Significance
EFA6, cytohesins, and BRAGs activate Arf GTPases in endocytic events. They carry a plasma membrane-binding PH domain in tandem with their catalytic Sec7 domain, which is autoinhibitory and mediates a positive feedback loop in cytohesins but not in BRAGs, and has an as-yet unknown role in EFA6 regulation. By reconstituting GDP/GTP exchange on membranes, we find that the PH domain of EFA6 is not autoinhibitory, but supports a negative feedback loop. This loop is controlled by interaction of Arf6-GTP with the PH-Ct domains of EFA6 and monitors Arf1 and Arf6 activation differentially. This suggests that EFA6 and cytohesins might be coupled in a mixed negative-positive feedback loop to shape the level and timing of Arf1 and Arf6 activation in endocytosis.
Keywords: endocytosis, membrane traffic, kinetics, autoinhibition
Abstract
Guanine nucleotide exchange factors (GEFs) of the exchange factor for Arf6 (EFA6), brefeldin A-resistant Arf guanine nucleotide exchange factor (BRAG), and cytohesin subfamilies activate small GTPases of the Arf family in endocytic events. These ArfGEFs carry a pleckstrin homology (PH) domain in tandem with their catalytic Sec7 domain, which is autoinhibitory and supports a positive feedback loop in cytohesins but not in BRAGs, and has an as-yet unknown role in EFA6 regulation. In this study, we analyzed how EFA6A is regulated by its PH and C terminus (Ct) domains by reconstituting its GDP/GTP exchange activity on membranes. We found that EFA6 has a previously unappreciated high efficiency toward Arf1 on membranes and that, similar to BRAGs, its PH domain is not autoinhibitory and strongly potentiates nucleotide exchange on anionic liposomes. However, in striking contrast to both cytohesins and BRAGs, EFA6 is regulated by a negative feedback loop, which is mediated by an allosteric interaction of Arf6-GTP with the PH-Ct domain of EFA6 and monitors the activation of Arf1 and Arf6 differentially. These observations reveal that EFA6, BRAG, and cytohesins have unanticipated commonalities associated with divergent regulatory regimes. An important implication is that EFA6 and cytohesins may combine in a mixed negative-positive feedback loop. By allowing EFA6 to sustain a pool of dormant Arf6-GTP, such a circuit would fulfill the absolute requirement of cytohesins for activation by Arf-GTP before amplification of their GEF activity by their positive feedback loop.
Guanine nucleotide exchange factors (GEFs), which activate small GTPases by stimulating their intrinsically very slow GDP/GTP exchange, are key players in the extraordinary diversity of small GTPases pathways (reviewed in ref. 1). Small GTPases carry little specificity determinants on their own to determine when and where they should be turned on and which pathway they should activate (2), which are instead largely monitored by their GEFs. Thus, understanding how different members of a GEF family activate an individual small GTPase in distinct patterns is a major issue in small GTPase biology in normal cells and in diseases.
An important contribution to the functional specificity of GEFs is how they themselves are regulated. Crystallographic studies combined with biochemical studies that reconstituted GEF-stimulated GDP/GTP nucleotide exchange have been instrumental in uncovering a growing complexity of regulatory mechanisms (reviewed in ref. 1). These include autoinhibitory elements outside the catalytic GEF domain that block access to the active site (3–7), large conformational changes that release autoinhibition in response to various stimuli (8–11), positive feedback loops in which freshly produced GTP-bound GTPases stimulate GDP/GTP exchange (10, 12–15), and potentiation of nucleotide exchange by colocalization on membranes (11, 13, 16, 17).
These previous studies demonstrated that a wide range of regulatory regimes can be achieved even at the scale of a single GEF family by regulatory mechanisms that combine in multiple ways. GEFs that activate small GTPases of the Arf family (ArfGEFs), which are major regulators of many aspects of membrane traffic and organelle structure in eukaryotic cells (reviewed in refs. 18 and 19), form one of the best-characterized GEF families to date (reviewed in ref. 1), making a comprehensive view of their regulatory repertoire within reach. ArfGEFs comprise two major groups: the BIG/GBF1 group, which functions at the Golgi, and a group composed of the exchange factor for Arf6 (EFA6), brefeldin A-resistant Arf guanine nucleotide exchange factor (BRAG), and cytohesin subfamilies, which activate Arf GTPases at the cell periphery and function in various aspects of endocytosis (reviewed in ref. 20). The actual substrates of these ArfGEFs have been difficult to establish, notably because the most abundant Arf isoform, Arf1, was long believed to be excluded from the plasma membrane where the Arf6 isoform is located. Accordingly, cytohesins and BRAGs have been described as Arf6-specific GEFs in cells but are now recognized as active Arf1-GEFs (16, 21, 22), whereas EFA6 remains the sole ArfGEF considered to be strictly Arf6-specific (23, 24).
Members of the EFA6, BRAG, and cytohesin subfamilies have divergent N-terminal domains but a related domain organization in their C terminus comprising a Sec7 domain, which stimulates GDP/GTP exchange, followed by a pleckstrin homology (PH) domain, which has multiple regulatory functions. In cytohesins, the PH domain recognizes signaling phosphoinositides by its canonical lipid-binding site (25), autoinhibits the Sec7 domain by obstructing its Arf-binding site (4), and amplifies nucleotide exchange by a positive feedback loop involving its direct interaction with Arf1-GTP or Arf6-GTP (10, 13, 21). In contrast, the PH domain of BRAG is not autoinhibitory and is not involved in a feedback loop, but instead strongly potentiates nucleotide exchange by binding to polyanionic membranes without marked phosphoinositides preference (16).
How members of the EFA6 subfamily are regulated is currently unknown. These ArfGEFs are found predominantly (although not exclusively) in the brain and function in the coordination of endocytosis and actin dynamics (23, 26, 27), in the maintenance of tight junctions (28), in microtubule dynamics in Caenorhabditis elegans embryos (29), and in the formation and maintenance of dendrites (30), although the molecular details of these functions remain largely unknown. Consistent with an important role in the brain, defects in EFA6 functions have been found in neurologic disorders (31) and in human gliomas (32). The PH domain of EFA6 subfamily members drives the localization of EFA6 members to the plasma membrane (26) and it binds to PIP2 lipids (33). It is followed by a 150-residue C-terminal (Ct) domain predicted to form a coiled coil, which massively induces actin-rich membrane protrusions when expressed with the PH domain (26). The divergence of regulatory mechanisms between Sec7-PH–containing cytohesins and BRAGs prompted us to undertake a quantitative biochemical investigation of EFA6 nucleotide exchange regulation. Our findings reveal an overlooked dual specificity of EFA6 for Arf1 and Arf6 and an unprecedented regulation by a negative feedback loop, with important potential implications for the activation of Arf GTPases in endocytic events.
Results
EFA6 Is Not Autoinhibited by Its PH Domain and Is Potentiated by Membranes.
To quantify the GEF efficiency of human EFA6, we performed nucleotide exchange kinetics analyses with EFA6A constructs encompassing the Sec7 domain (EFA6Sec7) or the entire C-terminal region (EFA6Sec7-PH-Ct) along with the cellular form of Arf6 that carries a myristate attached to its N-terminal glycine (myrArf6), full-length Arf6 lacking the myristate group (unmyrArf6), or a soluble construct lacking the N-terminal α-helix (Δ13Arf6). All recombinant proteins were highly pure (Fig. 1A and Fig. S1; for Arf proteins, see refs. 16 and 34), allowing accurate kinetics analysis using tryptophan fluorescence (Table 1).
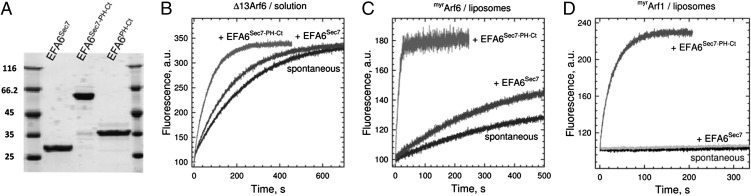
Fig. 1.
Regulation of Arf1 and Arf6 activation by EFA6 on membranes. (A) EFA6 constructs used in this study (12% SDS/PAGE, 3 μg/lane). (B) EFA6 is not autoinhibited by its PH-Ct domain. Representative tryptophan fluorescence kinetic traces for Δ13Arf6 (1 μM) activation in solution by EFA6 constructs (100 nM) as indicated. (C) EFA6 is strongly potentiated by membranes. Representative kinetic traces for myrArf6 (0.4 μM) activation by EFA6Sec7-PH-Ct (10 nM) or EFA6Sec7 (220 nM) in the presence of liposomes. (D) EFA6 is a potent Arf1GEF on membranes. Representative tryptophan fluorescence kinetic traces for myrArf1 activation by EFA6Sec7-PH-Ct (3.75 nM) or EFA6Sec7 (575 nM).
Table 1.
Catalytic efficiencies (kcat/KM, M−1s−1) of EFA6Sec7 or EFA6Sec7-PH-Ct toward Arf1 and Arf6 constructs in the absence (−) or presence (+) of 100 μM liposomes
| Substrate | EFA6Sec7 | EFA6Sec7-PH-Ct | |
| Δ13Arf6 | − | 1.70 ± 0.02 104 | 1.11 ± 0.02 105 |
| + | ND | 2.90 ± 0.07 105 | |
| unmyrArf6 | − | 6.87 ± 0.24 102 | 1.13 ± 0.07 103 |
| + | 7.80 ± 0.38 102 | 1.42 ± 0.05 106 | |
| myrArf6 | + | 5.07 ± 0.17 103 | 2.06 ± 0.08 107 |
| Δ17Arf1 | − | 3.18 ± 0.12 102 | 1.13 ± 0.05 103 |
| unmyrArf1 | + | ND | 1.67 ± 0.35 102 |
| myrArf1 | + | 5.36 ± 0.55 102 | 5.61 ± 0.13 106 |
ND, not determined.
All values are the average ± SD of three independent experiments.
We first analyzed the nucleotide exchange efficiency (kcat/KM) of the different EFA6 constructs in solution (Fig. 1B). EFA6Sec7 activated Δ13Arf6 with kcat/KM = 1.7 × 104 M−1s−1, which is in the same range as values reported for cytohesins (35) and BRAGs (16). EFA6Sec7-PH-Ct was more active than EFA6Sec7 by approximately sevenfold, indicating that the PH-Ct domain is not autoinhibitory but instead contributes a slight potentiation. EFA6Sec7 and EFA6Sec7-PH-Ct were essentially inactive toward unmyrArf6 (in the 103 M−1s−1 range), demonstrating that neither the Sec7 domain nor the PH-Ct domain carry determinants to displace the N-terminal helix of Arf6, which locks its GDP-bound form (36).
We next analyzed the efficiency of our EFA6 constructs in the presence of PIP2-containing anionic liposomes (Fig. 1C). EFA6Sec7 was insensitive to liposomes, whereas liposomes increased the catalytic efficiency of EFA6Sec7-PH-Ct toward unmyrArf6, which tethers weakly to membranes by its amphipathic N-terminal helix, but not toward Δ13Arf6, which lacks all membrane-tethering elements. The maximal kcat/KM value (2 × 107 M−1s−1) was reached for myrArf6, in which all membrane-tethering elements are present, which is approximately three orders of magnitude more efficient than EFA6Sec7/Δ13Arf6 in solution. These observations indicate that a major component of EFA6 full efficiency stems from its recruitment to membranes by its PH-Ct domain.
We next analyzed whether the efficiency of EFA6 would be sensitive to liposome characteristics, as has been shown recently for other human and bacterial ArfGEFs (11, 13, 16) (Table S1 and Fig. S2A). The addition of uncharged liposomes had essentially no effect compared with experiments carried out in solution, whereas all liposomes containing PIP2, PS, or both strongly potentiated nucleotide exchange. Surprisingly, the effects of PIP2 and PS were not additive, raising the possibility that EFA6 could bind to membranes by unspecific electrostatic interactions, as was recently reported for BRAG (16). EFA6Sec7-PH-Ct and EFA6PH-Ct bound IP3, the inositol-phosphate headgroup of PIP2, with KD values in the same range as those reported for GST-EFA6PH- and PIP2-containing vesicles (33) (Fig. S2 B and C), suggesting that EFA6 interacts with PIP2 in a specific manner. The addition of IP3 did not have a significant effect on the nucleotide exchange efficiency of EFA6Sec7-PH-Ct in solution (Fig. S2D), suggesting that binding of PIP2 to the PH domain does not exert an allosteric effect on the Sec7 domain. Finally, EFA6 did not appear to be sensitive to the curvature of liposomes (Fig. S2E) or to the presence of cholesterol (Table S1).
Taken together, the foregoing experiments suggest that membranes potentiate EFA6Sec7-PH-Ct efficiency through colocalization with myrArf6 and the resulting optimization of their relative orientations, and that the PH-Ct domain has specific interactions with PIP2 and possibly nonspecific electrostatic interactions with anionic lipids.
EFA6A Is an Efficient Arf1-GEF on Membranes.
Previous studies found that EFA6 is inactive toward Arf1 in vitro (23, 26, 37), consistent with the long-prevailing view that Arf1 is excluded from the plasma membrane. However, more recent studies established that Arf1 is activated at the plasma membrane (21, 38) and showed that both Arf1 and Arf6 are substrates for cytohesins (4, 13) and BRAGs (16, 22). This prompted us to analyze the exchange efficiency of EFA6Sec7 and EFA6Sec7-PH-Ct toward myrArf1, unmyrArf1, and Δ17Arf1 in solution and on liposomes of varying composition (Table 1 and Table S1). EFA6 did not activate Arf1 in most cases (kcat/KM in the 103 M−1s−1 range or less), confirming previous observations. However, when using EFA6 and Arf1 constructs competent for membrane binding (EFA6Sec7-PH-Ct and myrArf1) in the presence of liposomes (Fig. 1D), kcat/KM increased by more than 10,000-fold compared with Δ17Arf1/EFA6Sec7 in solution, reaching a catalytic efficiency of 5.6 ± 0.1 106 M−1s−1, which is only 3.6 lower than the catalytic efficiency toward myrArf6 on liposomes.
The insensitivity of Arf1 to activation by EFA6 in solution could be related to its switch 1 region, which is important for its interaction with ArfGEFs and has the same sequence in Arf1 and Arf6, but is structurally less dynamic in Arf1 (39) than in Arf6 (40). The Sec7 domain of EFA6 features an arginine residue (R625) that replaces a highly conserved valine that contacts the switch 1 of Arf1 in cytohesins (41), and thus would be well suited to sense this difference; however, the R625V mutation only slightly impaired Δ13Arf6 activation (Fig. S3A), and it did not restore activation of Δ17Arf1 by EFA6Sec7 (Fig. S3B). Thus, EFA6 is an efficient GEF for myrArf1 on liposomes, but why nucleotide exchange is not recapitulated with Δ17Arf1 in solution cannot be explained by a simple sequence difference.
EFA6A Is Regulated by a Negative Feedback Loop Mediated by Arf6-GTP, but Not by Arf1-GTP.
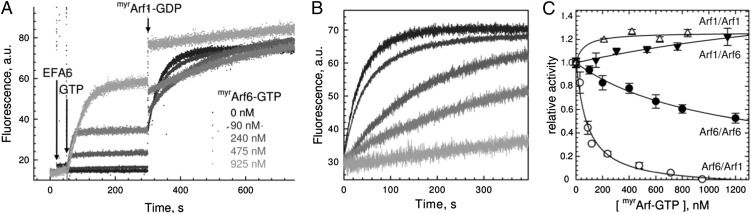
Positive feedback loops by freshly produced GTP-bound GTPases is emerging as a common trait of GEFs regulation (10, 12–15), although several examples have now been described that depart from this paradigm (7, 11, 16). To investigate the existence of a feedback mechanism in EFA6, we generated increasing amounts of myrArf-GTP on liposomes before measuring nucleotide exchange, using all four possible combinations between Arf1 and Arf6. In striking contrast with the positive feedback effects observed in other GEFs, preloading liposomes with myrArf6-GTP resulted in a significant decrease of nucleotide exchange from both myrArf1 (Fig. 2 A and B) and myrArf6 (Fig. S4A). myrArf6-GTP inhibited myrArf1 activation, with an IC50 of 81 ± 16 nM, and myrArf6 activation, with an IC50 of 944 ± 203 nM (Fig. 2C). In contrast, myrArf1-GTP had no effect on EFA6 exchange rate, regardless of whether the substrate was myrArf1 (Fig. 2C and Fig. S4B) or myrArf6 (Fig. S4C). These results identify a negative feedback loop never reported before for a GEF, and they suggest that this loop discriminates between the two Arf isoforms.
Fig. 2.
EFA6A is regulated by a negative feedback loop mediated by Arf6-GTP. (A) A representative feedback loop experiment. Increasing amounts of myrArf6-GTP (as indicated) were first generated on liposomes by EFA6Sec7-PH-Ct (6 nM). After the plateau was reached, a second exchange reaction was initiated by the addition of 0.4 μM myrArf1-GDP, from which kobs values were determined. (B) Alignment of the second part of the reaction shown in A. (C) Relative activities for the different Arf1/Arf6 combinations as a function of the initial myrArf-GTP amount generated on liposomes.
We next investigated the mechanism through which Arf6-GTP mediates its negative feedback effect. A simple possibility could be that Arf6-GTP competes with Arf-GDP as an inhibitor or a substrate of the Sec7 active site. The addition of increasing concentrations of Δ13Arf6-GTP had no effect on the activation of Δ13Arf6 by EFA6Sec7 in solution, indicating that Arf6-GTP does not act through a simple competition effect (Fig. S5A). Likewise, EFA6Sec-PH-Ct was unable to stimulate GTP-GDP exchange from myrArf6-GTP on liposomes (Fig. S5B), indicating that Arf6-GTP is not a substrate of EFA6.
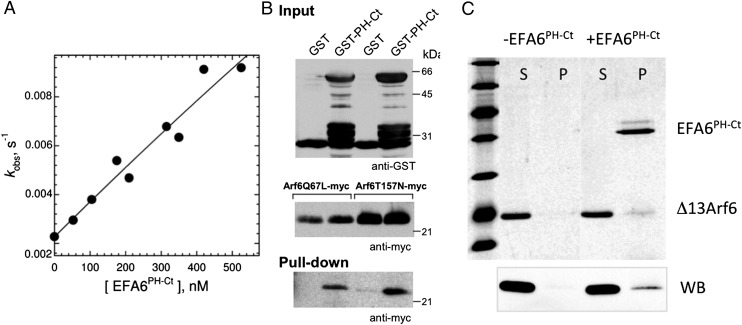
Alternatively, myrArf6-GTP could act through an allosteric interaction with the PH-Ct domain. To test this hypothesis, we added increasing amounts of purified EFA6PH-Ct to liposomes preloaded with a fixed amount of myrArf6-GTP before starting the exchange reaction with EFA6Sec-PH-Ct and myrArf1. EFA6PH-Ct relieved the feedback effect in a dose-dependent manner (Fig. 3A and Fig. S5C), suggesting that EFA6PH-Ct reverses the negative feedback loop by titrating myrArf6-GTP. Confirming that EFA6PH-Ct binds to Arf6-GTP, Arf6Q67L and Arf6T157N expressed in cells, both loaded predominantly with GTP (42), were efficiently pulled down by purified GST-EFA6PH-Ct (Fig. 3B). Finally, purified EFA6PH-Ct was able to recruit Δ13Arf6-GTP (which is soluble and does not bind to membranes) to liposomes (Fig. 3C), indicating that Arf6-GTP and EFA6PH-Ct interact directly and independently of the catalytic Sec7 domain.
Fig. 3.
The negative feedback effect is mediated by interaction of Arf6-GTP with EFA6PH-Ct. (A) EFA6PH-Ct reverses the negative feedback effect in a dose-dependent manner. myrArf1 activation was performed as in Fig. 2A, except with increasing amounts of EFA6PH-Ct added before the first exchange reaction (fixed myrArf6-GDP concentration of 400 nM). (B) Pull-down of myc-tagged GTP-bound Arf6 mutants from cellular extracts by purified GST-tagged EFA6PH-Ct. (C) Cosedimentation of Δ13Arf6-GTP with liposome-bound EFA6PH-Ct analyzed by SDS/PAGE stained with Sypro orange (Upper) and by Western blot analysis with an anti-Arf antibody (Lower). S, supernatant; P, pellet.
This ensemble of experiments suggests that the GEF activity of EFA6 is regulated by a negative feedback loop actuated specifically by myrArf6-GTP, and that the underlying mechanism involves an allosteric interaction of myrArf6-GTP with the PH-Ct domain of EFA6.
Discussion
Higher eukaryotes have three subfamilies of ArfGEFs involved in endocytic events— cytohesins, BRAGs, and EFA6—all of which carry a PH domain associated with a catalytic Sec7 domain. Previous studies highlighted that this organization determines different regulatory mechanisms in cytohesins (4, 10, 13, 21) and in BRAGs (16), but how the GEF activity of EFA6 is regulated by its PH domain remained unknown. By analyzing the regulation of EFA6A activity toward Arf1 and Arf6 using nucleotide exchange reactions reconstituted on artificial membranes, this study reveals unexpected commonalities and divergences among the three ArfGEF subfamilies, yielding a framework for envisioning how they work on peripheral membranes.
Our analysis shows that the Sec7 domain of EFA6 has a GEF efficiency toward Arf6 in the same range as that of cytohesins (35) and BRAGs (16). Likewise, the C-terminal region of EFA6, which includes the PH domain, supports strong potentiation of nucleotide exchange by membranes (by approximately three orders of magnitude), which is in the range reported for BRAGSec7-PH (16). Our study also reveals that EFA6Sec7-PH-Ct is almost as potent an activator of Arf1 as Arf6 on membranes, possibly explaining why EFA6 leads to Arf1 activation in cell cultures (21). Taken together, these observations reveal that EFA6 do not have major discrepancies with cytohesins and BRAGs in terms of intrinsic GEF efficiency or substrate preference.
In contrast, the regulatory modalities of EFA6 depart from those of cytohesins and BRAGs in various respects (Table 2). Notably, EFA6 is not autoinhibited by its PH domain and is regulated by a negative feedback loop mediated by interaction of its PH-Ct domains with Arf6-GTP. This interaction is reminiscent of the mechanism of feedback control of cytohesins by interaction of their PH domain with Arf-GTP (4, 10, 13, 21); however, the presence of a large C-terminal domain, which likely associates with the PH domain in EFA6 as suggested by small-angle X-ray scattering (SAXS) analysis (Fig. S1B), and the opposite regulatory effect of Arf-GTP suggest that the interaction probably is structurally very different. Furthermore, EFA6 discriminates between Arf1 and Arf6 as effectors of the negative feedback loop, an effect not seen in cytohesins, which are equally sensitive to activation by Arf1-GTP and Arf6-GTP (13). It should also be noted that EFA6 is potentiated equally well by PIP2-containing or anionic membranes but does not have a strict requirement for a specific phosphoinositide, and thus has membrane preferences distinct from those of both cytohesins (25) and BRAGs (16). Given the strong similarities between EFA6 subfamily members in their Sec7-PH-Ct regions (26), we predict that the regulatory modalities found for EFA6A in this study apply to all members of the subfamily. Whether their N-terminal regions, which are more divergent than their Sec7-PH-Ct regions (26) and have functions independent of Arf GTPases (29), add another layer of regulation remains to be investigated.
Table 2.
Regulatory regime of Sec7-PH–containing endocytic ArfGEFs
Negative and positive feedback loops, such as found in EFA6 and cytohesins, are crucial elements that can be coupled to determine the timing and amplitude of various cellular behaviors (reviewed in ref. 43), suggesting the attractive possibility that EFA6 and cytohesins can leverage their regulatory regimes to work in concert. Remarkably, the negative feedback loop in EFA6 would provide a simple solution to the absolute requirement of cytohesins for activation by Arf-GTP before their GEF activity is amplified by a positive feedback loop (13) (Fig. 4). Being devoid of constitutive autoinhibition by its PH domain or of a strict requirement for a signaling lipid for its recruitment to the plasma membrane, EFA6 could produce basal levels of Arf6-GTP, with its negative feedback loop working initially as a limiter (Fig. 4, Left). Cytohesins, on recruitment to the plasma membrane induced by an increase in PIP2 or PIP3, could then mobilize this dormant Arf6-GTP pool to trigger their own GEF activity (Fig. 4, Center). The resulting burst in Arf6-GTP and Arf1-GTP production would then concurrently sustain the GEF activity of cytohesins and turn off the GEF activity of EFA6, making cytohesins the major active ArfGEFs at that stage (Fig. 4, Right). In this model, EFA6 and Arf6 would have a general (although not necessarily unique) function in tuning the level of cytohesin activity.
Fig. 4.
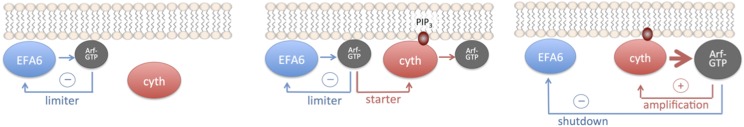
Model of coupled negative-positive feedback circuit between EFA6 and cytohesins.
The foregoing model involving an ArfGEF circuit with differential activation of Arf1 and Arf6 is supported by certain lines of evidence. Notably, a separation of Arf6 activation and effector functions has been observed in endocytic receptor recycling, in which a dormant pool of Arf6-GTP was observed in clathrin-coated pits that became mobilized only later in fast endocytic events (44). This finding would be consistent with the proposed role of EFA6 in sustaining a basal amount of Arf6-GTP. Likewise, it has been shown that Arf1 and Arf6 have different peaks of activation during phagocytosis, with Arf1 activation delayed and lasting longer than Arf6 activation, although the GEFs involved are unknown (38). Recent work also showed that Arf6 and Arf1 must be activated in sequence for WRC-mediated actin assembly, and that this requires a cytohesin and either EFA6 or BRAG working in concert (45).
It should be noted that a corollary of the feedback circuit model is that expression of dominant active or inactive Arf GTPases or of ArfGEFs and ArfGEF mutants is predicted to interfere with multiple, possibly opposing steps, which should be taken into account when interpreting the resulting phenotypes. Accordingly, the challenge to investigate such feedback circuit will be to find ways to perturb and monitor individual steps separately. Future investigations of the structural mechanism whereby Arf6-GTP and the PH-Ct domain of EFA6 establish the negative feedback loop should be an important step toward this goal.
Experimental Procedures
Expression and Purification of Recombinant Proteins.
Arf1 and Arf6 constructs were expressed in Escherichia coli, purified, and, when appropriate, modified by coexpression or in vitro myristoylation and/or loaded with GDP or GTP nucleotides as described previously (16, 34). Cloning, expression, and purification of human EFA6ASec7 (residues 527–727), EFA6ASec7-PH-Ct (residues 527–1024), and EFA6APH-Ct (residues 730–1024) are described in SI Experimental Procedures. EFA6PH-Ct and EFA6Sec7-PH-Ct were further characterized by synchrotron SAXS (Fig. S1A) and by size exclusion chromatography with multiangle light scattering (Fig. S1B), respectively, which demonstrated that the constructs are monomers, and thus the C-terminal domain does not form a homodimeric coiled coil.
Cell Culture, Reagents, and Antibodies.
Baby hamster kidney cells (BHK-21) were grown in BHK-21 medium (Gibco-BRL), containing 5% FCS, 10% Tryptose phosphate broth, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. Mouse mAb against the myc epitope (clone 9E10; Roche Diagnostics) was obtained from Jackson ImmunoResearch.
Liposomes.
All lipids were obtained from Avanti Polar Lipids. Liposomes were prepared as described previously (13) in a buffer containing 50 mM Hepes pH 7.4 and 120 mM potassium acetate. Unless specified otherwise, liposomes contained 34% phosphatidylcholine (PC), 14% phosphatidylethanolamine (PE), 21% phosphatidylserine (PS), 0.7% phosphatidylinositol-4,5-diphosphate (PIP2), and 30% cholesterol and were extruded through a 0.2-μm filter (Whatman).
Nucleotide Exchange Kinetics.
Nucleotide exchange kinetics were performed by monitoring tryptophan fluorescence (λexc = 298 nm; λexc = 340 nm) on a fluorimeter (Cary) equipped with stirring and thermostating devices. Experiments were performed in HKM buffer (50 mM Hepes buffer, pH 7.4, 120 mM potassium acetate, and 1 mM MgCl2) supplemented with 1 mM DTT at 37 °C. Pseudofirst-order rate constants (kobs) were obtained by fitting the kinetic traces with exponential functions, except for EFA6Sec7-PH-Ct and myrArf6, which had more complex kinetics, as was previously observed with other Arf6GEFs (16). In this case, kobs values were derived from initial velocities as described in SI Experimental Procedures. kcat/KM values were determined from kobs measured over a range of GEF concentrations, as described previously (16, 46). The liposome concentration was 100–200 μM, at which the protein/lipid ratio is sufficiently high for most of the lipid surface to remain accessible to the proteins (13).
Binding Experiments.
Equilibrium dissociation constants (Kd) of IP3 to EFA6PH-Ct or EFA6Sec7-PH-Ct were determined from the decrease in fluorescence emission at 340 nm by fitting the following equation: ΔF340nm= ΔFmax× [L]/(Kd + [L], where [L] is the concentration of IP3. For pull-down experiments, BHK-21 cells were transfected with plasmids encoding C-terminally myc-tagged Arf6Q67L or Arf6T157N, using Jet Pei reagent. After 24 h, cells were lysed in 50 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2, 10% glycerol, 1% Triton-X100, 2 mM DTT, and a mixture of protease inhibitors (Roche), and then centrifuged at 15,000 × g for 10 min at 4 °C. Supernatants were incubated with 2 μM GST constructs in the presence of 0.75% BSA and glutathione Sepharose beads overnight at 4 °C. The beads were then washed, and bound proteins were eluted using SDS sample buffer and separated on SDS/PAGE.
The presence of Arf6 in the eluate was detected by Western blot analysis using the anti-tag antibodies. For cosedimentation experiments, purified ∆13Arf6-GTP (1.5 μM) and EFA6PH-Ct proteins (0.5 μM) were incubated with sucrose-loaded fluorescent liposomes (34.3% PC, 14% PE, 21% PS, 0.7% PIP2, 30% cholesterol, and 0.2% NBD-PE) extruded on a 0.2-μm filter. Liposomes were sedimented for 20 min at 400,000 × g, checked by fluorescence using a FujiLAS 3000 imager equipped with a CCD camera, and analyzed on 15% SDS/PAGE stained with Sypro-orange and by Western blot analysis with the 1D9 anti-Arf monoclonal antibody (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We are grateful to Laetitia Cormier and Véronique Henriot (Laboratoire d’Enzymologie et Biochimie Structurales/IMAGIF, Centre National de la Recherche Scientifique) for cloning various plasmids, to the scientific staff at the SWING beamline (SOLEIL synchrotron, Gif-sur-Yvette, France) for SAXS data collection and analysis, and to Bruno Antonny (Centre National de la Recherche Scientifique, Valbonne, France) for a critical reading of the manuscript. This work was supported by grants from the Association pour la Recherche sur le Cancer (to J.C. and D.P.), from the Fondation pour la Recherche Médicale (to M.F.-K.), and from the Agence Nationale de la Recherche (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409832111/-/DCSupplemental.
References
- 1.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93(1):269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 2.Cherfils J, Zeghouf M. Chronicles of the GTPase switch. Nat Chem Biol. 2011;7(8):493–495. doi: 10.1038/nchembio.608. [DOI] [PubMed] [Google Scholar]
- 3.Rehmann H, Das J, Knipscheer P, Wittinghofer A, Bos JL. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature. 2006;439(7076):625–628. doi: 10.1038/nature04468. [DOI] [PubMed] [Google Scholar]
- 4.DiNitto JP, et al. Structural basis and mechanism of autoregulation in 3-phosphoinositide–dependent Grp1 family Arf GTPase exchange factors. Mol Cell. 2007;28(4):569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amor JC, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280(2):1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 6.Yu B, et al. Structural and energetic mechanisms of cooperative autoinhibition and activation of Vav1. Cell. 2010;140(2):246–256. doi: 10.1016/j.cell.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwig JS, et al. Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. eLife. 2013;2:e00813. doi: 10.7554/eLife.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gureasko J, et al. Role of the histone domain in the autoinhibition and activation of the Ras activator Son of Sevenless. Proc Natl Acad Sci USA. 2010;107(8):3430–3435. doi: 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sondermann H, et al. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell. 2004;119(3):393–405. doi: 10.1016/j.cell.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Malaby AW, van den Berg B, Lambright DG. Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors. Proc Natl Acad Sci USA. 2013;110(35):14213–14218. doi: 10.1073/pnas.1301883110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Folly-Klan M, et al. A novel membrane sensor controls the localization and ArfGEF activity of bacterial RalF. PLoS Pathog. 2013;9(11):e1003747. doi: 10.1371/journal.ppat.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margarit SM, et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112(5):685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 13.Stalder D, et al. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem. 2011;286(5):3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell. 2012;22(4):799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina F, et al. Activated RhoA is a positive feedback regulator of the Lbc family of Rho guanine nucleotide exchange factor proteins. J Biol Chem. 2013;288(16):11325–11333. doi: 10.1074/jbc.M113.450056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aizel K, et al. Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2. PLoS Biol. 2013;11(9):e1001652. doi: 10.1371/journal.pbio.1001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera M, et al. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci. 2014;127(Pt 5):1043–1051. doi: 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D’Souza-Schorey C, Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7(5):347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 19.Donaldson JG, Jackson CL. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12(6):362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casanova JE. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8(11):1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 21.Cohen LA, et al. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18(6):2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jian X, Gruschus JM, Sztul E, Randazzo PA. The pleckstrin homology (PH) domain of the Arf exchange factor Brag2 is an allosteric binding site. J Biol Chem. 2012;287(29):24273–24283. doi: 10.1074/jbc.M112.368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franco M, et al. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18(6):1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macia E, Partisani M, Paleotti O, Luton F, Franco M. Arf6 negatively controls the rapid recycling of the β2 adrenergic receptor. J Cell Sci. 2012;125(Pt 17):4026–4035. doi: 10.1242/jcs.102343. [DOI] [PubMed] [Google Scholar]
- 25.Cronin TC, DiNitto JP, Czech MP, Lambright DG. Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J. 2004;23(19):3711–3720. doi: 10.1038/sj.emboj.7600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Derrien V, et al. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J Cell Sci. 2002;115(Pt 14):2867–2879. doi: 10.1242/jcs.115.14.2867. [DOI] [PubMed] [Google Scholar]
- 27.Brown FD, Rozelle AL, Yin HL, Balla T, Donaldson JG. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J Cell Biol. 2001;154(5):1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein S, Partisani M, Franco M, Luton F. EFA6 facilitates the assembly of the tight junction by coordinating an Arf6-dependent and -independent pathway. J Biol Chem. 2008;283(44):30129–30138. doi: 10.1074/jbc.M803375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nat Cell Biol. 2010;12(12):1235–1241. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi S, et al. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J Neurosci. 2006;26(18):4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitaliani R, et al. Paraneoplastic encephalitis, psychiatric symptoms, and hypoventilation in ovarian teratoma. Ann Neurol. 2005;58(4):594–604. doi: 10.1002/ana.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M, et al. EFA6A enhances glioma cell invasion through ADP ribosylation factor 6/extracellular signal-regulated kinase signaling. Cancer Res. 2006;66(3):1583–1590. doi: 10.1158/0008-5472.CAN-05-2424. [DOI] [PubMed] [Google Scholar]
- 33.Macia E, et al. The pleckstrin homology domain of the Arf6-specific exchange factor EFA6 localizes to the plasma membrane by interacting with phosphatidylinositol 4,5-bisphosphate and F-actin. J Biol Chem. 2008;283(28):19836–19844. doi: 10.1074/jbc.M800781200. [DOI] [PubMed] [Google Scholar]
- 34.Padovani D, Zeghouf M, Traverso JA, Giglione C, Cherfils J. High-yield production of myristoylated Arf6 small GTPase by recombinant N-myristoyl transferase. Small GTPases. 2013;4(1):3–8. doi: 10.4161/sgtp.22895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeeh JC, et al. Dual specificity of the interfacial inhibitor brefeldin a for arf proteins and sec7 domains. J Biol Chem. 2006;281(17):11805–11814. doi: 10.1074/jbc.M600149200. [DOI] [PubMed] [Google Scholar]
- 36.Ménétrey J, Macia E, Pasqualato S, Franco M, Cherfils J. Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factor specificity. Nat Struct Biol. 2000;7(6):466–469. doi: 10.1038/75863. [DOI] [PubMed] [Google Scholar]
- 37.Macia E, Chabre M, Franco M. Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J Biol Chem. 2001;276(27):24925–24930. doi: 10.1074/jbc.M103284200. [DOI] [PubMed] [Google Scholar]
- 38.Beemiller P, Hoppe AD, Swanson JA. A phosphatidylinositol-3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcgamma receptor-mediated phagocytosis. PLoS Biol. 2006;4(6):e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buosi V, et al. Insight into the role of dynamics in the conformational switch of the small GTP-binding protein Arf1. J Biol Chem. 2010;285(49):37987–37994. doi: 10.1074/jbc.M110.134445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biou V, et al. SAXS and X-ray crystallography suggest an unfolding model for the GDP/GTP conformational switch of the small GTPase Arf6. J Mol Biol. 2010;402(4):696–707. doi: 10.1016/j.jmb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426(6966):525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 42.Klein S, Franco M, Chardin P, Luton F. Role of the Arf6 GDP/GTP cycle and Arf6 GTPase-activating proteins in actin remodeling and intracellular transport. J Biol Chem. 2006;281(18):12352–12361. doi: 10.1074/jbc.M601021200. [DOI] [PubMed] [Google Scholar]
- 43.Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322(5900):390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montagnac G, et al. Decoupling of activation and effector binding underlies ARF6 priming of fast endocytic recycling. Curr Biol. 2011;21(7):574–579. doi: 10.1016/j.cub.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc Natl Acad Sci USA. 2013;110(42):16880–16885. doi: 10.1073/pnas.1311680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Béraud-Dufour S, et al. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17(13):3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.