Significance
Vaccination is a powerful means of preempting or treating disease. The process depends on successful recognition of a foreign entity (an antigen) to elicit a strong immune response. The delivery of an antigen encoded as a DNA molecule (a genetic antigen) requires the assistance of a vector to facilitate the process of gene expression within immune system sentinels termed antigen-presenting cells (APCs). In this study, two normally distinct vectors (a bacterial cell and a synthetic polymer) were combined to generate a hybrid vector. The new vector coupled synergistic mechanisms to assist and improve gene delivery to APCs. Furthermore, the hybrid vector provides unique and complimentary engineering capabilities that were demonstrated to tailor and improve APC gene delivery further.
Keywords: nonviral vectors, cationic polymers, bactofection, vaccination, immunization
Abstract
Genetic vaccines offer a treatment opportunity based upon successful gene delivery to specific immune cell modulators. Driving the process is the vector chosen for gene cargo packaging and subsequent delivery to antigen-presenting cells (APCs) capable of triggering an immune cascade. As such, the delivery process must successfully navigate a series of requirements and obstacles associated with the chosen vector and target cell. In this work, we present the development and assessment of a hybrid gene delivery vector containing biological and biomaterial components. Each component was chosen to design and engineer gene delivery separately in a complimentary and fundamentally distinct fashion. A bacterial (Escherichia coli) inner core and a biomaterial [poly(beta-amino ester)]-coated outer surface allowed the simultaneous application of molecular biology and polymer chemistry to address barriers associated with APC gene delivery, which include cellular uptake and internalization, phagosomal escape, and intracellular cargo concentration. The approach combined and synergized normally disparate vector properties and tools, resulting in increased in vitro gene delivery beyond individual vector components or commercially available transfection agents. Furthermore, the hybrid device demonstrated a strong, efficient, and safe in vivo humoral immune response compared with traditional forms of antigen delivery. In summary, the flexibility, diversity, and potential of the hybrid design were developed and featured in this work as a platform for multivariate engineering at the vector and cellular scales for new applications in gene delivery immunotherapy.
Vaccines are often regarded as the most cost-effective advancement of modern medicine (1). Gene therapy has emerged as an alternative approach to the classical vaccine paradigm by using the delivery of specific therapeutic cargo for the purpose of altering gene expression (2, 3). In this context, the delivery of nucleic acids is accomplished by a variety of biological or synthetic vectors. Through specific engineering tools, each vector is designed to influence antigen-presenting cell (APC) gene expression levels to modulate an immune response toward the generation of antigen reactivity and memory. Vector-mediated gene delivery efficacy is strongly correlated with overcoming APC barriers, such as cellular uptake, phagosomal/lysosomal escape, nucleic acid unpackaging, nuclear translocation (excluding RNA-based therapeutics), and sustained gene expression (4). To be clinically relevant, vectors must also exert minimal to no cytotoxicity.
The ability to deliver nucleic acids and proteins to mammalian cells has been demonstrated independently by both biomaterial-based nanoparticles and bacteria. Of the biomaterial vectors, cationic polymers (CPs) generally feature simple and scalable synthetic schemes capable of being tailored to specific applications (5). Specifically, poly(beta-amino esters) (PBAEs) are a class of CPs recognized for their facile synthesis and elevated levels of gene delivery (compared with commercially available reagents) (6–10). CPs also typically demonstrate or can be designed to exhibit low cytotoxicity. Bacterial vectors provide an orthogonal set of engineering tools to influence gene delivery. In particular, Escherichia coli, a rod-shaped, Gram-negative bacterium that is ∼2 μm in length and 0.5 μm in diameter, has been identified as an ideal gene carrier due to simple and scalable culture protocols, a rapid growth rate, and a wealth of established molecular biology manipulation techniques (11). Nonpathogenic strains of E. coli have exhibited minimal toxicity and immunogenicity (12–15), further supported by in vivo and in vitro delivery of plasmid (12–14, 16, 17) and larger DNA constructs (18, 19), shRNA (20), and immunogenic peptides (17, 21, 22).
An alternative option to address the requirements of gene delivery is to combine the individual advantages of vectors, such as E. coli and CPs, and to leverage dual vector-specific toolsets to overcome APC gene delivery barriers. For example, E. coli, which inherently maintains plasmid DNA (pDNA) and natively promotes phagocytic uptake by APCs, can be engineered to escape the lysosome by expression of a pore-forming listeriolysin O (LLO) protein (16, 17, 21–24). Conversely, CPs facilitate uptake by generalized endocytosis mechanisms and instigate lysosomal escape by the “proton sponge effect” (4, 5). Combined, the innate and malleable features of these normally disparate vectors offer the potential to complement and enhance individual capabilities while simultaneously minimizing limitations.
Thus, we report the generation of a hybrid biosynthetic vector that combines the capabilities of both bacterial and CP components for targeted gene delivery to APCs (SI Appendix, Fig. S1). Ninety-one structurally diverse PBAEs were synthesized and screened after surface attachment to LLO-producing E. coli for in vitro gene delivery to murine macrophages. After multiple rounds of screening, an optimal PBAE and bacterial strain were identified that, when combined together, possessed gene delivery potency greater than either vector in isolation. To demonstrate the engineering potential of the hybrid device, each individual vector was modified using vector-associated tools. Specifically, the lethal lysis gene E (LyE) from bacteriophage ΦX174 was incorporated into bacterial strains, which resulted in significant improvements to APC gene delivery and cytotoxicity. Similarly, mannose was attached to the terminal end of the optimal PBAE before hybrid device formation, resulting in increased vector uptake and gene delivery. Finally, the hybrid vector was successfully tested in the context of in vivo humoral immune response. The results and combined features of this new vector offer a promising platform for future applications in genetic vaccination as a byproduct of the duality in vector composition and engineering capability.
Results and Discussion
PBAE Synthesis.
The synthesis of PBAEs proceeds via the conjugate (Michael) addition of amines to acrylate groups (6). Building upon high-throughput polymer synthesis schemes, we produced a library of 91 PBAEs from a diverse set of monomers (9, 10, 25, 26) (SI Appendix, Fig. S1A and Table S1). Our library was synthesized on a 1- to 2-g scale with increased monomer concentrations to provide greater control of stoichiometry, increase polymer molecular weight and end-group termination, and reduce intramolecular cyclization (25). Each polymer was analyzed by gel permeation chromatography (GPC), and library averages for molecular weight (Mw) and polydispersity index were 5,300 and 1.4, respectively, consistent with previous reports (6, 8, 10, 27) (SI Appendix).
Bacterial Strain Generation.
Vectors were first engineered to deliver a mammalian expression reporter plasmid (pCMV-Luc) and to test three expression parameters associated with LLO: gene dosage, promoter strength, and gene expression regulation (SI Appendix, Table S2). An inducible T7 promoter (28) and a constitutive tet promoter were selected in this study to accommodate expression variation. Both LLO expression cassettes were either maintained on multicopy plasmids or integrated into the chromosome of BL21(DE3) at the clpP gene location (16).
High-Throughput Gene Delivery Screen.
The hybrid biosynthetic gene delivery vector formation process is driven by electrostatic interactions between positively charged polymers and the negatively charged E. coli outer membrane. To expedite the combinatorial screening process, a high-throughput assay was developed to investigate the surface addition of 91 structurally diverse PBAEs (SI Appendix, Fig. S1B). The screen was segmented into three sequential stages of selection. First, all 91 PBAEs were added in three dosages (0.1, 1.0, and 10 mg/mL) to YWT7-hly/pCMV-Luc [SI Appendix, Table S2; strain 1 (S1)], chosen initially because of greater relative gene delivery compared with strains 2–5 (16, 23). Upon coincubation at 22 °C for 15 min, hybrid devices were added to 96-well plated RAW264.7 cells at a 10:1 multiplicity of infection (MOI; ratio of hybrid devices to macrophages). Gene delivery was evaluated by measuring the amount of luciferase expression, standardizing by total protein content, and comparing with untreated bacterial vectors. Numerous improvements in gene delivery beyond bactofection alone were observed across PBAE types and dosage levels, with zeta potential readings providing further support for the surface modification of the bacterial component of the hybrid vector (SI Appendix, Figs. S2 and S3). From these results, 20 PBAEs (A5, A11, B6, B9, B11, C2, C5, C9, C10, C13, D1, D7, D9, D13, E1, E7, F1, F7, F11, and G2) were selected for further analysis in a secondary screen. The monomers occurring with the highest frequency were C diacrylates (five of 20 PBAEs) and four amines (1, 7, 9, and 11; three of 20 PBAEs each). Interestingly, monomers 1 and 7 contain two amine groups, whereas monomers 9 and 11 contain two alcohol groups. Both functionalities have been previously associated with increased gene delivery (25, 26, 29).
The secondary screen expanded the polymer dosage range (0.1, 0.25, 0.5, and 1.0 mg/mL) and evaluated MOI dependencies. Selection was predicated upon hybrid vectors exceeding gene delivery levels of a positive control (FuGENE 6; Promega) and both the polymer and bacterial vectors in isolation. Hybrid device gene delivery efficacy was influenced by the MOI, and within each MOI, different polymer dosage trends were observed (SI Appendix, Fig. S4). Specifically, at the lowest MOI (1:1), most hybrid devices (14 of 20) demonstrated positively correlated dosage-dependent increases in gene delivery. Each of these combinations surpassed the bacterial control but, with the exception of D9, failed to meet or surpass the FuGENE 6 control, possibly because of the low dosage of pDNA delivered at this MOI. At a 1:1 MOI, there are 3 × 104 hybrid vectors per well, and each bacterially-maintained pCMV-Luc plasmid (regulated by the pBR322 origin) is held at 20–40 copies per bacterium (30), resulting in a final pDNA delivery dosage of 0.008 ng per well (Table 1). Taking the amount of pDNA into account, a standardized delivery efficiency metric, determined by normalizing luminescence readings by pDNA amount (23), was used to allow better comparison between vectors. Across the three MOIs and PBAE dosages, the hybrid vector containing polymer D9 was the only vector to exceed bacterial and synthetic controls consistently. Interestingly, substantially different D9 dosage trends were observed across the three MOIs. At the lowest MOI (1:1), hybrid vectors demonstrated a positive dosage-dependent correlation with respect to both gene delivery and delivery efficiency. However, at 10:1 and 100:1 MOIs, optimal gene delivery and delivery efficiency were observed at the lower D9 dosages of 0.1 and 0.25 mg/mL, respectively, and gene delivery and delivery efficiency were reduced with continued D9 addition. In the third stage of assessment, D9 hybridization was tested across E. coli strains producing different levels of LLO (SI Appendix, Fig. S5 and Table S2). Gene delivery across all strains was improved with the YWT7-hly/pCMV-Luc:D9 hybrid demonstrating the greatest relative transfection values. To assess gene delivery metrics further, additional scoring criteria were introduced to the vectors of Table 1. In particular, population-based delivery efficiency was analyzed by introducing a separate EGFP plasmid to the S1 strain (SI Appendix, Table S2) and monitoring the percentage of GFP-positive cells. When combined with APC viability, a more comprehensive delivery efficiency metric, which we term the total gene delivery performance (TGDP), emerges as the heading of the last column of Table 1. Using TGDP, it is clear that the hybrid vector outperforms an expanded set of synthetic controls that included seven commercially available transfection agents in addition to D9 and mannosylated D9 (to be described later).
Table 1.
Vectors used during transfection studies and gene delivery comparison
| Vector | MOI | D9 dosage, mg | Vector size, nm | No. of vectors per well | pDNA per vector, ng | Total pDNA, ng | Gene delivery (GD), luminescence per μg of protein | Delivery efficiency, GD per total ng of pDNA | GFP-positive cells | Delivery efficiency × % of GFP cells | Viable cells | Delivery efficiency × % of GFP cells × % of viable cells (TGDP) |
| S1: D9 hybrid | 1:1 | 0 | ∼2,000 | 30,000 | 2.7 × 10−7* | 0.008 | 8 | 970 | 8% | 78 | 99% | 77 |
| 0.1 | 11 | 1,400 | ||||||||||
| 0.25 | 15 | 1,880 | ||||||||||
| 0.4 | 19 | 2,370 | 10% | 240 | 100% | 240 | ||||||
| 0.5 | 19 | 2,410 | ||||||||||
| 1 | 22 | 2,740 | ||||||||||
| 10:1 | 0 | ∼2,000 | 300,000 | 2.7 × 10−7* | 0.08 | 110 | 1,360 | 32% | 430 | 82% | 350 | |
| 0.1 | 180 | 2,250 | ||||||||||
| 0.25 | 180 | 2,280 | ||||||||||
| 0.4 | 220 | 2,730 | 27% | 730 | 88% | 650 | ||||||
| 0.5 | 170 | 2,090 | ||||||||||
| 1 | 130 | 1,630 | ||||||||||
| 100:1 | 0 | ∼2,000 | 3,000,000 | 2.7 × 10−7* | 0.8 | 1,190 | 1,480 | 79% | 1,170 | 55% | 640 | |
| 0.1 | 2,350 | 2,930 | ||||||||||
| 0.25 | 2,460 | 3,070 | ||||||||||
| 0.4 | 2,360 | 2,940 | 67% | 1,970 | 58% | 1,140 | ||||||
| 0.5 | 1,820 | 2,260 | ||||||||||
| 1 | 1,580 | 1,960 | ||||||||||
| D9 polyplex | N/A | 0.018 | 150 | 8.3 × 109† | 7.2 × 10−8 | 600 | 30 | 0.051 | 7% | 0.003 | 94% | 0.003 |
| D9-Man polyplex | N/A | 0.018 | 150 | 8.3 × 109† | 7.2 × 10−8 | 600 | 40 | 0.067 | 23% | 0.015 | 95% | 0.015 |
| FuGENE 6 (Promega) | N/A | N/A | N/A | N/A | N/A | 200 | 44 | 0.219 | 31% | 0.068 | 100% | 0.068 |
| FuGENE HD (Promega) | N/A | N/A | N/A | N/A | N/A | 200 | 100 | 0.521 | 55% | 0.285 | 100% | 0.285 |
| ViaFECT (Promega) | N/A | N/A | N/A | N/A | N/A | 100 | 22 | 0.225 | 16% | 0.035 | 100% | 0.035 |
| OmniFect (TransOMIC Technologies) | N/A | N/A | N/A | N/A | N/A | 100 | 38 | 0.375 | 20% | 0.075 | 100% | 0.075 |
| Xfect (Clontech Laboratories) | N/A | N/A | N/A | N/A | N/A | 500 | 33 | 0.067 | 57% | 0.038 | 78% | 0.030 |
| JET-PEI (Polyplus transfection) | N/A | N/A | N/A | N/A | N/A | 250 | 130 | 0.511 | 30% | 0.152 | 85% | 0.129 |
| GeneJuice (EMD Millipore) | N/A | N/A | N/A | N/A | N/A | 100 | 3 | 0.027 | 1% | 0.000 | 100% | 0.000 |
N/A, not applicable.
Amount of bacterially maintained pDNA was estimated using a pBR322 origin, which is regulated at 20–40 copies per bacterium, and the molecular weight of the plasmid [estimated as plasmid base pairs × 650 (daltons per base pair)].
Polyplex number was calculated by dividing the total mass of D9 added per well at a 30:1 polymer-to-pDNA ratio by an estimated mass of one polyplex. Individual polyplex mass was estimated using the measured effective diameter to calculate volume and a density of 1.22 g/mL (31).
Hybrid Vector Characterization.
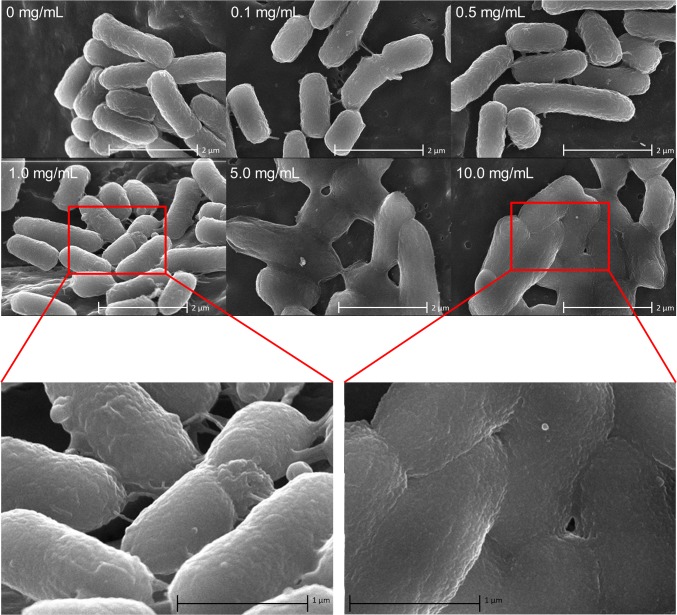
To confirm D9 addition visually, hybrid and bacterial vectors were analyzed by SEM, revealing dosage-dependent surface modification (Fig. 1). Interestingly, synergistic dosage ranges (<1 mg/mL) contain individualized cells with polymeric extensions (Fig. 1, Left). Treatment with excess polymer (5 and 10 mg/mL), however, results in heavy surface coating and systemic adherence between hybrid vectors (Fig. 1, Right). Coalescence of hybrid vectors is a likely cause for the decline in gene delivery at elevated polymer dosages (SI Appendix, Fig. S4), due to the inability of APCs to phagocytose resulting conglomerates (31, 32). Additional hybrid device characterization studies, including in vitro APC interaction responses and physical property measurements, are presented in SI Appendix, Figs. S6–S8.
Fig. 1.
SEM of bacterial and hybrid vectors. Images of YWT7-hly/pCMV-Luc treated with various concentrations of D9 are shown.
Bacteriophage ΦX174 LyE Incorporation.
Previously, we identified a positive correlation between bacterial cell wall weakening and gene delivery (12). To investigate this relationship further and demonstrate the biological engineering potential of the hybrid device, we introduced and conditionally expressed bacteriophage ΦX174 LyE. Production of the resulting endolysin leads to the formation of transmembrane tunnels through the bacterial cellular envelope (33, 34). Attenuated bacterial vectors will presumably experience further membrane destabilization upon lysosomal entrapment, facilitating increased release of LLO and pDNA. Elevated concentrations of LLO will instigate additional lysosomal rupture, which is expected to increase the effective concentration of cytosolic pDNA for nuclear translocation.
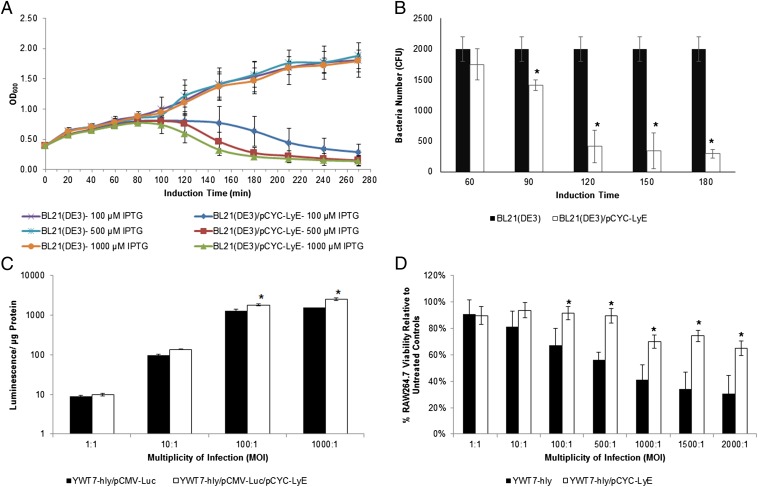
Initially, pCYC-LyE was tested in growth studies at three isopropyl-β-d-thiogalactopyranoside (IPTG) induction concentrations (Fig. 2A). Bacterial strains containing LyE demonstrated IPTG concentration-dependent lysis. Critical point lysis occurred at 150, 120, and 100 min for 100, 500, and 1,000 μM IPTG, respectively. Taking these observations into account, shear studies with 5-s sonication times were performed with 1,000 μM IPTG-induced LyE-containing bacterial strains (Fig. 2B). Noticeable viability reduction occurs after a 90-min induction period.
Fig. 2.
Bacteriophage LyE incorporation into bacterial vectors and resulting characterization. (A) Growth studies at three different IPTG induction concentrations. (B) Shear studies at 1,000 μM IPTG induction over time before sonication and viability analysis. Gene delivery (C) and (4, 5-dimethylthiazol-2-yl)-diphenyltetrazolium bromide assay (D) of 1,000 μM IPTG-induced bacterial vector strains. Untreated controls are reported as 100% viability. *Statistical significance (95% confidence) for samples with and without pCYC-LyE compared at induction time points (B) or MOIs (C and D).
To extend the properties of the bacterial gene delivery vectors further, pCYC-LyE was incorporated into the previously best-performing strain, YWT7-hly/pCMV-Luc (S1). Using 1,000 μM IPTG induction for 1 h, gene delivery experiments were conducted at 1:1, 10:1, 100:1, and 1,000:1 MOIs (Fig. 2C). LyE-containing S1 demonstrated improved gene delivery in relation to increasing MOI. Based upon previous observations (12), membrane destabilization results in elevated gene delivery due to increased pDNA and protein release coupled to decreased APC cytotoxicity. Thus, to evaluate the impact of LyE upon cytotoxicity, bacterial strains were induced with 100, 500, and 1,000 μM IPTG for 1 h and transfected at 1:1, 10:1, 100:1, 500:1, 1,000:1, 1,500:1, and 2,000:1 MOIs. At 100 and 500 μM IPTG induction, LyE expression significantly improves cytotoxicity compared with the bacterial controls (SI Appendix, Fig. S9). At excessive MOI levels, such as 2,000:1, LyE-containing S1 exhibits 46% and 53% viability at 100 μM and 500 μM IPTG, respectively. Upon higher induction (1,000 μM IPTG), 2,000:1 MOI viability is further elevated to 65% (Fig. 2D). Using the highest IPTG concentration, gene delivery is approximately doubled, with ∼80% viability at 1,000:1 MOI.
Mannose-Mediated Targeted Delivery of Hybrid Devices.
To improve upon delivery specificity, mannose, an antagonist of CD206 (primarily expressed on APCs) (35, 36), was grafted onto D9 (8, 29, 37, 38). In addition to promoting APC delivery specificity, CD206 is a type 1 membrane immune receptor that mediates uptake of bacteria (39). Studies using CD206-mediated targeting have demonstrated the ability to deliver adjuvant-free antigens, resulting in concomitant increases in immune responses and cross-presentation (39–41). Thus, D9 was end-capped by d-mannose (SI Appendix, Fig. S10). This was accomplished by synthesizing acrylate-capped D9 (D9-ac) and further modifying with an end-capping amine (D9-am). In parallel, allyl-α-d-mannopyranoside was synthesized and reacted with D9-am to produce mannose-terminated D9 (D9-Man). Synthesis was confirmed using GPC and NMR (SI Appendix, Fig. S11).
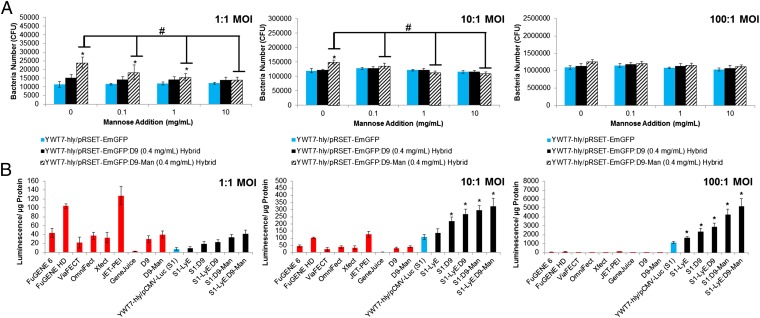
Mannosylated hybrids (0.4 mg/mL) were prepared with bacterial strain YWT7-hly/pRSET-emerald green GFP and evaluated for uptake (Fig. 3A) and gene delivery (Fig. 3B). Additional d-mannose was included to test competitive inhibition of the CD206 receptor. At 1:1 and 10:1 MOIs, uptake of mannosylated hybrids was significantly higher than that of either the bacterial or unmannosylated hybrid vectors in the absence of exogenously added mannose. However, upon addition of free mannose, uptake of only the mannosylated hybrid vector decreases proportionately. At the highest MOI, 100:1, uptake of mannosylated particles is slightly, but insignificantly, elevated compared with controls and decreases upon mannose addition.
Fig. 3.
Uptake and gene delivery studies of mannosylated D9 hybrid vectors. (A) Uptake of bacterial and hybrid vectors in the presence of increasing concentrations of mannose. (B) Gene delivery of hybrid vectors at three MOIs. Positive controls included FuGENE 6 (Promega), FuGENE HD (Promega), ViaFECT (Promega), OmniFect (TransOMIC Technologies), Xfect (Clontech Laboratories), JET-PEI (Polyplus transfection), and GeneJuice (EMD Millipore). *Statistical significance (95% confidence) compared with associated YWT7-hly/pRSET-emerald green GFP (EmGFP) and YWT7-hly/pRSET-EmGFP:D9 datasets (A) or S1 and synthetic controls (B). #Statistical significance (95% confidence) indicates reduced bacterial uptake at each mannose concentration.
Next, hybrid devices were combined with previous LyE strains to test the feasibility of a dual-engineered hybrid vector (Fig. 3B). Hybrid vectors were prepared and transfected using D9 and D9-Man at 0.4 mg/mL with S1 and S1/pCYC-LyE (S1-LyE). Gene delivery was improved across all MOIs by the separate molecular biology (introduction of LyE) and polymer chemistry (mannosylation of D9) toolsets uniquely afforded by the hybrid vector. The final approach and results clearly indicate the synergistic and engineering potential of the hybrid vector design. Complementary flow cytometry population data were collected for all samples in Fig. 3B (SI Appendix, Fig. S12), with the S1-LyE:D9-Man vector showing the best combination of the highest percentage of GFP-positive cells with no associated APC cellular toxicity.
Mouse Model Immunization.
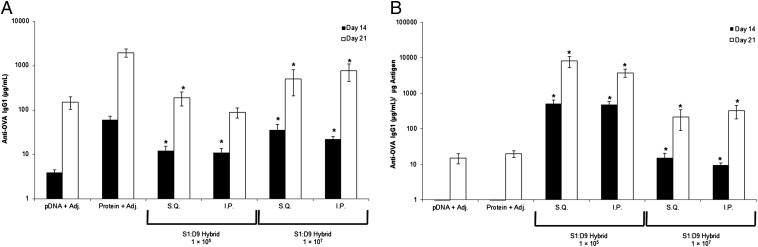
Finally, to test the hybrid vector approach in an in vivo setting, we completed mouse model assessment of antibody response against the model ovalbumin (OVA) antigen (Fig. 4). No toxicity was observed among the mouse subjects, and total IgG1 titers obtained were comparable to the recombinant OVA-positive control. The results emphasize the following: (i) the immune response potential of the hybrid vector without the need for adjuvant inclusion and before optimization studies and (ii) hybrid device signal strength when normalized to the amount of genetic antigen administered, highlighting the efficiency of the vector to invoke a response compared with positive controls. The results further support the potential of the hybrid vector technology toward APC-mediated immune modulation and eventual full-vaccination strategies.
Fig. 4.
Mouse model immunization. Total (A) and normalized (B) antibody responses are compared between genetic (pDNA) and protein OVA antigens delivered with adjuvant (Adj.) and genetic antigen delivery using the hybrid vector. *Statistical significance (95% confidence) compared with pDNA (A, respective time points) or pDNA and protein (B, respective time points) OVA controls. I.P., intraperitoneal; S.Q., subcutaneous.
Conclusions
The hybrid biosynthetic vector developed in this study combined the innate and engineering capabilities of individual biological and biomaterial components. The new vector thus allowed for a significantly expanded set of variables available to influence APC gene delivery. Notably, the native gene-carrying capacity of the E. coli component of the hybrid vector allowed for a substantial improvement in gene delivery efficiency. Results were extended by leveraging the engineering capabilities of the hybrid vector to address the cellular barriers to APC gene delivery in vitro further and to test model antigen delivery in vivo. The vector and approach offer a broader set of features and capabilities through which APC gene delivery can be carefully modulated to elicit desired immune responses.
Materials and Methods
Bacterial and hybrid vectors were prepared from bacterial cultures inoculated at 2% (vol/vol) from overnight starter cultures. Plasmid selection antibiotics were used as needed during bacterial culture within lysogeny broth medium. Following incubation at 36 °C with shaking until 0.4–0.5 OD600, samples were induced with 0.1 mM IPTG at 30 °C for 1 h. Bacterial vectors were then washed once and standardized to 0.5 OD600 in PBS, whereas bacterial strains to be used in hybrid vector formation were washed once and standardized to 1.0 OD600 in 25 mM NaOAc (pH 5.15). Polymer dosages dissolved in chloroform were desiccated and resuspended in 25 mM NaOAc (pH 5.15) before equal volume addition to 1.0 OD600 bacterial strains. Hybrid vectors (final 0.5 OD600) and bacterial vectors in PBS were allowed to incubate at 22 °C for 15 min before being diluted into RPMI medium to produce desired MOIs.
Polyplexes were generated following previously described protocols (7). Briefly, polymer dosages (chosen to ensure complete pDNA complexation) in chloroform were added to Eppendorf tubes before desiccation and resuspension in 25 mM NaOAc buffer (pH 5.15). An equal volume of pDNA in 25 mM NaOAc buffer (pH 5.15) was added and mixed by vortexing on setting 5 (Analog Vortex Mixer; Fisher Scientific) for 10 s. Polymer/pDNA self-assembly was allowed to continue for 15 min.
All remaining experimental details, including characterization and assays, are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Randall Smith, C. K. Chen, and Chong Cheng (University at Buffalo) for assistance with flow cytometry and polymer synthesis and characterization. We recognize support from National Institutes of Health Awards AI088485 (to B.A.P.) and DC013554 (to A.P.H.) and a State University of New York at Buffalo Schomburg Fellowship (to C.H.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411355111/-/DCSupplemental.
References
- 1.Schmidt C. Vaccines for pandemics. Nat Biotechnol. 2013;31(11):957–960. doi: 10.1038/nbt.2733. [DOI] [PubMed] [Google Scholar]
- 2.Wirth T, Parker N, Ylä-Herttuala S. History of gene therapy. Gene. 2013;525(2):162–169. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- 3.Leboulch P. Gene therapy: Primed for take-off. Nature. 2013;500(7462):280–282. doi: 10.1038/500280a. [DOI] [PubMed] [Google Scholar]
- 4.Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming nonviral gene delivery barriers: Perspective and future. Mol Pharm. 2013;10(11):4082–4098. doi: 10.1021/mp400467x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 6.Lynn DM, Langer R. Degradable poly(β-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122(44):10761–10768. [Google Scholar]
- 7.Green JJ, Zugates GT, Langer R, Anderson DG. Poly(β-amino esters): Procedures for synthesis and gene delivery. Methods Mol Biol. 2009;480:53–63. doi: 10.1007/978-1-59745-429-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of base polymer hydrophobicity and end-group modification on polymeric gene delivery. Biomacromolecules. 2011;12(10):3592–3600. doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sunshine JC, Peng DY, Green JJ. Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharm. 2012;9(11):3375–3383. doi: 10.1021/mp3004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunshine JC, Sunshine SB, Bhutto I, Handa JT, Green JJ. Poly(β-amino ester)-nanoparticle mediated transfection of retinal pigment epithelial cells in vitro and in vivo. PLoS ONE. 2012;7(5):e37543. doi: 10.1371/journal.pone.0037543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsa S, Pfeifer B. Engineering bacterial vectors for delivery of genes and proteins to antigen-presenting cells. Mol Pharm. 2007;4(1):4–17. doi: 10.1021/mp0600889. [DOI] [PubMed] [Google Scholar]
- 12.Jones CH, et al. Polymyxin B treatment improves bactofection efficacy and reduces cytotoxicity. Mol Pharm. 2013;10(11):4301–4308. doi: 10.1021/mp4003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsen MD, et al. Bactofection of lung epithelial cells in vitro and in vivo using a genetically modified Escherichia coli. Gene Ther. 2008;15(6):434–442. doi: 10.1038/sj.gt.3303090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castagliuolo I, et al. Engineered E. coli delivers therapeutic genes to the colonic mucosa. Gene Ther. 2005;12(13):1070–1078. doi: 10.1038/sj.gt.3302493. [DOI] [PubMed] [Google Scholar]
- 15.Chart H, Smith HR, La Ragione RM, Woodward MJ. An investigation into the pathogenic properties of Escherichia coli strains BLR, BL21, DH5alpha and EQ1. J Appl Microbiol. 2000;89(6):1048–1058. doi: 10.1046/j.1365-2672.2000.01211.x. [DOI] [PubMed] [Google Scholar]
- 16.Parsa S, Wang Y, Rines K, Pfeifer BA. A high-throughput comparison of recombinant gene expression parameters for E. coli-mediated gene transfer to P388D1 macrophage cells. J Biotechnol. 2008;137(1-4):59–64. doi: 10.1016/j.jbiotec.2008.07.1815. [DOI] [PubMed] [Google Scholar]
- 17.Critchley RJ, et al. Potential therapeutic applications of recombinant, invasive E. coli. Gene Ther. 2004;11(15):1224–1233. doi: 10.1038/sj.gt.3302281. [DOI] [PubMed] [Google Scholar]
- 18.Laner A, et al. Bacterial transfer of large functional genomic DNA into human cells. Gene Ther. 2005;12(21):1559–1572. doi: 10.1038/sj.gt.3302576. [DOI] [PubMed] [Google Scholar]
- 19.Cheung W, et al. Bacterial delivery of large intact genomic-DNA-containing BACs into mammalian cells. Bioeng Bugs. 2012;3(2):86–92. doi: 10.4161/bbug.18621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang S, Fruehauf J, Li CJ. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat Biotechnol. 2006;24(6):697–702. doi: 10.1038/nbt1211. [DOI] [PubMed] [Google Scholar]
- 21.Radford KJ, et al. A recombinant E. coli vaccine to promote MHC class I-dependent antigen presentation: Application to cancer immunotherapy. Gene Ther. 2002;9(21):1455–1463. doi: 10.1038/sj.gt.3301812. [DOI] [PubMed] [Google Scholar]
- 22.Higgins DE, Shastri N, Portnoy DA. Delivery of protein to the cytosol of macrophages using Escherichia coli K-12. Mol Microbiol. 1999;31(6):1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 23.Parsa S, Wang Y, Fuller J, Langer R, Pfeifer BA. A comparison between polymeric microsphere and bacterial vectors for macrophage P388D1 gene delivery. Pharm Res. 2008;25(5):1202–1208. doi: 10.1007/s11095-008-9563-x. [DOI] [PubMed] [Google Scholar]
- 24.Grillot-Courvalin C, Goussard S, Courvalin P. Wild-type intracellular bacteria deliver DNA into mammalian cells. Cell Microbiol. 2002;4(3):177–186. doi: 10.1046/j.1462-5822.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- 25.Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(β-amino esters) Mol Ther. 2005;11(3):426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew Chem Int Ed Engl. 2003;42(27):3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 27.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc Chem Res. 2008;41(6):749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 29.Zugates GT, et al. Gene delivery properties of end-modified poly(β-amino ester)s. Bioconjug Chem. 2007;18(6):1887–1896. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]
- 30.Bolivar F, et al. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- 31.Doshi N, Mitragotri S. Macrophages recognize size and shape of their targets. PLoS ONE. 2010;5(4):e10051. doi: 10.1371/journal.pone.0010051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champion JA, Walker A, Mitragotri S. Role of particle size in phagocytosis of polymeric microspheres. Pharm Res. 2008;25(8):1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eko FO, et al. New strategies for combination vaccines based on the extended recombinant bacterial ghost system. Vaccine. 1999;17(13-14):1643–1649. doi: 10.1016/s0264-410x(98)00423-x. [DOI] [PubMed] [Google Scholar]
- 34.Witte A, Wanner G, Sulzner M, Lubitz W. Dynamics of PhiX174 protein E-mediated lysis of Escherichia coli. Arch Microbiol. 1992;157(4):381–388. doi: 10.1007/BF00248685. [DOI] [PubMed] [Google Scholar]
- 35.Daigneault M, Preston JA, Marriott HM, Whyte MK, Dockrell DH. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS ONE. 2010;5(1):e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Devey L, et al. Tissue-resident macrophages protect the liver from ischemia reperfusion injury via a heme oxygenase-1-dependent mechanism. Mol Ther. 2009;17(1):65–72. doi: 10.1038/mt.2008.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eltoukhy AA, et al. Effect of molecular weight of amine end-modified poly(β-amino ester)s on gene delivery efficiency and toxicity. Biomaterials. 2012;33(13):3594–3603. doi: 10.1016/j.biomaterials.2012.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zugates GT, et al. Rapid optimization of gene delivery by parallel end-modification of poly(β-amino ester)s. Mol Ther. 2007;15(7):1306–1312. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- 39.He LZ, et al. Antigenic targeting of the human mannose receptor induces tumor immunity. J Immunol. 2007;178(10):6259–6267. doi: 10.4049/jimmunol.178.10.6259. [DOI] [PubMed] [Google Scholar]
- 40.Keler T, He L, Graziano RF. Development of antibody-targeted vaccines. Curr Opin Mol Ther. 2005;7(2):157–163. [PubMed] [Google Scholar]
- 41.He LZ, et al. A novel human cancer vaccine elicits cellular responses to the tumor-associated antigen, human chorionic gonadotropin β. Clin Cancer Res. 2004;10(6):1920–1927. doi: 10.1158/1078-0432.ccr-03-0264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.