidence of orientation and recognition of the two transferring phosphoryl moieties, while ligating the inert phosphate to amino acid residues whose positions are invariant. Three high-resolution crystal structures of TSA complexes for step 1 of the reaction identify direct recognition of the glucopyranose moiety by five amino acid residues. This contrasts strongly with step 2, where the βG16BP intermediate released after step 1 undergoes conformational rotation of the C5–C6 bond to rebind with largely indirect recognition, mediated by additional waters. Substrate recognition in step 2 thus is made subservient to accurate conservation of a common TS for concerted phosphoryl transfer. This result further endorses the necessity for phosphate esters to organize substrate binding in a fashion that delivers prealignment of the scissile O–P bond with the nucleophile in the tbp TS complex.
Materials and Methods
Full details of the chemical synthesis of the two key analogs of βG1P, βG1CFSP and βG1CP, with appropriate analytical data are given in SI Appendix. Both βG1CFSP and βG1CP formed well-populated TSA complexes with MgF3− and with AlF4− ligands as shown by solution 19F NMR. βG1CFSP and βG1CP were screened in crystallization trials for MgF3− or AlF4− complexes, and crystalline TSA complexes for step 1 were obtained in three cases: βPGM-MgF3−-βG1CFSP, βPGM-AlF4−-βG1CFSP, and βPGM-MgF3−-βG1CP complexes. Full details of their X-ray analysis and all further experiments are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. N. H. Williams (University of Sheffield) and Dr. C. E. Webster (University of Memphis) for valuable discussions. This work is supported by the Biotechnology and Biological Sciences Research Council (Y.J.), a European Synchrotron Radiation Facility research studentship (to E.P.), the Canadian Institutes of Health Research, and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates for the following crystal structures have been deposited in the Protein Data Bank www.pdb.org: βPGM-MgF3−-βG1CP (PDB ID code 4C4R), βPGM-MgF3−-βG1CFSP (PDB ID code 4C4S), βPGM-AlF4−-βG1CFSP (PDB ID code 4C4T), and βPGM-AlF4−-G6CP (PDB ID code 2WF7).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402850111/-/DCSupplemental.
References
- 1.Lad C, Williams NH, Wolfenden R. The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases. Proc Natl Acad Sci USA. 2003;100(10):5607–5610. doi: 10.1073/pnas.0631607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray WJ, Jr, Burgner JW, 2nd, Post CB. Characterization of vanadate-based transition-state-analogue complexes of phosphoglucomutase by spectral and NMR techniques. Biochemistry. 1990;29(11):2770–2778. doi: 10.1021/bi00463a021. [DOI] [PubMed] [Google Scholar]
- 3.Mehra-Chaudhary R, Mick J, Tanner JJ, Henzl MT, Beamer LJ. Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens. Proteins. 2011;79(4):1215–1229. doi: 10.1002/prot.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter NJ, et al. Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF3− rather than by phosphoranes. Proc Natl Acad Sci USA. 2010;107(10):4555–4560. doi: 10.1073/pnas.0910333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxter NJ, et al. Anionic charge is prioritized over geometry in aluminum and magnesium fluoride transition state analogs of phosphoryl transfer enzymes. J Am Chem Soc. 2008;130(12):3952–3958. doi: 10.1021/ja078000n. [DOI] [PubMed] [Google Scholar]
- 6.Griffin JL, et al. Near attack conformers dominate β-phosphoglucomutase complexes where geometry and charge distribution reflect those of substrate. Proc Natl Acad Sci USA. 2012;109(18):6910–6915. doi: 10.1073/pnas.1116855109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jedrzejas MJ, Chander M, Setlow P, Krishnasamy G. Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus. EMBO J. 2000;19(7):1419–1431. doi: 10.1093/emboj/19.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regni C, Naught L, Tipton PA, Beamer LJ. Structural basis of diverse substrate recognition by the enzyme PMM/PGM from P. aeruginosa. Structure. 2004;12(1):55–63. doi: 10.1016/j.str.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Nishitani Y, et al. Crystal structures of N-acetylglucosamine-phosphate mutase, a member of the α-D-phosphohexomutase superfamily, and its substrate and product complexes. J Biol Chem. 2006;281(28):19740–19747. doi: 10.1074/jbc.M600801200. [DOI] [PubMed] [Google Scholar]
- 10.Shackelford GS, Regni CA, Beamer LJ. Evolutionary trace analysis of the alpha-D-phosphohexomutase superfamily. Protein Sci. 2004;13(8):2130–2138. doi: 10.1110/ps.04801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Z, Dunaway-Mariano D, Allen KN. HAD superfamily phosphotransferase substrate diversification: structure and function analysis of HAD subclass IIB sugar phosphatase BT4131. Biochemistry. 2005;44(24):8684–8696. doi: 10.1021/bi050009j. [DOI] [PubMed] [Google Scholar]
- 12.Golicnik M, et al. Kinetic analysis of β-phosphoglucomutase and its inhibition by magnesium fluoride. J Am Chem Soc. 2009;131(4):1575–1588. doi: 10.1021/ja806421f. [DOI] [PubMed] [Google Scholar]
- 13.Dai J, Wang L, Allen KN, Radstrom P, Dunaway-Mariano D. Conformational cycling in β-phosphoglucomutase catalysis: reorientation of the β-D-glucose 1,6-(Bis)phosphate intermediate. Biochemistry. 2006;45(25):7818–7824. doi: 10.1021/bi060136v. [DOI] [PubMed] [Google Scholar]
- 14.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. Caught in the act: the structure of phosphorylated β-phosphoglucomutase from Lactococcus lactis. Biochemistry. 2002;41(26):8351–8359. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- 15.Baxter NJ, et al. MgF3− and α-galactose 1-phosphate in the active site of β-phosphoglucomutase form a transition state analogue of phosphoryl transfer. J Am Chem Soc. 2009;131(45):16334–16335. doi: 10.1021/ja905972m. [DOI] [PubMed] [Google Scholar]
- 16.Cliff MJ, et al. Transition state analogue structures of human phosphoglycerate kinase establish the importance of charge balance in catalysis. J Am Chem Soc. 2010;132(18):6507–6516. doi: 10.1021/ja100974t. [DOI] [PubMed] [Google Scholar]
- 17.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science. 2003;299(5615):2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 18.Berkowitz DB, Bose M, Pfannenstiel TJ, Doukov T. α-fluorinated phosphonates as substrate mimics for glucose 6-phosphate dehydrogenase: The CHF stereochemistry matters. J Org Chem. 2000;65(15):4498–4508. doi: 10.1021/jo000220v. [DOI] [PubMed] [Google Scholar]
- 19.Forget SM, et al. Synthesis and enzymatic evaluation of ketose phosphonates: the interplay between mutarotation, monofluorination and acidity. Chem Sci. 2012;3(6):1866–1878. [Google Scholar]
- 20.Barton DH, McCombie SW. A new method for the deoxygenation of secondary alcohols. J Chem Soc Perkin 1. 1975;(16):1574–1585. [PubMed] [Google Scholar]
- 21.Norris AJ, Toyokuni T. A concise and stereoselective synthesis of C-glycosyl analogues of β-L-fucopyranosyl phosphate and β-L-rhamnopyranosyl phosphate. J Carbohydr Chem. 1999;18(9):1097–1105. [Google Scholar]
- 22.Lecomte V, Stéphan E, Rager MN, Jaouen G. Versatile use of hindered oxalates for the stereoselective preparation of novel 11-modified androst-5-ene derivatives. J Org Chem. 2004;69(9):3216–3219. doi: 10.1021/jo0401016. [DOI] [PubMed] [Google Scholar]
- 23.Shibata N, Sato H, Sakaki S, Sugita Y. Theoretical study of magnesium fluoride in aqueous solution. J Phys Chem B. 2011;115(35):10553–10559. doi: 10.1021/jp2053647. [DOI] [PubMed] [Google Scholar]
- 24.Baxter NJ, et al. A Trojan horse transition state analogue generated by MgF3− formation in an enzyme active site. Proc Natl Acad Sci USA. 2006;103(40):14732–14737. doi: 10.1073/pnas.0604448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray WJ, Jr, Post CB, Puvathingal JM. Reaction of the isosteric methylenephosphonate analog of α-D-glucose 1-phosphate with phosphoglucomutase. Induced-fit specificity revisited. Biochemistry. 1993;32(1):38–47. doi: 10.1021/bi00052a007. [DOI] [PubMed] [Google Scholar]
- 26.Blackburn GM, Kent DE, Kolkmann F. The synthesis and metal binding characteristics of novel, isopolar phosphonate analogues of nucleotides. J Chem Soc Perkin 1. 1984;(0):1119–1125. [Google Scholar]
- 27.Bowler MW, Cliff MJ, Waltho JP, Blackburn GM. Why did nature select phosphate for its dominant roles in biology? New J Chem. 2010;34(5):784–789. [Google Scholar]
- 28.Ray WJ, Jr, Roscelli GA. A kinetic study of the phosphoglucomutase pathway. J Biol Chem. 1964;239(4):1228–1236. [PubMed] [Google Scholar]
- 29.Ray WJ, Jr, Long JW. Thermodynamics and mechanism of the PO3 transfer process in the phosphoglucomutase reaction. Biochemistry. 1976;15(18):3993–4006. doi: 10.1021/bi00663a014. [DOI] [PubMed] [Google Scholar]
- 30.Lin Z, et al. The structure of rabbit muscle phosphoglucomutase at intermediate resolution. J Biol Chem. 1986;261(1):264–274. [PubMed] [Google Scholar]
- 31.Ray WJ, Jr, Long JW, Owens JD. An analysis of the substrate-induced rate effect in the phosphoglucomutase system. Biochemistry. 1976;15(18):4006–4017. doi: 10.1021/bi00663a015. [DOI] [PubMed] [Google Scholar]
- 32.Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci USA. 1981;78(7):4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amyes TL, Richard JP, Tait JJ. Activation of orotidine 5′-monophosphate decarboxylase by phosphite dianion: the whole substrate is the sum of two parts. J Am Chem Soc. 2005;127(45):15708–15709. doi: 10.1021/ja055493s. [DOI] [PubMed] [Google Scholar]
- 34.Amyes TL, Richard JP. Enzymatic catalysis of proton transfer at carbon: activation of triosephosphate isomerase by phosphite dianion. Biochemistry. 2007;46(19):5841–5854. doi: 10.1021/bi700409b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhai X, Amyes TL, Richard JP. Enzyme architecture: remarkably similar transition states for triosephosphate isomerase-catalyzed reactions of the whole substrate and the substrate in pieces. J Am Chem Soc. 2014;136(11):4145–4148. doi: 10.1021/ja501103b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Ray WJ, Jr, Baranidharan S. Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 4):392–405. doi: 10.1107/S0907444997000875. [DOI] [PubMed] [Google Scholar]
- 37.Regni C, Schramm AM, Beamer LJ. The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme. J Biol Chem. 2006;281(22):15564–15571. doi: 10.1074/jbc.M600590200. [DOI] [PubMed] [Google Scholar]
- 38.Dundas J, et al. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucl Acids Res. 2006;34(web server issue):W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.