Significance
In this study, we found that human endogenous retoriviruses type-H (HERV-Hs) are transiently hyperactivated during reprogramming toward induced pluripotent stem cells (iPSCs) and play important roles in this process. However, when reprogramming is complete and cells acquire full pluripotency, HERV-H activity should decrease to levels comparable with those in embryonic stem cells because failure to resilence this activity leads to the differentiation-defective phenotype in neural lineage. We also found that during reprogramming, reprogramming factors, including POU class 5 homeobox 1 (OCT3/4), sex determining region Y-box 2 (SOX2), and Krüppel-like factor 4 (KLF4) (OSK) bind to and activate long-terminal repeats of HERV-Hs. KLF4 possibly precludes Tripartite motif containing 28 and recruits not only OCT3/4 and SOX2, but also E1A binding protein p300 (p300) histone acethyltransferase on HERV-H loci. Therefore, OKSM-induced HERV-H activation constitutes an unanticipated and critical mechanism for iPSC formation.
Keywords: retrotransposon, epigenetics, evolution
Abstract
Pluripotency can be induced in somatic cells by overexpressing transcription factors, including POU class 5 homeobox 1 (OCT3/4), sex determining region Y-box 2 (SOX2), Krüppel-like factor 4 (KLF4), and myelocytomatosis oncogene (c-MYC). However, some induced pluripotent stem cells (iPSCs) exhibit defective differentiation and inappropriate maintenance of pluripotency features. Here we show that dynamic regulation of human endogenous retroviruses (HERVs) is important in the reprogramming process toward iPSCs, and in re-establishment of differentiation potential. During reprogramming, OCT3/4, SOX2, and KLF4 transiently hyperactivated LTR7s—the long-terminal repeats of HERV type-H (HERV-H)—to levels much higher than in embryonic stem cells by direct occupation of LTR7 sites genome-wide. Knocking down LTR7s or long intergenic non-protein coding RNA, regulator of reprogramming (lincRNA-RoR), a HERV-H–driven long noncoding RNA, early in reprogramming markedly reduced the efficiency of iPSC generation. KLF4 and LTR7 expression decreased to levels comparable with embryonic stem cells once reprogramming was complete, but failure to resuppress KLF4 and LTR7s resulted in defective differentiation. We also observed defective differentiation and LTR7 activation when iPSCs had forced expression of KLF4. However, when aberrantly expressed KLF4 or LTR7s were suppressed in defective iPSCs, normal differentiation was restored. Thus, a major mechanism by which OCT3/4, SOX2, and KLF4 promote human iPSC generation and reestablish potential for differentiation is by dynamically regulating HERV-H LTR7s.
Human pluripotent stem cells can be generated through two paths: (i) embryonic stem cells (ESCs) can be derived from embryos (1), and (ii) induced pluripotent stem cells (iPSCs) can be generated from differentiated cells through factor-mediated reprogramming (2). Most iPSCs are highly similar to ESCs, but we recently showed that ∼10% of iPSC clones have a differentiation-defective phenotype, such that 20% of cells were undifferentiated, even after in vitro-directed neural differentiation (3). These differentiation-defective (DD)-iPSC clones exhibited high expression levels of ∼10 genes—including abhydrolase domain containing 12B (ABHD12B), HERV-H LTR-associating 1 (HHLA1) and chromosome 4 open reading frame 51 (C4ORF51)—driven by the long-terminal repeats (LTRs) of human endogenous retroviruses (HERVs).
HERVs constitute ∼8% of the human genome as a result of their transposon activity, but they can no longer perform transposition (4). HERV type-H (HERV-H) transcripts are expressed in ESCs/iPSCs at higher levels than in differentiated cells (5). Approximately 80% of the LTRs belonging to the 50 most highly expressed HERV-H proviruses are occupied by core transcription factors involved in pluripotency, including POU class 5 homeobox 1 (OCT3/4), sex determining region Y-box 2 (SOX2), and NANOG homeobox (NANOG). Furthermore, HERV-H proviruses are expressed less in some iPSCs than in other iPSCs and ESCs, suggesting that HERV-H expression may be a barometer of pluripotency (5). Species-specific transposable elements, including HERVs, contribute up to 25% of the core transcription-factor binding sites in mouse and human pluripotent stem cells, wiring new genes into the core regulatory network of pluripotency in each species (6). These observations suggest that transposable elements may be important determinants of pluripotency. However, little is known about the roles of HERVs in reprogramming during iPSC generation.
In the present study, we found that during reprogramming of somatic cells toward iPSCs, HERV-H LTR7s were transiently activated to levels much higher than in ESCs, and this transient activation was required for efficient reprogramming. When reprogramming was complete, HERV-H expression decreased to levels comparable with those in ESCs. However, in DD-iPSC clones, HERV-H LTR7s remained aberrantly activated, leading to the defective differentiation. Thus, transient hyperactivation of HERVs is important in reprogramming somatic cells toward pluripotency and establishment of differentiation potential, revealing a previously unrecognized mechanism critical to cellular reprogramming technology.
Results
Characteristics of DD-iPSCs.
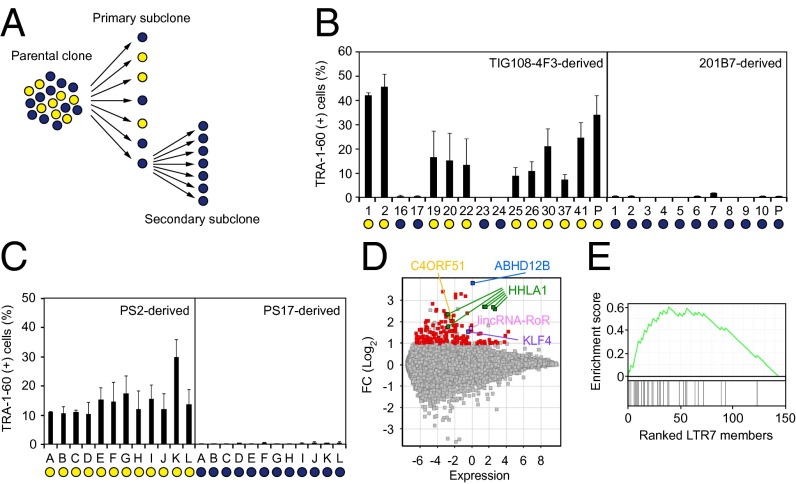
To better understand the nature of DD-iPSCs, we performed single-cell subcloning with four defective iPSC lines established using retroviral vectors, such as TKCBV5-6 (7), TIG108-4F3 (3), and TIG118-4F1 (3), and integration-free episomal vectors, such as 451F3 (8) (Fig. 1A). The subclones were identical to their parental clones regarding patterns of integrated retroviral vectors and short tandem repeats (Fig. S1A and Dataset S1). Based on marker-gene expression and neural differentiation potential, each DD-iPSC subclone had a normal or DD phenotype, whereas all subclones derived from ESCs and normal iPSCs exhibited a normal phenotype (3, 9) (Fig. 1B and Fig. S1B). The primary DD subclones derived from TIG108-4F3 DD-iPSCs were then used to produce secondary subclones, all of which showed the DD phenotype. Similarly, all normal primary subclones produced only normal secondary subclones (Fig. 1C). These data demonstrate that each DD-iPSC parental clone is monoclonal but consists of both DD and normal iPSCs. However, the DD phenotype is stable once subclones are isolated.
Fig. 1.
Enrichment of LTR7s in subcloned DD-iPSCs. (A) Summary of single-cell subcloning. (B) Differentiation potential of primary subclones. Shown are the percentages of TRA-1-60 (+) cells 14 d after neural induction of each primary subclone analyzed by flow cytometry. Blue and yellow circles indicate normal and DD-iPSC subclones/parents, respectively. n = 3. Error bars are SDs. (C) Differentiation potential of secondary subclones. Shown are the percentages of TRA-1-60 (+) cells 14 d after neural induction of TIG108-4F3-PS2- and PS17-derived secondary subclones. Blue and yellow circles indicate normal and DD-iPSC subclones, respectively. n = 3. Error bars are SDs. (D) Differential expression of genes between normal and DD-iPSCs. MA plot comparing global gene expression in normal (n = 18) and DD (n = 37) primary subclones derived from four DD-iPSCs parental clones (TIG108-4F3, TIG118-4F1, 451F3, and TKCBV5-6). Red and colored dots indicate genes with significantly higher expression in DD-iPSCs (FC > 2, FDR < 0.05). (E) Correlation between DD-marker expression and the presence of LTR7 elements. GSEA plot showing enrichment of LTR7 elements in 144 DD-iPSC markers. DD-iPSC markers are displayed in order of their fold-changes between normal- (n = 18) and DD- (n = 37) iPSC subclones in expression levels determined by a microarray.
The subcloning experiments allowed us to compare DD-iPSCs and their normal counterparts under the same genetic background. Microarray comparison of global gene expression in normal and DD-iPSC subclones identified 144 marker genes that were enriched in DD-iPSCs (Fig. 1D and Dataset S2), including the three previous reported genes ABHD12B, HHLA1, and C4ORF51. We also identified long intergenic non-protein coding RNA, regulator of reprogramming (lincRNA-RoR), an HERV-H LTR7-related large intergenic noncoding RNA (lincRNA), and KLF4 as DD-iPSC marker genes (3, 10). Of the DD-iPSC markers, 21.5% (31 of 144) were located within 30 kb downstream of LTR7s. Gene set enrichment analysis (GSEA) exhibited a significant correlation between DD-iPSC marker expression and the existence of HERV-H LTR7s [enrichment score = 0.59, false-discovery rate (FDR) q-value < 0.01] (Fig. 1E), showing that aberrant activation of LTR7s is a characteristic feature of the DD phenotype.
Similarity Between DD-iPSCs and Partially Reprogrammed Cells.
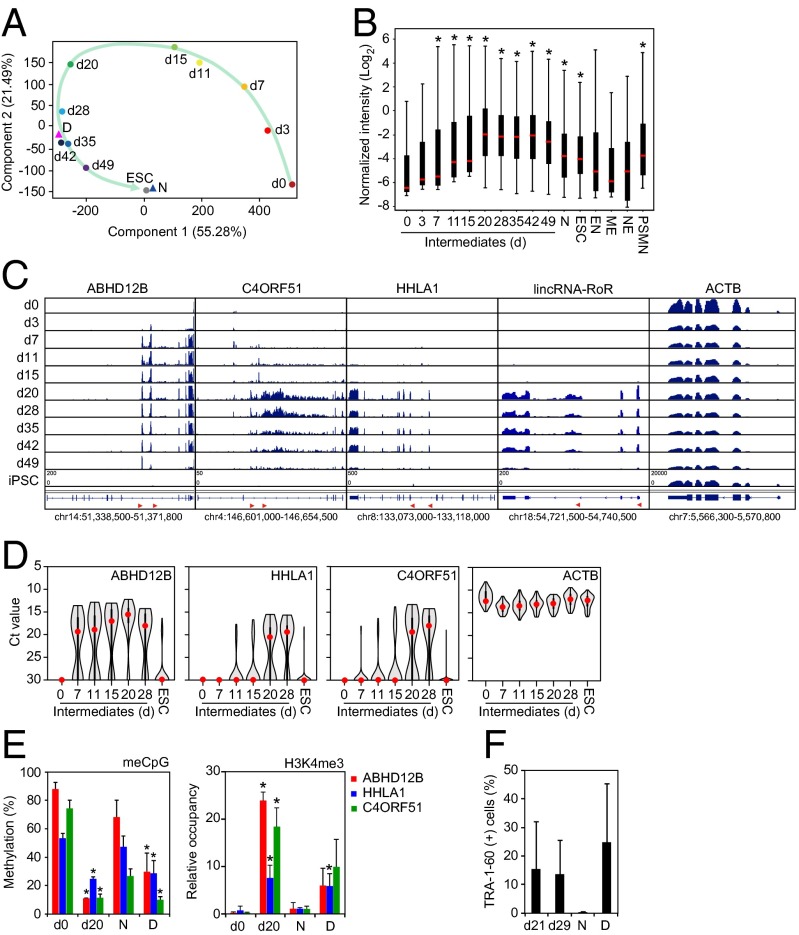
Next, we tried to understand why and how LTR7s were aberrantly activated in DD-iPSCs. To this end, we examined LTR7 activities during the course of iPSC generation. We sorted TRA-1-60–positive (+) reprogrammed cells on various days after retroviral transduction of OCT3/4, SOX2, KLF4, and myelocytomatosis oncogene (c-MYC) (subsequently referred to as OSKM) and analyzed their global gene expression by microarrays (11, 12). Using principle component analyses (PCA) with the 144 DD-iPSC markers, we found similarities between DD-iPSC subclones and TRA-1-60 (+) intermediate reprogrammed cells (Fig. 2A and Fig. S2). During reprogramming, TRA-1-60 (+) cells showed transiently enhanced expression of the DD-iPSC markers (including those driven by LTR7s), which reached significantly higher levels than in ESCs and normal iPSCs (Fig. 2B). When ESCs and normal iPSCs differentiated into endoderm (EN), mesoderm (ME), and neuroectoderm (NE), the expression of these markers significantly decreased. However, expression remained high in primitive streak-like mesendoderm (PSMN) (12). Deep sequencing of RNA (RNA-seq) from TRA-1-60 (+) cells exhibited the chimeric transcripts of ABHD12B, HHLA1, C4ORF51, and lincRNA-RoR with LTR7 sequences that meant transcription from intragenic LTR7s of HERV-Hs (Fig. 2C) (3, 10). Single-cell quantitative RT-PCR (qRT-PCR) showed that virtually all TRA-1-60 (+) cells—but not human dermal fibroblasts (HDFs) or ESCs—expressed the DD-iPSC marker genes related to HERV-H LTR7s (Fig. 2D). Furthermore, in both TRA-1-60 (+) intermediate cells on day 20 and DD-iPSCs, we observed less CpG dinucleotide methylation and more trimethylation of lysine 4 on histone H3 (H3K4me3) in the LTR7-driven DD-iPSC marker genes (Fig. 2E). LTR7-driven DD-iPSC marker genes were highly expressed in TRA-1-60 (+) cells derived from HDFs as well as from adipose tissue-derived mesenchymal stem cells (mesoderm), astrocytes (ectoderm), and bronchial epithelium (endoderm) (Fig. S3). Furthermore, we found that on days 21 and 29, TRA-1-60 (+) cells showed defective neural differentiation, in that they still contained TRA-1-60 (+) cells even after in vitro directed neural differentiation (Fig. 2F). Overall, these data show that DD-iPSC clones are similar to TRA-1-60 (+) intermediate reprogrammed cells in both gene expression and neural differentiation ability.
Fig. 2.
Resemblance of DD-iPSC and partially reprogrammed cells. (A) Principal component analysis of DD-iPSC marker genes. Comparison of expression of 144 DD-iPSC marker genes in HDFs (day 0, n = 4), intermediate reprogrammed cells derived from HDFs induced by OSKM [EGFP (+) cells on day 3 and TRA-1-60 (+) cells on d7-49, n = 3–4 in each time point], ESCs (n = 4), and normal (N, n = 18) and DD (D, n = 37)-iPSC subclones. The green arrow indicates the route of reprogramming. (B) Distribution of DD-iPSC marker gene expression. The box plot shows expression of 144 DD-iPSC marker genes in microarray data and their distribution in intermediate reprogrammed cells [EGFP (+) cells on day 3 and TRA-1-60 (+) cells on days 7–49], normal iPSCs, ESCs, and ESC/normal iPSC-derived differentiated progenies such as EN, ME, and NE, and PSMN. Red and black boxes indicate the median and quartile, respectively. Post hoc pairwise comparisons were performed by Tukey’s test (*P < 0.01 vs. day 0). (C) Transcription of DD-iPSC markers from LTR7 during reprogramming. Expression of ABHD12B, HHLA1, C4ORF51, lincRNA-RoR, and ACTB in HDFs (day 0), intermediate reprogrammed cells [EGFP (+) cells on day 3 and TRA-1-60 (+) cells on days 7–49] and iPSCs were revealed by RNA-seq. Red arrowheads indicate the LTR7 position and direction in each locus. (D) All TRA-1-60 (+) cells transiently express DD-iPSC markers. Ct values plotted by single-cell qRT-PCR for ABHD12B, HHLA1, C4ORF51, and ACTB in intermediate reprogrammed cells (days 0–28 in the x axis) and ESCs. At least 42 single cells were analyzed for each sample. Red dots indicate median values. Gray hourglass shapes represent the distribution of Ct value. Ct 30 indicates undetectable expression, which was indicated by Ct values >26. (E) Epigenetic statuses of LTR7s in TRA-1-60 (+) cells. The percentages of CpG methylation (Left) and H3K4me3 statuses (Right) in LTR7s on each locus including ABHD12B, HHLA1, and C4ORF51 revealed by bisulfite conversion/pyrosequencing and ChIP-qPCR, respectively. Day 0, HDFs (n = 3); day 20, TRA-1-60 (+) cells (n = 3); N, normal iPSCs (n = 3); D, DD-iPSCs (n = 3). Error bars are SD. *P < 0.05 vs. N was calculated by t test. (F) Neural differentiation-defective phenotype of TRA-1-60 (+) cells during reprogramming. Proportions of TRA-1-60 (+) cells after SFEBq neural inducing culture for 14 d. n = 3. Error bars are SDs.
Genome-Wide LTR7 Activation During Reprogramming.
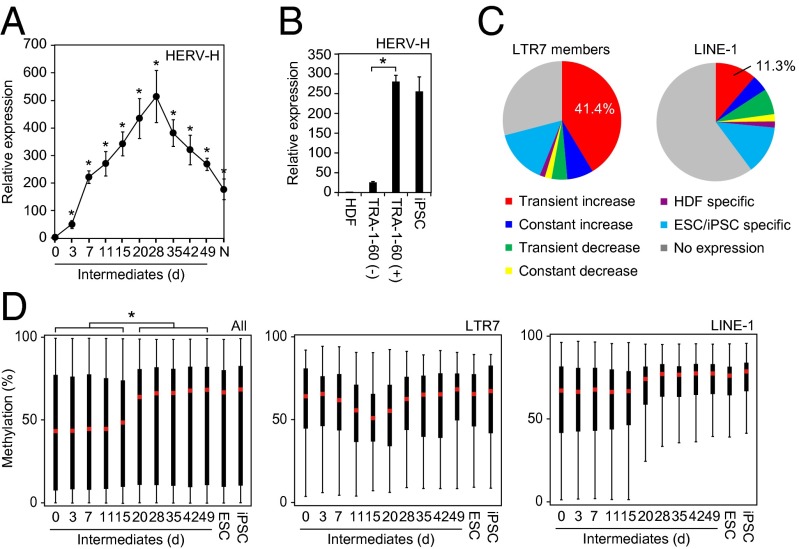
This last observation prompted us to examine the genome-wide LTR7 activity during reprogramming. qRT-PCR using a primer set for a conserved sequence of HERV-H LTR7 (13, 14) revealed that HERV-H transcripts transiently increased in TRA-1-60 (+) cells during reprogramming (Fig. 3A). The expression level of HERV-H in TRA-1-60 (+) intermediates on day 7 was significantly higher than those in TRA-1-60 (−) cells (Fig. 3B). RNA-seq showed that more than 40% of 3,771 LTR7 members in the human genome were transiently activated in TRA-1-60 (+) intermediate reprogrammed cells (Fig. 3C), whereas another transposable element, long-interspersed element-1 (LINE-1), showed varying expression patterns. Array-based analyses revealed that CpG methylation of LTR7 regions in TRA-1-60 (+) cells transiently decreased (Fig. 3D) (15). In contrast, global CpGs and those around LINE-1 elements gradually became methylated during reprogramming. Therefore, LTR7s were activated in a genome-wide manner during OSKM-mediated reprogramming.
Fig. 3.
Transient hyperactivation of LTR7s during iPSC generation. (A) Transition of total LTR7 transcription level during reprogramming. The plot shows the relative expression of total HERV-H in intermediate reprogrammed cells [EGFP (+) cells on day 3 and TRA-1-60 (+) cells on days 7–49] and normal iPSCs (N) compared with HDFs (day 0) revealed by qRT-PCR. Each value was normalized to that of G3PDH. n = 3. Error bars are SD. *P < 0.05 vs. HDF was calculated by Dunnett test. (B) Abundant HERV-H expression in TRA-1-60 (+) intermediates. Shown are relative expression of HERV-H in HDFs, TRA-1-60 (−) or (+) cells on day 7 and normal iPSCs analyzed by qRT-PCR. Each value was normalized to that of G3PDH. n = 3. Error bars are SD. *P < 0.05 was calculated by t test. (C) Expression patterns of the LTR7 family during reprogramming. Data are shown as LTR7 members and LINE-1 reads per kilobase of exon per million mapped reads (RPKM) in HDFs, TRA-1-60 (+) cells on day 20, and ESCs/normal iPSCs (n = 8). (D) Distribution of CpG methylation during reprogramming. The box plots show the distribution of methylation level at CpGs on all probes (Left), LTR7 (Center), and LINE-1 (Right) regions with overhang sequences (250 bp) in HDFs (day 0), intermediate reprogrammed cells [EGFP (+) cells on d3 and TRA-1-60 (+) cells on days 7–49], ESCs, and normal iPSCs. Red and black bars indicate the median and quartile, respectively. n = 3. Post hoc pairwise comparisons were performed by Tukey’s test (*P < 0.01).
Role of OSK in LTR7 Activation.
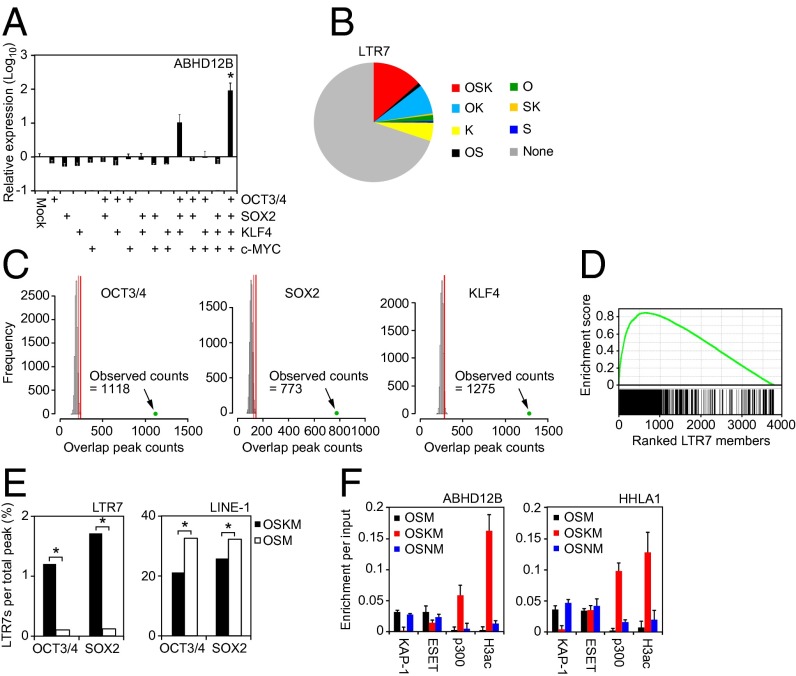
We next examined how OSKM helps activate the LTR7s during reprogramming. In day 7-transduced HDFs, we found that forced expression of OSK or OSKM, but not any single reprogramming factor or another combination, induced expression of the LTR7-driven gene ABHD12B (Fig. 4A). Thus, OCT3/4, SOX2, and KLF4 are all required for LTR7 activation. ChIP and sequencing (ChIP-seq) analyses showed that ∼15% of 3,771 LTR7s had cobinding of OCT3/4, SOX2, and KLF4 (OSK), ∼8% had cobinding of OCT3/4 and KLF4 (OK), and ∼5% had binding of KLF4 alone (K) (Fig. 4B). Compared with random binding, the concentrations of OCT3/4, SOX2, or KLF4 binding in LTR7s was highly significant (Fig. 4C). In addition, GSEA exhibited a significant correlation between OSK binding and HERV-H LTR7 expression in TRA-1-60 (+) cells (enrichment score = 0.85, P = 2.4 × 10−155) (Fig. 4D). The number of OCT3/4- and SOX2-bound LTR7s markedly decreased in the absence of KLF4 (P = 2.2 × 10−16 for both OCT3/4 and SOX2) (Fig. 4E), but such drastic decreases were not observed in OCT3/4- or SOX2-binding to LINE-1. Two proteins, KAP-1 (KRAB-associated protein 1) and histone methyltransferase SET domain bifurcated 1 (ESET), have been shown to be critical in suppression of endogenous retroviruses (16). ChIP experiments revealed that in HDFs transduced with OSKM, the binding of KAP-1 to LTR7s significantly decreased in ABHD12B and HHLA1 loci, but this decrease was not observed with OSM or OSNM (OSM with NANOG instead of KLF4) (Fig. 4F). In addition, the interaction between p300 and acetylated histone H3 was enriched by OSKM transduction, but not when KLF4 was absent (Fig. 4F). Therefore, KLF4 activates LTR7s by promoting OSK binding, recruiting the coactivator p300, and excluding KAP-1.
Fig. 4.
Role of OSK in LTR7 activation. (A) OSK is required for activation of ABHD12B expression. Relative expression level of ABHD12B on day 7 posttransduction for all combinations of OSKM. Error bars are SDs. n = 3. *P < 0.05 vs. Mock was calculated by Dunnett test. (B) Distribution of reprogramming factor occupancy on all LTR7s loci revealed by ChIP-seq. (C) Significance of the interaction of reprogramming factors to LTR7s. Histograms show counts of peaks for OCT3/4, SOX2, or KLF4 overlapped with randomly selected regions (10,000 random trials). The 95th percentile count of distribution is marked by red lines. Green dots show counts of ChIP-seq peaks on LTR7 regions with overhang sequences (250 bp). (D) GSEA plot showing enrichment of OSK occupancies in expressed LTR7s. Expressed LTR7 family members in TRA-1-60 (+) cells on day 15 are enriched in the set of LTR7s that show full-array OSK binding (P = 2.4e-155). (E) KLF4-dependent binding of OCT3/4 and SOX2 to LTR7s. Bars show the percentage of OCT3/4- or SOX2-bound LTR7 family members and LINE-1 in HDFs transduced with OSKM (closed) or OSM (open) on day 3 posttransduction. χ2 tests were performed between the proportions (*P < 0.05). (F) Interaction between HERV-H loci and chromatin modifiers. ChIP assays were performed to analyze the interaction of ABHD12B and HHLA1 loci with KAP-1, ESET, p300, and pan-acetyl histone H3 (H3ac) occupancy in HDFs transduced with OSM, OSKM, or OSNM on day 3 were analyzed by ChIP-qPCR. n = 3. Error bars are SD.
KLF4, a DD-iPSC Marker, Activates LTR7.
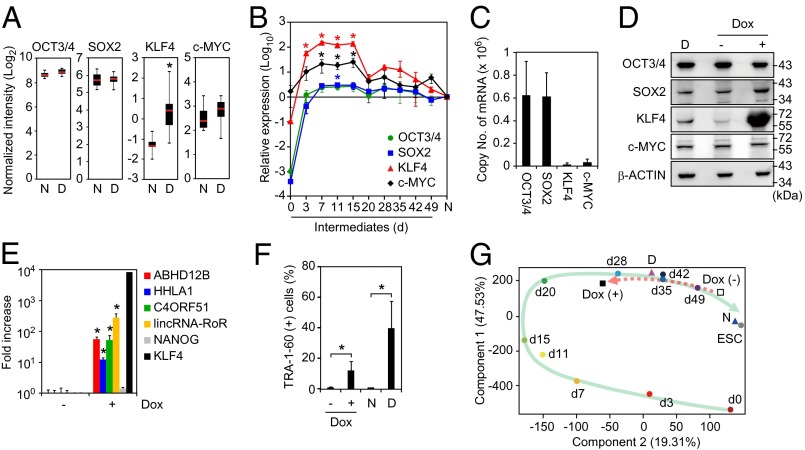
In addition to LTR7-driven transcripts, we identified KLF4 as a marker gene associated with the DD phenotype (Fig. 1D). Among the OSKM reprogramming factors, only KLF4 was enriched in DD-iPSC subclones (Fig. 5A). Whether KLF4 expression was derived from transgene or endogenous locus differed among clones (Fig. S4A). In the subclones derived from TIG118-4F1 and 451F3, the expression of endogenous KLF4 highly correlated to the DD phonotype (Fig. S4B). On the other hand, there was no significant correlation between neural differentiation potentials and endogenous KLF4 expression in the subclones derived from TIG108-4F3 and TKCBV5-6, which mainly expressed exogenous KLF4 (Fig. S4B). These data suggest that KLF4 expression including both aberrant activation of endogenous genes and insufficient silencing of retroviral vectors could be associated with the DD phenotype. We therefore analyzed the expression of KLF4, together with the remaining reprogramming factors, during iPSC generation. Total expression levels of OCT3/4 and SOX2 (from both endogenous genes and transgenes) increased more than 1,000-fold within 3 d after retroviral transduction and approached the levels in ESCs/iPSCs (Fig. 5B). After retroviral transgenes were silenced, the expression of OCT3/4 and SOX2 remained high because the endogenous genes were induced. Conversely, overexpression of KLF4 was transient and decreased once the retroviral transgenes were silenced (Fig. 5 A and B). Accordingly, the copy number of KLF4 mRNA was less than 1/40 of those for OCT3/4 and SOX2 in ESCs and normal iPSCs (Fig. 5C). This fact develops the transient increase of KLF4 expression during reprogramming toward iPSCs. On the other hand, the expression of OCT3/4 and SOX2 are constant even after transgene silencing occurred between days 15 and 20 posttransduction. Overall, increased expression of KLF4 correlated with aberrant activation of LTR7s in both the reprogramming process and in DD-iPSCs.
Fig. 5.
Role of KLF4 in the DD phenotype. (A) High expression of KLF4 in DD-iPSCs. Expression levels of total OCT3/4, SOX2, KLF4, and c-MYC in normal- (N; n = 18) and DD- (D; n = 37) iPSC primary subclones in microarray analysis. *FDR < 0.05 vs. N was calculated by t test. (B) Relative expression of total OSKM in intermediate reprogrammed cells were quantified by qRT-PCR and compared with those in iPSC. Each value was normalized to that of G3PDH. n = 3. Error bars are SDs. *P < 0.05 vs. iPSC (N) was calculated by Dunnett test. (C) Copy number of OSKM mRNAs in iPSCs. Data are shown as copy numbers of mRNA per 50 ng of total RNA calculated using a plasmid encoding each factor as a standard in qRT-PCR. n = 23. Error bars are SDs. (D) Expression of KLF4 protein. Western blot analyses of expression of OCT3/4, SOX2, KLF4, c-MYC, and β-ACTIN proteins in DD-iPSCs (D) and normal iPSCs (N) transduced with Dox-inducible KLF4 maintained with (+) or without (−) Dox. (E) KLF4 induces DD-iPSC marker expression in iPSCs. Bars show the relative expression levels of ABHD12B, HHLA1, C4ORF51, lincRNA-RoR, NANOG, and KLF4 in KLF4-overexpressing iPSCs analyzed by qRT-PCR. Each value was normalized to that of G3PDH. n = 3. Error bars are SDs. *P < 0.05 vs. Dox (−) were calculated by t test. (F) KLF4 prevents neural differentiation. Normal iPSCs transduced with Dox-inducible KLF4 were differentiated into neural cells using the SFEBq method with (+) or without (−) Dox. Bars show the percentages of TRA-1-60 (+) cells after a SFEBq neural inducing culture for 14 d. N and D represent normal and DD-iPSCs, respectively. n = 3. Error bars are SDs. *P < 0.05 was calculated by t test. (G) KLF4 changes the fate of iPSCs. PCA of microarray data from HDFs (day 0), TRA-1-60 (+) intermediate reprogrammed cells, normal iPSC subclones (N), DD-iPSC subclones (D), and Dox-inducible KLF4-transduced iPSCs with (+) or without (−) Dox for the 144 DD-iPSC marker genes. The green arrow indicates the route of reprogramming. The red broken arrow indicates the fate transition after induction of the KLF4 transgene.
To further examine the role of KLF4 in LTR7 activation, we introduced a doxycycline (Dox)-inducible KLF4 expression cassette into normal iPSCs using a PiggyBac transposon system (17) (Fig. 5D). Dox-induced KLF4 expression activated the LTR7-related transcripts ABHD12B, HHLA1, C4ORF51, and lincRNA-RoR but did not affect non-LTR–related genes, such as NANOG (Fig. 5E). Furthermore, overexpression of KLF4 in normal iPSCs produced the DD phenotype (Fig. 5F) (18). In contrast to neural lineage commitment, we observed no effects of KLF4 on the differentiation potentials of iPSCs into EN, ME, and PSMN (Fig. S5). This tendency was common between DD-iPSCs and KLF4-overexpressing iPSCs. PCA on the 144 DD-iPSC markers showed that KLF4-overexpressing iPSCs are quite similar to TRA-1-60 (+) intermediate reprogrammed cells (Fig. 5G), which confirms that KLF4 helps establish the DD phenotype.
To clarify the specificity by which KLF4 activates HERV-Hs in HDFs, we replaced KLF4 in the OSKM induction mixture with the reprogramming factor NANOG (referred to as OSNM). OSNM induced a few TRA1-60 (+) cells on day 7 and ESC-like colonies on day 28 (Fig. S6A). In TRA-1-60 (+) cells induced by OSNM, the expressions of KLF4 (Fig. S6B), HERV-Hs (Fig. S6C), and LTR7-driven genes (Fig. S6D) were only slightly activated. Therefore, overexpression of KLF4 and hyperactivation of HERV-H LTR7s are strongly correlated with efficient reprogramming in iPSC generation.
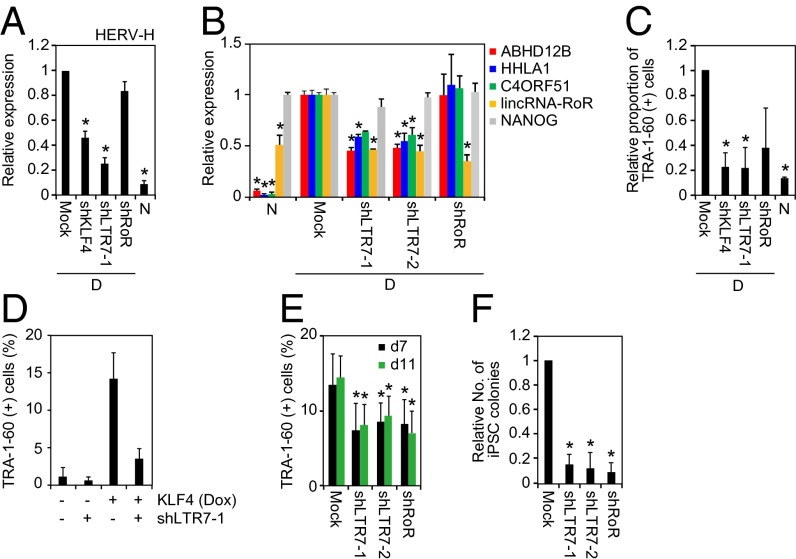
We next performed loss-of-function experiments to further investigate the roles of KLF4 and LTR7 in reprogramming and the DD phenotype. We designed four sets of short hairpin RNAs (shRNAs): one targeted KLF4 (shKLF4); two targeted LTR7 sequences conserved among ABHD12B, HHLA1, C4ORF51, and lincRNA-RoR (shLTR7-1 and shLTR7-2); and one targeted lincRNA-RoR (shRoR). In DD-iPSCs, shKLF4 and shLTR7-1, but not shRoR, significantly suppressed the total expression of HERV-Hs (Fig. 6A). The two shRNAs targeting the conserved LTR7 sequences effectively suppressed ABHD12B, HHLA1, C4ORF51, and lincRNA-RoR, but did not suppress NANOG (Fig. 6B). shRoR specifically repressed lincRNA-RoR expression but did not affect ABHD12B, HHLA1, C4ORF51, or NANOG (Fig. 6B). Suppressing KLF4 or HERV-H LTR7s in DD-iPSCs effectively reversed the DD phenotype and made the cells comparable to normal iPSCs (Fig. 6C). We observed a similar trend for shRoR, but the change was not statistically significant (P = 0.09). In addition, shLTR7-1 canceled the DD phenotype of KLF4-overexpressing iPSCs (Fig. 6D). Transducing these shRNAs with OSKM reduced the number of TRA-1-60 (+) cells on days 7 and 11 (Fig. 6E) and almost completely inhibited the generation of iPSC colonies (Fig. 6F). These data confirmed the important roles of KLF4 and LTR7 in reprogramming and the DD phenotype.
Fig. 6.
Loss of function experiments to test roles of KLF4 and LTR7s in reprogramming and the DD phenotype. (A) KLF4 is responsible for HERV-H expression. Shown are relative expressions of HERV-H in DD-iPSCs (D) transduced with KLF4 shRNA (shKLF4), LTR7 shRNA-1 (shLTR7-1), or shRoR, and normal iPSCs (N) compared with those of Mock-transduced DD-iPSCs. Each value was normalized to that of G3PDH. n = 3. Error bars are SDs. n = 3. *P < 0.05 vs. Mock was calculated by Dunnett test. (B) Knockdown of LTR7 expression. Bars show relative expression of ABHD12B, HHLA1, C4ORF51, lincRNA-RoR, and NANOG in normal iPSCs (N) and DD-iPSCs (D) transduced with empty vector (Mock), LTR7 shRNA-encoding vectors (shLTR7-1 and -2), or shRoR compared with Mock analyzed by microarray. Error bars are SDs. N =2. *P < 0.05 vs. Mock was calculated by Dunnett test. (C) Suppression of KLF4/HERV-H LTR7 rescues the DD phenotype. Shown are the relative proportions of residual TRA-1-60 (+) cells on day 14 after neural differentiation of DD-iPSCs (D) carrying empty vector (Mock), LTR7 shRNAs (shLTR7-1), or shRoR, compared with normal iPSCs (N). n = 3. *P < 0.05 vs. Mock was calculated by Dunnett test. (D) Suppression of LTR7 rescues the KLF4-induced DD phenotype. Shown are the percentages of residual TRA-1-60 (+) cells on day 14 after neural differentiation of normal iPSCs carrying dox-inducible KLF4, and empty vector or LTR7 shRNA (shLTR7-1). Differentiation was performed in the presence (+) or absence (−) of Dox. Error bars are SDs. n = 2. (E) LTR7 activity enhances reprogramming efficiency. Shown are the percentages of TRA-1-60 (+) cells on days 7 (black) and 11 (green) posttransduction of OSKM with empty vector (Mock), LTR7 shRNA-encoding vectors (shLTR7-1 and -2), or shRoR vector. n = 3. Error bars are SDs. *P < 0.05 vs. Mock was calculated by Dunnett test. (F) LTR7 activity facilitates iPSC generation. Shown are the relative numbers of iPSC colonies on day 25 posttransduction of OSKM with empty vector (Mock), LTR7 shRNA-encoding vectors (shLTR7-1 and -2), or shRoR vector. Error bars are SDs. n = 4. *P < 0.05 vs. Mock was calculated by Dunnett test.
Discussion
In this study, we found that genome-wide HERV-Hs, including lincRNA-RoR, are transiently hyperactivated during reprogramming toward iPSCs and play important roles in this process. However, when reprogramming is complete and cells acquire full pluripotency, HERV-H LTR7 activity should decrease to levels comparable with those in ESCs. Failure to resilence this activity leads to the DD phenotype. This observation resembles NANOG, which promotes induction and maintenance of pluripotency, but suppress differentiation when aberrantly expressed (19). We also found that during reprogramming, OSK factors bind to and activate LTR7s. Therefore, a major mechanism by which OSK reprogramming factors promote human iPSC generation is by transiently hyperactivating HERV-H LTR7s. Noteworthy, our findings suggest the significance of the transition state of intermediate reprogrammed cells, including hyperactivation of HERV-Hs induced by reprogramming factors. Among these cells, KLF4 particularly plays important roles for the activation of HERV-Hs. Our data also revealed that NANOG as a replacer of KLF4 in iPSC generation can induce less HERV-H activity during reprogramming (11). Therefore, the reason why the significant difference of reprogramming activity between KLF4 and NANOG can be explained with our data.
Recent study by Lu et al. showed that HERV-H activity regulated by OCT3/4 and p300 is important for generation and self-renewal of iPSCs (20). Among OSKM reprogramming factors, we showed that KLF4 levels are the most important for activating and resuppressing LTR7s. First, the binding of OCT3/4 and SOX2 to LTR7s was highly dependent on the presence of KLF4. Corroborating this finding, we and others have previously demonstrated that the KLF4 protein binds to OCT3/4 and SOX2 proteins. Second, we detected a surge in KLF4 expression during reprogramming, which was correlated with the transient hyperactivation of LTR7s. At around 15 d after transduction, overexpression of OSKM from retroviral transgenes is silenced. However, OCT3/4 and SOX2 maintained high expression levels because of the activation of their endogenous genes. In contrast, the endogenous KLF4 gene was only weakly activated, thus its total expression level rapidly decreased. In DD-iPSC clones and subclones, KLF4 is expressed at higher levels than in normal iPSCs, in agreement with the expression level of KLF4 helping determine LTR7 activity. Furthermore, we found that KLF4, together with OCT3/4 and SOX2, increased the binding of coactivator p300 to LTR7s and decreased KAP-1 binding to LTR7s. It has been shown that both KLF4 and KAP-1 bind to methylated DNA (21), suggesting a competition between the two proteins. Overall, we found that KLF4 strongly promotes LTR7 activity.
Among LTR7-driven transcripts, we found that lincRNA-RoR importantly influenced reprogramming and the DD phenotype. This result is consistent with a report from Loewer et al., who showed that lincRNA-RoR promoted iPSC generation (22). The authors identified lincRNA-RoR as one of 10 lincRNAs whose expression levels were higher in iPSCs than in ESCs (22). In contrast, the levels of lincRNA-RoR within most iPSC clones in our study were comparable to those of ESCs. Only DD-iPSCs showed higher expression levels. The functions of lincRNA-RoR remain elusive, but it may serve as a microRNA (miRNA) sponge that protects SOX2 and NANOG from miRNA-mediated degradation by sharing the binding sites of miRNAs that suppress the core transcription factors (23). Alternatively, lincRNA-RoR may suppress p53 (24), which inhibits reprogramming (25–29). Other LTR7-driven transcripts besides lincRNA-RoR likely also contribute to reprogramming and the DD phenotype, given that shRoR only weakly reversed the DD phenotype compared with shKLF4 or shLTR7s. Further studies, including genetic deletion of lincRNA-RoR, are required to fully understand how the activation of LTR7s contributes to reprogramming and the DD phenotype.
Our results suggest that reprogramming processes may use unique transposable elements in each species. Because neither HERV-H sequences nor lincRNA-RoR are conserved in mice, their activation cannot contribute to mouse reprogramming. Bourque and colleagues compared the binding sites of OCT3/4 and NANOG in their target genes and showed that species-specific transposable elements have substantially altered the transcriptional circuitry of pluripotent stem cells (6). Thus, ERV-1, including HERV-H, plays a major role in reprogramming human cells, whereas ERV-K, which is enriched in Oct3/4- and Nanog-binding sites in mice (6), may be involved in reprogramming mouse cells. Another study showed that a small portion of mouse ESCs and iPSCs express ERV-L retroviruses and possess the ability to differentiate not only into embryonic lineages but also into extraembryonic cells (30). Recently, Friedli et al. showed that aberrant activation of intracisternal A particle, a member of ERV-K, occurred during reprogramming of mouse embryonic fibroblasts toward iPSCs, as well as HERV-H behavior in a human case, which may suggest the importance of ERV activity in reprogramming beyond species (31). An important future task will involve examining the roles of species-specific ERVs in reprogramming and pluripotency.
Materials and Methods
Detailed descriptions of materials and methods are available in SI Materials and Methods. See Dataset S3 for primer sequences used in this study. Plasmids are available from Addgene (www.addgene.org).
Supplementary Material
Acknowledgments
We thank D. Srivastava for critical reading of the manuscript; G. Howard for editorial assistance; S. Arai, S. Ando, Y. Inoue, and N. Amano for technical assistance; M. Koyanagi-Aoi for sharing data; A. Morizane and J. Takahashi for guidance regarding the cellular differentiation; and H. Suemori, T. Kitamura, K. Okita, K. Eto, N. Takayama, and K. Woltjen for providing important materials. We are also grateful to Y. Miyake, R. Kato, E. Minamitani, S. Takeshima, R. Fujiwara, Y. Higuchi and K. Nakahara for administrative support. This work was supported in part by Grants-in-Aid for Scientific Research from the Japanese Society for the Promotion of Science (JSPS) and from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT); a grant from the Leading Project of the MEXT; a grant from the Funding Program for World-Leading Innovative Research and Development in Science and Technology (First Program) of the JSPS; a grant from Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine; a grant from World Premier International Research Center Initiative (WPI), MEXT; a grant from Japan Foundation for Applied Enzymology; and iPS Cell Research Fund. M.O. was supported as a JSPS fellow.
Footnotes
Conflict of interest statement: S.Y. is a member without salary of the scientific advisory board of iPS Academia Japan.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE54848 and GSE56569).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413299111/-/DCSupplemental.
References
- 1.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Koyanagi-Aoi M, et al. Differentiation-defective phenotypes revealed by large-scale analyses of human pluripotent stem cells. Proc Natl Acad Sci USA. 2013;110(51):20569–20574. doi: 10.1073/pnas.1319061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Santoni FA, Guerra J, Luban J. HERV-H RNA is abundant in human embryonic stem cells and a precise marker for pluripotency. Retrovirology. 2012;9:111. doi: 10.1186/1742-4690-9-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42(7):631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 7.Takayama N, et al. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J Exp Med. 2010;207(13):2817–2830. doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 9.Morizane A, Doi D, Kikuchi T, Nishimura K, Takahashi J. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J Neurosci Res. 2011;89(2):117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- 10.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13(11):R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanabe K, Nakamura M, Narita M, Takahashi K, Yamanaka S. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci USA. 2013;110(30):12172–12179. doi: 10.1073/pnas.1310291110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi K, et al. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat Commun. 2014;5:3678. doi: 10.1038/ncomms4678. [DOI] [PubMed] [Google Scholar]
- 13.Jern P, Sperber GO, Ahlsén G, Blomberg J. Sequence variability, gene structure, and expression of full-length human endogenous retrovirus H. J Virol. 2005;79(10):6325–6337. doi: 10.1128/JVI.79.10.6325-6337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang Q, Xu Z, Xu R, Wu L, Zheng S. Expression patterns of non-coding spliced transcripts from human endogenous retrovirus HERV-H elements in colon cancer. PLoS ONE. 2012;7(1):e29950. doi: 10.1371/journal.pone.0029950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471(7336):68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowe HM, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140(3):519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H, et al. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell. 2011;8(6):695–706. doi: 10.1016/j.stem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Darr H, Mayshar Y, Benvenisty N. Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development. 2006;133(6):1193–1201. doi: 10.1242/dev.02286. [DOI] [PubMed] [Google Scholar]
- 20.Lu X, et al. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21(4):423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 21.Quenneville S, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44(3):361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loewer S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25(1):69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Zhang A, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23(3):340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banito A, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23(18):2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedli M, et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014 doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.