Significance
As primary components of the Staphylococcus aureus cell envelope, teichoic acids play important but poorly understood roles in cell division and pathogenesis. Using an unbiased screen combined with targeted gene knockouts, we constructed an interaction network connecting wall teichoic acids to several other cellular pathways, including peptidoglycan biosynthesis, surface protein display, cell envelope D-alanylation, and lipoteichoic acid synthesis. We showed that lipoteichoic and wall teichoic acids play redundant roles in initiating cell division, but have distinct genetic interactions that reveal unique functions. Our findings provide insight into essential cell envelope processes in a clinically important pathogen as well as targets for compound combinations to kill antibiotic-resistant S. aureus.
Keywords: synthetic lethality, TraDIS, Tn-seq
Abstract
Staphylococcus aureus contains two distinct teichoic acid (TA) polymers, lipoteichoic acid (LTA) and wall teichoic acid (WTA), which are proposed to play redundant roles in regulating cell division. To gain insight into the underlying biology of S. aureus TAs, we used a small molecule inhibitor to screen a highly saturated transposon library for cellular factors that become essential when WTA is depleted. We constructed an interaction network connecting WTAs with genes involved in LTA synthesis, peptidoglycan synthesis, surface protein display, and D-alanine cell envelope modifications. Although LTAs and WTAs are synthetically lethal, we report that they do not have the same synthetic interactions with other cell envelope genes. For example, D-alanylation, a tailoring modification of both WTAs and LTAs, becomes essential when the former, but not the latter, are removed. Therefore, D-alanine–tailored LTAs are required for survival when WTAs are absent. Examination of terminal phenotoypes led to the unexpected discovery that cells lacking both LTAs and WTAs lose their ability to form Z rings and can no longer divide. We have concluded that the presence of either LTAs or WTAs on the cell surface is required for initiation of S. aureus cell division, but these polymers act as part of distinct cellular networks.
Bacteria are surrounded by complex cell envelopes that perform many different functions important for survival. In Gram-positive organisms, the major components of the cell envelope are peptidoglycan (PG) and teichoic acids (TAs) (Fig. 1). PG, a polymer of highly crosslinked carbohydrate chains that encapsulates the cell, is a well-established target for clinically useful antibiotics due to its essential role in providing structural support and protection from osmotic lysis. TAs are also critical for envelope integrity, but their functions are not as well understood. In Staphylococcus aureus there are two main types of TA: lipoteichoic acids (LTAs), which are embedded in the cell membrane and comprise a poly(glycerolphosphate) backbone tailored with D-alanine esters, and wall teichoic acids (WTAs), which are covalently attached to PG and possess a poly(ribitolphosphate) backbone functionalized with both D-alanine and N-acetylglucosamine (GlcNAc) moieties (1, 2). Deconvoluting the functions of these polymers has been challenging because they play several different roles. Moreover, because they have structurally similar backbones and cannot be simultaneously deleted without loss of viability (3, 4), they are proposed to have redundant functions. This view is consistent with the observation that deletion of either pathway results in similar cell division defects (3, 5). Furthermore, the same D-alanylation machinery is used to tailor both polymers (6). D-alanine modification of TAs modulates the net charge of the cell envelope, decreases susceptibility to cationic antimicrobial peptides, and is proposed to affect activity of cell surface enzymes, but whether D-alanylation is more important for LTA or WTA function has not been addressed (7, 8).
Fig. 1.
TAs are a major component of the S. aureus cell envelope. WTAs are attached to PG, whereas LTAs are linked to membrane-bound lipid carriers (diacylglycerol, DAG). Tunicamycin is a selective inhibitor of the first step of WTA biosynthesis, catalyzed by TarO (10, 18, 19).
A better understanding of S. aureus TAs is necessary because these polymers play critical roles in pathogenesis and their biosynthetic pathways are possible targets for antibiotics (9–11). We reasoned that insight into distinct functions of TAs could be gleaned by identifying their genetic interactions. The recent merging of classical transposon mutagenesis with high-throughput next-generation sequencing (e.g., TraDIS, Tn-seq) has enabled parallel genotype-phenotype profiling and functional mapping for large collections of mutants (12, 13). These methods have been used to identify genetic factors that influence susceptibility to lethal antibiotics (14), but can also be applied to map cell envelope networks. Here, we apply a nonlethal small molecule inhibitor of WTA biosynthesis to identify synthetic lethal interactions with the WTA pathway. We uncovered several genes/pathways that became essential in the absence of WTAs, but were dispensable when LTAs were deleted. Thus, although WTAs and LTAs share redundant functions in cell division, they have different genetic interactions and act in distinct cellular networks.
Results
Design of an Unbiased Screen for Cellular Factors that Interact with WTAs.
To understand TA function better, we sought to design a genome-wide synthetic lethal screen for factors that interact with WTAs. Transposon mutagenesis is the standard way to generate a diverse mutant collection for genome-wide interrogations, but it is not possible to construct a transposon library in a WTA-deficient strain because bacteria lacking WTAs are temperature-sensitive as well as phage-resistant, which makes them recalcitrant to available techniques for making transposon mutant libraries (5, 15–17). In principle, it is possible to overcome this issue by constructing the transposon library in a wildtype strain and then using an inhibitor to turn off WTA expression. We have previously shown that the natural product tunicamycin is a potent and highly selective inhibitor of S. aureus TarO, the first enzyme in the WTA pathway (Fig. 1) (10, 18, 19). To use tunicamycin to comprehensively probe the S. aureus genome for interactions with WTAs, we needed a highly saturated transposon mutant library. Using a mariner-based transposon system (16), we constructed a mutant pool in S. aureus HG003. Transposon-directed insertion site sequencing (TraDIS, similar to Tn-seq), a PCR-based next generation sequencing technique that determines the location and abundance of transposon insertions in a population, showed that the library contained 60,000 independent insertion sites (Fig. 2A), which represents approximately 20-fold coverage of the S. aureus genome (12, 13). The high saturation of the transposon library enabled us to screen for genes that interact with WTAs by using tunicamycin to turn off WTA synthesis.
Fig. 2.
The transposon screen identified 24 genes that are synthetically lethal with WTA depletion. (A) Schematic showing how genes that become essential (purple) with WTA depletion (i.e., TarO inhibition) are identified by comparing the reads for tunicamycin-treated and untreated transposon libraries. (B) The 10 most depleted genes are plotted by −log P values; the area of the circles reflects the extent of growth depletion, with the smallest circles denoting ∼90% depletion. (C) Dilution series of selected wild-type and mutant strains grown in the absence and presence of 0.4 μg/mL tunicamycin. One wild-type control is shown; the full set of control and complemented strains are provided in Fig. S3.
Identification of Transposon Mutants Sensitive to WTA Depletion.
We grew replicate cultures of the pooled mutant library in the presence and absence of tunicamycin, sequenced the transposon insertion sites, and quantified PCR reads for each transposon mutant under each condition (Fig. 2A, Fig. S1, and Dataset S1). The differences in PCR reads (normalized by total reads obtained) between the two conditions, expressed as growth ratios, were assigned statistical significance based on the coverage of transposon insertions within the boundaries of a particular ORF (Mann–Whitney U test). Using this method, we were able to assess mutants in 2,535 ORFs (85% of the genome) for differential growth. Forty-nine ORFs showed a difference in growth upon tunicamycin treatment (P < 0.05), and of these 2 were enriched and 47 were depleted from the population. To identify genes that were synthetically lethal with WTA deletion, we established a stringent cutoff of >90% growth depletion in tunicamycin with a P value of <0.002. Only 10 genes met these criteria (Fig. 2B). Decreasing the stringency of the cutoff to 85% growth depletion with P < 0.04 yielded 14 additional genes (Fig. S2). Hence, a very small subset of ∼2,500 disrupted genes in the mutant library showed a strong synthetic phenotype with WTA depletion. Notably, the majority of the genes in the list of 24, including 8 of the top 10 genes, were predicted to localize to the membrane or to have a cell envelope-related function. Because WTAs reside in the cell envelope, one would expect their synthetic lethal interactions to be weighted toward factors that contribute to envelope biogenesis and integrity.
Targeted Knockouts of Top Candidate Genes Confirm Sensitivity to WTA Depletion.
Transposons often inactivate the ORF into which they insert, but may allow transcription of a truncated mRNA and can also have indirect effects such as polar disruption of nearby genes. Because the top genes identified in the screen were well saturated with transposon insertions in the control cultures (no tunicamycin), but had few or no insertions across multiple sites in the treated samples, it seemed likely that they were indeed synthetically lethal with WTA depletion (Table S1). To confirm that growth depletion was the result of gene inactivation, we selected a subset of the top 10 genes spanning a range of growth depletion ratios and P values and prepared targeted knockouts to confirm susceptibility to tunicamycin. Knockouts tested included ΔgraR, ΔvraFG, ΔSAOUHSC_00965 (hereafter known as Δ965), and ΔlyrA. Because simultaneous deletion of LTAs and WTAs was predicted to be lethal, we also examined a strain that lacks LTAs (ΔltaS4S5) to examine its tunicamycin susceptibility (3, 4, 20). (Because library preparation involved a high-temperature step and strains lacking LTAs are thermosensitive, we were unable to assess the ltaS locus from the growth depletion analysis as it was not well represented in the untreated control; ref. 3.) All strains were tested in a dilution series on agar plates with and without tunicamycin. Tunicamycin had no effect on the growth of wild-type S. aureus colonies, but prevented growth of the ΔltaS4S5 strain, consistent with a synthetic lethal interaction between the LTA and WTA pathways. The other mutant strains also exhibited tunicamycin sensitivity (Fig. 2C). Complementation with plasmid-borne copies of deleted genes restored growth in the presence of tunicamycin (Fig. S3). These experiments showed that the genome-wide growth depletion analysis accurately identified genes that become essential when WTA synthesis is inhibited.
D-Alanylation Becomes Essential When WTA Synthesis Is Inhibited.
Four of the top 10 genes identified in the transposon screen, graR, graS, vraF, and vraG, encode a regulatory protein complex comprising a two component signaling system and a two-component ABC transporter (21–23). The GraRSVraFG complex is predicted to regulate expression of lyrA, also in the top 10 list, but it is best known for regulating the dltABCD operon (Fig. 3A), which encodes the machinery that installs D-alanines on both LTAs and WTAs (6). Because Dlt null mutants are temperature-sensitive, there were almost no insertions in the dlt operon in the untreated library even though this modification is not essential in a wild-type background (7, 24). Therefore, the genome-wide growth depletion analysis did not provide information about possible synthetic interactions between the dltABCD genes and WTAs. To assess whether the D-alanylation pathway itself becomes essential when WTAs are depleted, we constructed two Dlt null strains, ΔdltA and ΔdltD, and tested their sensitivity to WTA-inhibitory concentrations of tunicamycin (Fig. 3B and Figs. S3 and S4). Neither of the knockout strains grew on plates or in liquid media containing tunicamycin, but complementation with a plasmid expressing dltABCD restored growth. Hence, the D-alanylation machinery is required when WTA expression is inhibited, implying that D-alanylated LTAs are essential if WTAs are not produced.
Fig. 3.
Identification and validation of Dlt− mutants as synthetically lethal with WTA depletion. (A) Four of the 10 most depleted genes encode the GraRSVraFG complex, which regulates expression of lyrA (purple) and the dlt genes (black). (B) Dilution series of wild-type and dlt knockout strains grown in the absence and presence of 0.4 μg/mL tunicamycin (Tun) shows that the dlt deletion is synthetically lethal with WTA depletion. Complementation with the dltABCD operon restored viability in the presence of tunicamcyin. (C) Linkage analysis confirms compound-gene synthetic lethality. The ΔtarO::tetM locus could be transduced with the kanR gene (aphA3) into a wild-type background ∼50% of the time; it could not be transduced with the kanR gene into any of the tested mutant strains. WTA null recipients were identified by assaying for tetracycline resistance (TetR) and Congo Red susceptibility (Congo RedS). Because the ΔgraR strain was tet-marked, it could only be tested for Congo Red susceptibility.
Linkage Analysis Provided Gene–Gene Validation of Compound-Gene Synthetic Lethal Interactions.
The primary target of tunicamycin in S. aureus is TarO, the first enzyme in the WTA biosynthetic pathway, but there is also a secondary target, MraY (19). Although our screen and follow up validation used tunicamycin concentrations that are two orders of magnitude below those that inhibit MraY, we wanted to confirm key results using a fully genetic approach that did not rely on a small molecule (10). This step seemed necessary because some of the knockouts, including ΔdltA, ΔgraR, and ΔvraFG, are known to have increased susceptibility to cationic antimicrobial peptides (CAMPs) and certain other antibiotics with cell envelope targets (7, 8, 25). Increased susceptibility is proposed to occur because the cell surface lacks the D-alanine modifications that normally repel positively charged toxins at the envelope interface (26). Although tunicamycin is not charged at physiological pH, we wanted to rule out increased access to MraY as an explanation for the lethal phenotypes we observed. We made strains with regulated, ectopic expression of TarO, but observed leaky expression in the absence of inducer. Therefore, we used linkage analysis to test synthetic lethality of double mutant combinations. We constructed a ΔtarO::tetR strain with a kanamycin-resistance gene installed in an intergenic region 4.3 kbp downstream. This strain contained a copy of tarO integrated elsewhere in the chromosome to enable the preparation of phage lysates. We then measured linkage of the two antibiotic resistance markers after phage transduction into either the wild-type or mutant backgrounds (Fig. 3C). Transductants were selected on kanamycin plates and then scored for growth in media containing kanamycin, tetracycline, or Congo Red, an azo dye to which S. aureus strains lacking WTAs are highly sensitive (27). For the wild-type strain, approximately half the kanR transductants were both tetracycline resistant and Congo Red sensitive, indicating that the tarO deletion moves in tandem with the kanR marker 50% of the time. For the mutant strains, the compound sensitivity profiles showed that none of the kanR transductants contained the tarO deletion (Fig. 3C). (We were unable to assess linkage in ΔdltD and ΔltaS4S5, as the former deletion was marked with kanamycin resistance, and ltaS and tarO are within close proximity of each other on the chromosome.) This linkage disequilibrium confirmed synthetic lethality between ΔtarO and the other gene deletions, validating the screening results obtained using tunicamycin to inhibit WTA synthesis.
WTAs and LTAs Have Different Genetic Interactions.
WTAs and LTAs are proposed to play redundant cellular roles related to their anionic backbones. Having identified and validated a number of gene knockouts that are synthetically lethal with WTA removal, we next tested several of these in combination with LTA removal to determine whether LTAs and WTAs have similar genetic interactions. We were able to transduce marked deletions of ΔdltD, Δ965, ΔlyrA, and ΔgraR into the ΔltaS4S5 strain and each of the double mutants was viable. Because WTAs and LTAs have distinct sets of genetic interactions, they function as part of different cellular networks/pathways.
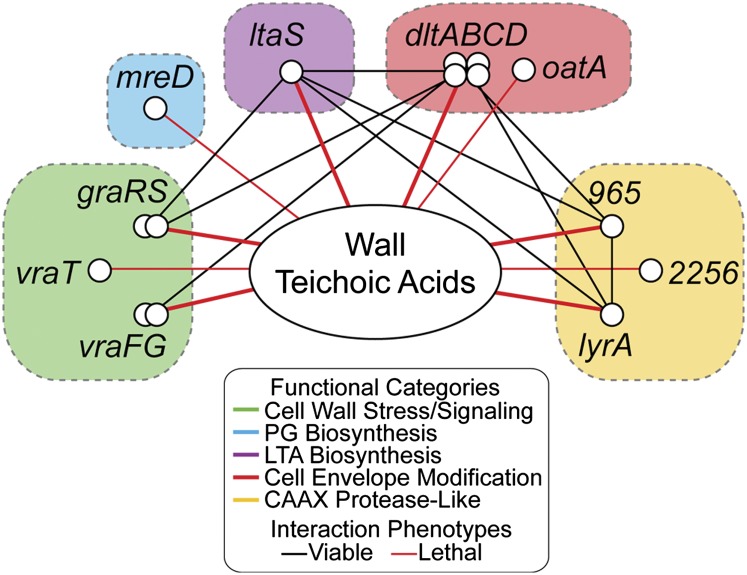
The network in Fig. 4 summarizes the interactions between WTAs, LTAs, and several other genes (Fig. 4 and Fig. S2). The WTA synthetic lethal interaction network includes, in addition to the LTA pathway, five signaling system components that regulate genes involved in modifying the cell envelope (VraFG, GraRS, YvqF/VraT) and two sets of genes that modify cell envelope components (dltABCD and oatA) (21–23, 28, 29). Three other genes in the WTA interaction network (lyrA, 965, and SAOUHSC_02256) encode structurally related membrane proteins that resemble type II CAAX prenyl endopeptidases, a family of proteins well known in eukaryotes because key members are involved in the proteolytic processing of prenylated Ras and other lipoproteins (30). There are five genes encoding CAAX protease-like (CPL) homologs in S. aureus, including the three identified in the growth depletion analysis, but only ΔlyrA mutants have been characterized (31, 32). These mutants have altered crosswall structures and are deficient in a subset of surface proteins, but as yet there is no evidence that LyrA has a proteolytic function. Although a molecular understanding of CPL homologs in prokaryotes is still lacking, the results reported here emphasize the importance of this family of proteins for S. aureus cell envelope physiology. The screen also identified mreD, which encodes a broadly conserved membrane protein implicated in regulating peptidoglycan synthesis (33), providing, to our knowledge, the first genetic connection between two pathways, PG and WTA biosynthesis, that are proposed to be functionally coupled (10, 24, 34).
Fig. 4.
An interaction network showing cell envelope-related genes connected to the WTA pathway. Genes (nodes) group into several distinct pathways or clusters encoding similar proteins (colored boxes). Black lines connect viable double deletion mutants. Thin red lines depict selected synthetic lethal interactions identified only from the genome-wide growth depletion analysis (growth depletion > 85%; P < 0.04); thick red lines depict synthetic lethal interactions that were confirmed in one or more follow up experiments. The full list of 24 growth-depleted genes is provided in Fig. S2.
LTA and Dlt Null Strains Depleted of WTAs Have Distinct Terminal Phenotypes.
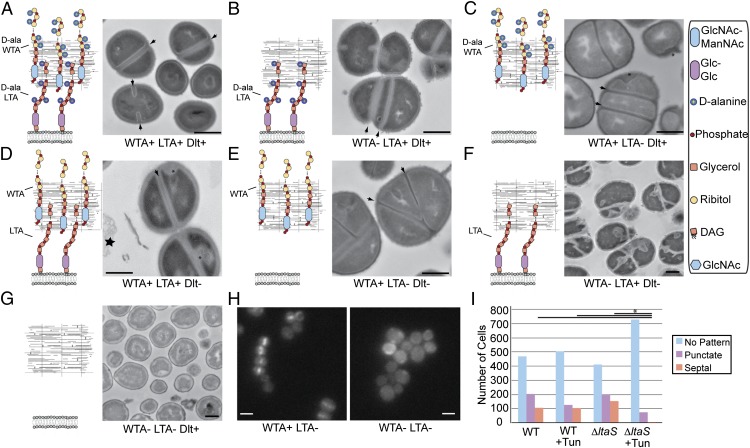
We were surprised that D-alanylation becomes essential when WTAs, but not LTAs, are deleted because D-alanylation is a shared tailoring modification of LTAs and WTAs. As LTAs are themselves essential in the absence of WTAs, we wondered whether mutants lacking the entire LTA backbone would, upon WTA depletion, have a terminal phenotype similar to mutants lacking only the D-alanine modification. To address this question, we used electron microscopy to examine strains deficient in LTAs or D-alanylation after treatment with tunicamycin to deplete cells of WTAs. Compared with untreated wild-type cells (Fig. 5A and Figs. S5A and S6 A and B), tunicamycin-treated cells were enlarged, had thickened crosswalls, and were impaired in daughter cell separation. Moreover, successive division planes were frequently placed at nonorthogonal angles and duplicated septa were common (note parallel crosswalls enclosing a narrow band of cytoplasmic material in Fig. 5B and Fig. S5B). Cells lacking LTAs due to ltaS deletion showed similar defects in cell separation and septal placement (Fig. 5C and Figs. S5C and S6A). These phenotypes, described previously, show that WTAs and LTAs help regulate S. aureus cell division (3, 10, 35, 36). The dlt null cells resembled untreated wild-type cells except for irregularities in peptidoglycan thickness and membrane contour both at crosswalls and along the cell periphery. In addition, lysed cells (“ghosts”) were commonly observed (Fig. 5D and Figs. S5 D and E and S6A). These phenotypes imply that D-alanylation contributes to cell envelope integrity, either directly or through regulation of cell wall biosynthetic or hydrolytic enzymes. The ΔdltDΔltaS4S5 mutant appeared identical to the ΔltaS4S5 mutant (Fig. 5E and Fig. S5F); however, dlt null strains in which WTAs were depleted showed defects characteristic of deficiencies in both pathways, but exaggerated. For example, crosswalls were mispositioned and daughter cells failed to separate as in WTA-deficient cells, but there were also marked aberrations in cell envelope ultrastructure characteristic of cells lacking D-alanylation (Fig. 5F and Figs. S5 G and H and S6 A and B). Within 5 h of treatment, nearly complete cell lysis was evident in treated samples, suggesting that the combined defects so weakened the cell envelope that rupture resulted (Fig. S5I). Remarkably, the ΔltaS4S5 and ΔdltDΔltaS4S5 mutants depleted of WTAs did not resemble cells lacking only one type of TA. Whereas we frequently observed duplicated and misplaced crosswalls in images of cells lacking either WTAs or LTAs, we saw almost no crosswalls in fields of cells that had both polymers removed (Fig. 5G and Figs. S5 J and K and S6A). Because FtsZ is the first intracellular protein to move to the division site during cell division, and is essential for recruiting other components of the division machinery (37), we transduced wild-type and ΔltaS4S5 cells with FtsZ-GFP, grew them in the absence or presence of tunicamycin to deplete WTAs, and used fluorescence microscopy to assess Z ring formation. We observed rings of fluorescent FtsZ in wild-type cells and cells lacking only one TA polymer, but saw no Z rings in ΔltaS4S5 cells depleted of WTAs (Fig. 5 H and I and Fig. S6 C–F). Thus, simultaneous removal of both types of TAs results in an inability to assemble Z rings and cells can no longer divide. When nondividing, tunicamycin-treated ΔltaS4S5 cells are plated on tunicamycin-free agar, they form colonies, showing that the phenotype is reversible (Fig. S7).
Fig. 5.
Effect of removing TAs and D-alanylation on cell morphology. Electron micrographs of S. aureus strains, grown in the presence and absence of 0.4 μg/mL tunicamycin. To the left of each electron micrograph is a schematic showing the type of TAs and their D-alanylation status. (A) Wlid-type cells, with division septa (black arrow). (B) Wild-type cells treated with tunicamycin to remove WTAs. The dark line along the crosswall disappears and septal positioning is dysregulated (black arrows). Cells also fail to separate efficiently. (C) ΔltaS4S5 cells exhibit defects similar to strains lacking WTAs, with multiple septa, larger cell size, and deficiency in separation. (D) Both ΔdltA (shown) and ΔdltD (Fig. S5) mutants retain the dark midline at the crosswall (black arrow), but shows several defects, including concavities in the cell membrane and irregular thickening of peptidoglycan (black asterisks) and ghost cells (black star). (E) The ΔdltDΔltaS4S5 mutant exhibits defects similar to ΔltaS4S5, including nonorthogonal septation, larger cells, and deficiency in cell separation; (F) Cells lacking D-alanylation and treated with tunicamycin exhibit exacerbated morphological defects, including impaired cell separation, increased incidence of membrane concavities and irregular peptidoglycan thickening (black asterisks), and improper placement and organization of septa (black arrows). (G) ΔltaS4S5 cells treated with tunicamycin show near complete abolition of cell septation. (H) Localization of FtsZ-GFP in untreated (left) and tunicamycin-treated (right) ΔltaS4S5 cells. (I) ΔltaS4S5 treated with tunicamycin lacked septal fluorescence in comparison with the other conditions (P < 10−9 by two-tailed t test). EM and light microscopy. (Scale bars: 500 nm and 1 μm, respectively.) Additional fields of view are provided in Figs. S5 and S6.
Discussion
Dissecting the functions of teichoic acids presents numerous challenges. These polymers play many different roles and deletion phenotypes are pleiotropic. Because LTAs and WTAs have similar, highly charged backbone structures and identical tailoring modifications, it is also difficult to identify distinct sets of physicochemical interactions for the two polymers using biochemical approaches. Using a TarO inhibitor, we carried out an unbiased screen of a large transposon mutant library for cellular factors that become essential when WTAs are depleted and identified approximately two dozen genes. We followed up on a subset of the top 10 candidates, and all of them confirmed. These results are a testament to the selectivity of the chemical probe for TarO and the high saturation of the transposon mutant library, which provided statistical power in the analysis. We have used the data to construct a synthetic lethal interaction network for WTAs, which can guide further investigations on S. aureus cell wall physiology. Although pharmacological interrogation of transposon mutant libraries has been performed previously, all earlier studies have focused on identifying factors implicated in tolerance or resistance to compounds that have essential targets, i.e., antibiotics. Our work shows that small molecules that inhibit nonessential targets have tremendous value for elucidating pathway interactions.
The LTA synthetic lethal network has not yet been mapped, but we tested several mutants that were not viable in the absence of WTAs for synthetic lethality with LTA deletion. All double mutant strains were fully viable. Although the ΔltaS4S5 mutant harbors a suppressor mutation that attenuates function of gdpP, deletion of this gene is not synthetically lethal with tunicamycin treatment and does not suppress the lethal effects of WTA depletion in the presence of Congo Red (Fig. S7 A and B). Hence, WTAs and LTAs have different genetic interactions, and the differences are not due to the suppressor mutation. In Bacillus subtilis, LTAs are proposed to coordinate septal cell envelope synthesis, whereas WTAs play a role in elongation (4). S. aureus does not have two distinct modes of cell wall synthesis, but the network analysis described here suggests that LTAs and WTAs act in different pathways to accomplish distinct as well as redundant functions.
A function for D-alanylation that is unique to the LTA pathway was also uncovered in our studies. We found that D-alanylation became essential on LTAs when WTAs were removed, but was not essential on WTAs when LTAs were removed. The D-alanine deficient cells had defects in cell wall ultrastructure and displayed increased lysis, showing that the D-alanine modification on LTAs contributes to membrane integrity. It has been proposed that D-alanylation influences LTA conformations, changing the density and rigidity of the cell wall (26). The presence of positively charged D-alanine esters on TAs may also negatively regulate hydrolytic enzymes, and removal of WTAs is also known to result in dysregulated autolysin activity (6, 8, 10, 35). The combined defects from removal of WTAs and D-alanylation evidently lead to catastrophic cell envelope damage during ongoing cell division.
An unexpected result to emerge from our studies was that simultaneous removal of LTAs and WTAs prevents FtsZ ring assembly and causes cell division to cease. FtsZ polymers serve as a scaffold for the recruitment and organization of division proteins at the septum. How extracellular TA polymers can influence the intracellular organization of division machinery remains unclear. Several proteins such as the nucleoid occlusion factor (Noc), FtsA, and EzrA are known regulators of FtsZ polymerization, and factors such as DNA damage, membrane potential, and the stringent response can affect Z ring assembly (37, 38). We are following up on possible mechanisms for how the absence of TAs inhibits Z ring formation.
The work reported here highlights an important use for small molecule probes in mapping bacterial networks. In addition to revealing previously unidentified biology, these interaction networks may provide target combinations for synthetically lethal antimicrobial compounds. From this study, we suggest that compound combinations that target both the WTA pathway and either D-alanylation or LTA synthesis are attractive because these factors are absent from humans and have previously been implicated in virulence/in vivo survival (9, 39). Synthetically lethal compound combinations that target these different cell envelope components may have potential as therapeutic agents against methicillin-resistant S. aureus (MRSA), which continues to be a serious threat to public health.
Materials and Methods
Bacterial strains, plasmids, and primers are listed in Table S1–S3. More detailed methods can be found in SI Materials and Methods.
Reagents and General Methods.
Plasmids were constructed in Escherichia coli XL-1 Blue (Stratagene) or Stellar Gold (Clontech) competent cells. Plasmids were electroporated or transduced into S. aureus via ϕ85. S. aureus was primarily grown in tryptic soy broth (TSB) medium at 30 °C. Antibiotic markers were selected with erythromycin (Em) (10 μg/mL), tetracycline (Tc) (2.5 μg/mL), carbenicillin (Carb) (100 μg/mL), kanamycin and neomycin (Km) (50 μg/mL, each), chloramphenicol (Cm) (10 μg/mL), and Congo Red (15 μg/mL). Tunicamycin was dissolved in DMSO and diluted to a final concentration of 0.4 μg/mL Novagen KOD Hot Start Polymerase was used for all PCRs and all restriction enzymes were purchased from New England Biolabs. A Clontech In-Fusion HD Cloning kit was used for isothermal assembly.
Transposon Library and Screen.
The library was constructed in S. aureus HG003 using the bursa aurealis system (16). Frozen aliquots of library were thawed and diluted 1:1,000 into 10 mL of BHI overnight at 37 °C. The grown culture was then diluted 1:100 into 100 mL of BHI with/without tunicamycin, then grown at 37 °C for 24 h. DNA was extracted, sheared, appended with a polyC tail (TdT, Promega), amplified and barcoded via PCR, and sequenced using an Illumina Hi-Seq2000. Insertions were mapped to ORFs using the sequenced NCTC 8325 genome and the Galaxy webserver (40, 41). The Mann–Whitney U test was used to identify mutants in genes for which tunicamycin treatment led to significant depletion compared with the number of identified reads for the untreated control.
Construction of Mutant Strains.
Gene deletions were made using the pMAD and pKFC allelic replacement systems (42, 43). Complementation was performed using pLOW (44), and double mutants were created through ϕ85 transduction of a marked deletion into unmarked deletion backgrounds.
Linkage Analysis.
Strain JSM009 was created by installing a aphA3 (KmR) marker ∼4.5 kb downstream of ΔtarO::tetR in TCM011 using pKFCtarOlink. Donor phage ϕ85 was prepared from this strain and used to transduce aphA3 into wild-type and mutant backgrounds. KmR colonies were obtained at 37 °C, then assayed for TetR and Congo Red sensitivity in 96-well format, overnight at 37 °C.
Phase Contrast and Fluorescence Microscopy.
Cells from exponential phase culture were pelleted and resuspended in 1× PBS, then mounted on slides covered with a 1% agarose in 1× PBS pad. Images were acquired with a Hamamatsu digital camera (ORCA-ER) connected to a Nikon Eclipse TE2000-U microscope with an X-cite 120 illumination system. Image manipulation was limited to changing brightness and contrast via ImageJ (45).
Supplementary Material
Acknowledgments
We thank O. Schneewind for the bursa aurealis transposon system; A. Gründling for strains ΔltaS4S5 and SEJ1; T. Meredith for strain TCM011; Tufts University Core Facility for Genomic Sequencing for processing of TnSeq samples; and L. Trakimas, M. Ericcson, and E. Benecchi of the Harvard Medical School Electron Microscopy Facility for processing of EM samples. This work was funded by National Institutes of Health Grants 1P01AI083214 (to S.W. and M.S.G.) and 1R01AI099144 (to S.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404099111/-/DCSupplemental.
References
- 1.Reichmann NT, Gründling A. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol Lett. 2011;319(2):97–105. doi: 10.1111/j.1574-6968.2011.02260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown S, Santa Maria JP, Jr, Walker S. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol. 2013;67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oku Y, et al. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol. 2009;191(1):141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28(7):830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chatterjee AN, Mirelman D, Singer HJ, Park JT. Properties of a novel pleiotropic bacteriophage-resistant mutant of Staphylococcus aureus H. J Bacteriol. 1969;100(2):846–853. doi: 10.1128/jb.100.2.846-853.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67(4):686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274(13):8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 8.Peschel A, Vuong C, Otto M, Götz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother. 2000;44(10):2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weidenmaier C, et al. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat Med. 2004;10(3):243–245. doi: 10.1038/nm991. [DOI] [PubMed] [Google Scholar]
- 10.Campbell J, et al. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol. 2011;6(1):106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasquina LW, Santa Maria JP, Walker S. Teichoic acid biosynthesis as an antibiotic target. Curr Opin Microbiol. 2013;16(5):531–537. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Opijnen T, Camilli A. Transposon insertion sequencing: A new tool for systems-level analysis of microorganisms. Nat Rev Microbiol. 2013;11(7):435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langridge GC, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19(12):2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher LA, Shendure J, Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio. 2011;2(1):e00315–e10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergara-Irigaray M, et al. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology. 2008;154(Pt 3):865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bae T, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA. 2004;101(33):12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Claveau D, Vaillancourt JP, Roemer T, Meredith TC. High-frequency transposition for determining antibacterial mode of action. Nat Chem Biol. 2011;7(10):720–729. doi: 10.1038/nchembio.643. [DOI] [PubMed] [Google Scholar]
- 18.Ward JB, Wyke AW, Curtis CAM. The effect of tunicamycin on wall-polymer synthesis in Bacilli. Biochem Soc Trans. 1980;8(2):164–166. doi: 10.1042/bst0080164. [DOI] [PubMed] [Google Scholar]
- 19.Pooley HM, Karamata D. Incorporation of [2-3H]glycerol into cell surface components of Bacillus subtilis 168 and thermosensitive mutants affected in wall teichoic acid synthesis: Effect of tunicamycin. Microbiology. 2000;146(Pt 4):797–805. doi: 10.1099/00221287-146-4-797. [DOI] [PubMed] [Google Scholar]
- 20.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falord M, Mäder U, Hiron A, Débarbouillé M, Msadek T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS ONE. 2011;6(7):e21323. doi: 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56(2):1047–1058. doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SJ, et al. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun. 2012;80(1):74–81. doi: 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atilano ML, et al. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci USA. 2010;107(44):18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meehl M, Herbert S, Götz F, Cheung A. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(8):2679–2689. doi: 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saar-Dover R, et al. D-alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog. 2012;8(9):e1002891. doi: 10.1371/journal.ppat.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, et al. Wall teichoic acid protects Staphylococcus aureus from inhibition by Congo red and other dyes. J Antimicrob Chemother. 2012;67(9):2143–2151. doi: 10.1093/jac/dks184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(1):83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bera A, Herbert S, Jakob A, Vollmer W, Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55(3):778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 30.Pei J, Mitchell DA, Dixon JE, Grishin NV. Expansion of type II CAAX proteases reveals evolutionary origin of γ-secretase subunit APH-1. J Mol Biol. 2011;410(1):18–26. doi: 10.1016/j.jmb.2011.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gründling A, Missiakas DM, Schneewind O. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol. 2006;188(17):6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol. 2010;78(1):238–252. doi: 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Land AD, Winkler ME. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J Bacteriol. 2011;193(16):4166–4179. doi: 10.1128/JB.05245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qamar A, Golemi-Kotra D. Dual roles of FmtA in Staphylococcus aureus cell wall biosynthesis and autolysis. Antimicrob Agents Chemother. 2012;56(7):3797–3805. doi: 10.1128/AAC.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlag M, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010;75(4):864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 36.Cole RM, Chatterjee AN, Gilpin RW, Young FE. Ultrastructure of teichoic acid-deficient and other mutants of staphylococci. Ann N Y Acad Sci. 1974;236(0):22–53. doi: 10.1111/j.1749-6632.1974.tb41480.x. [DOI] [PubMed] [Google Scholar]
- 37.Adams DW, Errington J. Bacterial cell division: Assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7(9):642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 38.Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107(27):12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collins LV, et al. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186(2):214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 40.Gillaspy AF, et al. 2006. The Staphylococcus aureus NCTC8325 genome. Gram Positive Pathogens eds. Fischetti V, Novick R, Ferretti J, Portnoy D (ASM Press, Washington, DC), pp 381–412.
- 41.Goecks J, Nekrutenko A, Taylor J. Galaxy Team Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70(11):6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato F, Sugai M. A simple method of markerless gene deletion in Staphylococcus aureus. J Microbiol Methods. 2011;87(1):76–81. doi: 10.1016/j.mimet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 44.Liew ATF, et al. A simple plasmid-based system that allows rapid generation of tightly controlled gene expression in Staphylococcus aureus. Microbiology. 2011;157(Pt 3):666–676. doi: 10.1099/mic.0.045146-0. [DOI] [PubMed] [Google Scholar]
- 45.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.