Significance
A long-standing question in the field of microbial pathogenesis is how virulence factors are regulated within host cells and how their activity is specifically directed toward a particular host cell compartment. Legionella pneumophila resolves this dilemma by tightly coupling the phospholipase A1 activity of one of its effectors, vacuolar protein sorting inhibitor protein D (VipD), to this protein’s interaction with endosomal host GTPases. We now present the crystal structure of VipD in complex with host cell Rab5c, providing a detailed look into the ingenious molecular mechanisms underlying the allosteric activation of a virulence factor by a host protein and its spatiotemporal regulation. These results open the path for the development of novel therapeutics aimed at blocking the VipD activation process rather than the enzyme’s active site.
Keywords: pathogenic bacteria, allosteric modulation, membrane composition, X-ray crystallography
Abstract
A challenge for microbial pathogens is to assure that their translocated effector proteins target only the correct host cell compartment during infection. The Legionella pneumophila effector vacuolar protein sorting inhibitor protein D (VipD) localizes to early endosomal membranes and alters their lipid and protein composition, thereby protecting the pathogen from endosomal fusion. This process requires the phospholipase A1 (PLA1) activity of VipD that is triggered specifically on VipD binding to the host cell GTPase Rab5, a key regulator of endosomes. Here, we present the crystal structure of VipD in complex with constitutively active Rab5 and reveal the molecular mechanism underlying PLA1 activation. An active site-obstructing loop that originates from the C-terminal domain of VipD is repositioned on Rab5 binding, thereby exposing the catalytic pocket within the N-terminal PLA1 domain. Substitution of amino acid residues located within the VipD–Rab5 interface prevented Rab5 binding and PLA1 activation and caused a failure of VipD mutant proteins to target to Rab5-enriched endosomal structures within cells. Experimental and computational analyses confirmed an extended VipD-binding interface on Rab5, explaining why this L. pneumophila effector can compete with cellular ligands for Rab5 binding. Together, our data explain how the catalytic activity of a microbial effector can be precisely linked to its subcellular localization.
Microbial pathogens have evolved numerous ways to subvert and exploit normal host cell processes and to cause disease. Intravacuolar pathogens use specialized translocation devices such as type IV secretion systems (T4SS) to deliver virulence proteins, so-called effectors, across the bacterial and host cell membrane into the cytosol of the infected cell (1–3). Many of the translocated effectors studied to date alter cellular events such as vesicle trafficking, apoptosis, autophagy, protein ubiquitylation, or protein synthesis, among others, thereby creating conditions that support intracellular survival and replication of the microbe (4, 5). Bacteria with a nonfunctional T4SS are often avirulent and degraded along the endolysosomal pathway, thus underscoring the importance of translocated effectors for microbial pathogenesis.
Although T4SS-mediated effector translocation may be a convenient way for pathogens to manipulate host cells from within the safety of their membrane-enclosed compartment, it also creates a challenging dilemma: how can the bacteria ensure that their translocated effectors reach the correct host cell target for manipulation, and how can they prevent them from indiscriminately affecting bystander organelles or proteins that may otherwise be beneficial for intracellular survival and replication of the microbe? It is reasonable to expect that regulatory mechanisms have evolved that restrain the catalytic activity of effectors. Although detailed insight into these processes is scarce, an emerging theme among effectors is that their enzymatic activity is functionally coupled to their interaction with a particular host factor. For example, SseJ from Salmonella enterica serovar Typhimurium displays glycerophospholipid-cholesterol acyltransferase activity only on binding to the active GTPases RhoA, RhoB, or RhoC (6–8). Likewise, Pseudomonas aeruginosa ExoU requires mono- or poly-ubiquitinated proteins for the activation of its phospholipase A2 (PLA2) domain (9), whereas Yersinia YpkA exhibits kinase activity only in the presence of host cell actin (10). Exactly how binding to host ligands results in the activation of these translocated effectors remains unclear because no structural information for these protein complexes is available.
VipD is a T4SS-translocated substrate of Legionella pneumophila, the causative agent of a potentially fatal pneumonia known as Legionnaires' disease, and another example of an effector whose catalytic activity depends on the presence of a host factor (11–14). Following uptake by human alveolar macrophages, L. pneumophila translocates VipD together with more than 250 other effector proteins through its Dot/Icm T4SS into the host cell cytoplasm (15). These effectors act on numerous host processes to mediate evasion of the endolysosomal compartment and to establish a Legionella-containing vacuole (LCV) that supports bacterial growth (16). Although the precise biological role of most L. pneumophila effectors remains unclear, we recently showed that VipD is important for endosomal avoidance by LCVs. The protein localizes to endosomes presumably by binding to the small GTPases Rab5 or Rab22, key regulators of endosomal function (13, 14). Rab GTPase binding to the C-terminal domain of VipD triggers robust phospholipase A1 (PLA1) activity within the N-terminal domain, resulting in the removal of phosphatidylinositol 3-phosphate [PI(3)P] and potentially other lipids from endosomal membranes (14). Without PI(3)P, endosomal markers such as early endosomal antigen 1 (EEA1) are lost from these membranes, most likely rendering the endosomal compartment fusion incompetent (17). L. pneumophila mutants lacking vipD are attenuated in avoiding endosomal fusion, and their LCVs acquire the endosomal marker Rab5 more frequently than LCVs containing the parental strain producing VipD (14). Thus, by coupling PLA1 activity to Rab5 binding, the catalytic activity of VipD is directed specifically against the endosomal compartment without visibly affecting neighboring cell organelles.
VipD was originally identified in a screen for L. pneumophila effectors that interfere with the vacuolar sorting pathway in yeast (11). The N-terminal half of VipD possesses high homology to patatin, a lipid acyl hydrolase present in the potato tuber (12, 13). Analogous to other patatin-like proteins, VipD harbors a conserved serine lipase motif Gly-x-Ser-x-Gly (x = any amino acid) as part of a Ser-Asp catalytic dyad that, together with two consecutive glycine residues (Asp-Gly-Gly motif), is expected to stabilize the oxyanion intermediate during the acyl chain cleavage (13). Mutation of these conserved catalytic residues in VipD results in loss of PLA1 activity (14), confirming their role in substrate hydrolysis.
The recently reported crystal structure of VipD confirmed the predicted bimodular organization (13) and, in addition, revealed a surface loop, called “lid” in other phospholipases, that shields the entry to the catalytic site. The inhibitory lid may explain why purified recombinant VipD alone exhibits little or no PLA1 activity in vitro. However, given that binding of Rab5 or Rab22 to VipD activates the PLA1 activity within the N-terminal region (14), we wondered if and how this binding event causes the inhibitory lid to be removed to render the active site substrate accessible.
Using an integrative approach involving X-ray crystallography, molecular dynamics, biochemistry, and cellular imaging, we now deciphered at a molecular level the mechanism that stimulates the intrinsic PLA1 activity of VipD and determined the underlying specificity for the VipD–Rab5 interaction and endosomal targeting.
Results
Overall Structure of the VipD–Rab5 Complex.
To determine the molecular basis underlying VipD binding and activation by Rab5, we initiated a crystallographic analysis of this complex. For that, we used a truncated form (residues 18–182) of constitutively active Rab5c(Q80L) lacking the N- and C-terminal hypervariable regions, and a VipD fragment [amino acid (aa) 19–564; VipD19–564] that was designed based on a previously solved structure of full-length VipDFL–Rab5c18–182 at lower resolution in which the terminal residues (1–18 and 565–621) of VipD were not structured. We obtained well-diffracting crystals of VipD19–564 in complex with Rab5c18–182(Q80L) bound to nonhydrolyzable guanosine 5′-[β,γ-imido]triphosphate (GppNHp) and solved the structure by molecular replacement (Fig. 1). Only the last seven C-terminal residues of VipD19–564 and a connecting loop formed by residues 345–354 could not be modeled because of poor electron density in these regions. The final model for the VipD19–564-Rab5c18–182(Q80L)-GppNHp structure was refined at 3.1 Å, with values for Rfactor and Rfree of 0.23 and 0.28, respectively (Table 1 and Fig. S1A).
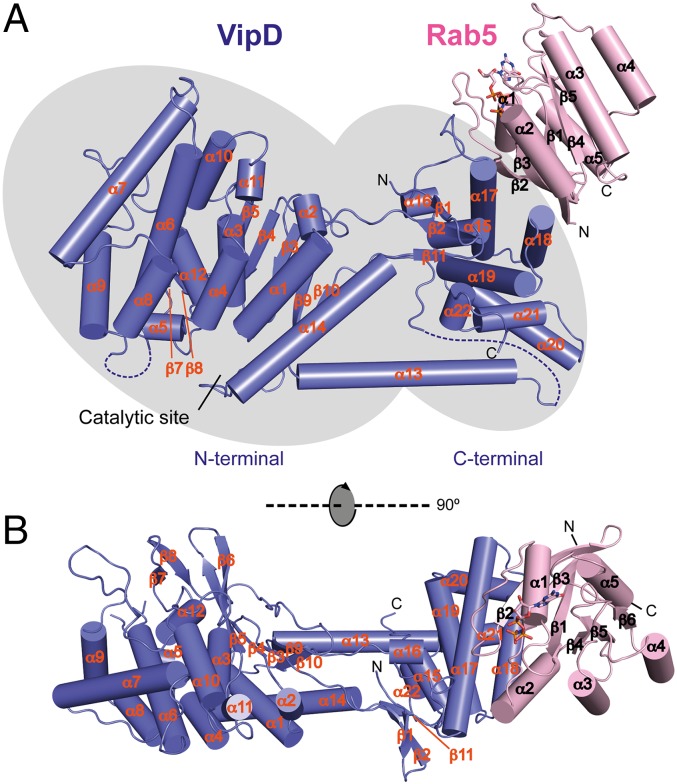
Fig. 1.
The sites for substrate catalysis and Rab5 binding are situated at opposite ends of VipD. Two orthogonal tube drawing representations of the crystal structure of VipD19–564 (slate) in complex with GppNHp-Rab5c18–182 (pink). (A) Side view. (B) Top view. VipD comprises two distinguishable but interconnected domains highlighted by gray elliptical shadows. The N-terminal half of VipD comprises a patatin-like phospholipase domain, whereas the C-terminal domain interacts with Rab5c. Note that the catalytic site and the Rab5 binding interface are located at opposite ends of VipD.
Table 1.
Data collection and refinement statistics for the VipD19–564–Rab5c18–182(Q80L):GppNHp complex
| VipD19–564–Rab5c18–182(Q80L) | |
| Data collection | |
| Space group | P1 |
| Cell dimensions | |
| a, b, c (Å) | 94.3, 98.0, 109.9 |
| α, β, γ (°) | 76.6, 80.8, 78.9 |
| Resolution (Å) | 30–3.07 (3.26–3.07)* |
| Rmeas | 0.07 (0.74) |
| I/σ | 17.0 (2.1) |
| Completeness (%) | 97.4 (92.6) |
| Redundancy | 3.5 (3.6) |
| Refinement | |
| Resolution (Å) | 3.07 |
| No. reflections | 67,479 |
| Rwork/Rfree | 0.23/0.28 |
| No. atoms | |
| Protein | 21,659 |
| Ligand/ion | 132 |
| B-factors | |
| Protein | 54 |
| Ligand/ion | 74 |
| RMSDs | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.631 |
Highest resolution shell is shown in parentheses.
The crystallographic asymmetric unit contained four VipD19–564–Rab5c18–182(Q80L) heterodimers with almost identical interaction modes (Fig. S1B). Superposition of the atomic coordinates showed a root mean square deviation (RMSD) of 0.65–0.69 Å among the four VipD19–564–Rab5c18–182(Q80L) complexes. Rab5c18–182(Q80L) was in its active conformation and bound to one molecule of GppNHp and one Mg2+ ion (Fig. 1). It adopted the classical GTPase fold consisting of a central six-stranded β-sheet surrounded by five α-helices (18). The structure of VipD19–564 exhibited two discernible but interconnected domains. Rab5c18–182(Q80L) interacted extensively with a helical hairpin situated at the C-terminal domain of VipD19–564, and, thus, at the distal end relative to the N-terminal catalytic site (Fig. 1). It is worth noting that, although the structure of active Rab5c18–182(Q80L) remained essentially unaltered, VipD19–564 exhibited several dramatic conformational rearrangements compared with the uncomplexed crystallographic model (13), as discussed next.
Rab5 Binding to VipD Induces Conformational Changes That Expose the Active Site.
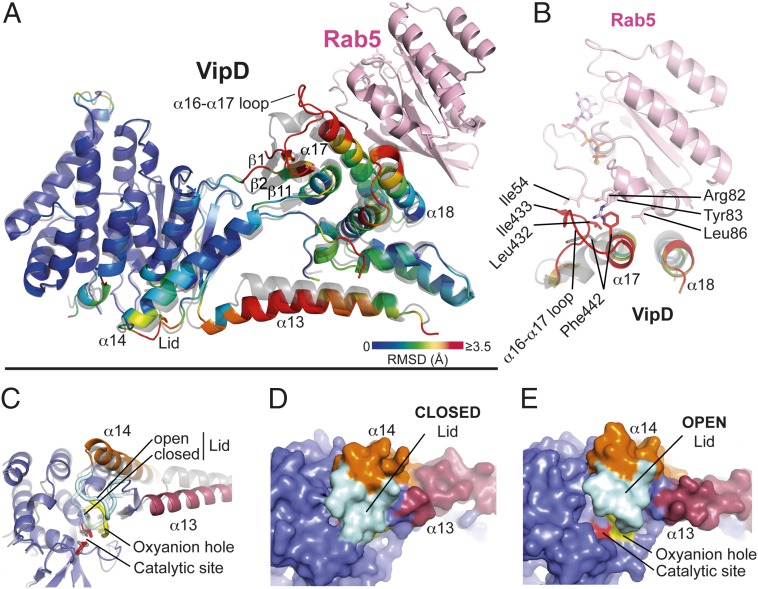
On Rab5, binding the largest RMSD in VipD19–564 occurred in its C-terminal domain and in the structural elements that connect it to the N-terminal phospholipase domain (Fig. 2A). Residue Phe442 located in helix α17 of the C-terminal domain of VipD19–564 undergoes a 90° rotation and enters a hydrophobic pocket in Rab5c18–182(Q80L) formed by Arg82, Tyr83, and Leu86 (Fig. 2B). This rotation pulls the adjacent α16-α17 loop of VipD19–564 toward Rab5c18–182(Q80L), thereby facilitating the hydrophobic interaction of Ile433 of VipD19–564 with Ile54 in the switch I region of Rab5c18–182(Q80L) (Fig. 2B). The displacement of loop α16–α17 in VipD19–564 induces a partial reorientation of the adjacent β-sheet formed by β1, β2, and β11, together with small shifts in helices of the C-terminal domain of VipD19–564. These cumulative movements cause helices α13 and α14 of VipD19–564 to swing out 14.5° and 6.6°, respectively, which is coupled with a coil-helix transition of the β9-α13 loop to adjoin helix α13 (Fig. 2A). This hinge motion of helices α13 and α14 (“chop-stick” mechanism) facilitates an outward displacement of the adjacent β10–α14 loop (subsequently named lid), resulting in the eventual opening of the active site (Fig. 2 C–E). Notably, there were no mayor crystallographic contacts in the areas corresponding to α13, α14, and the lid, making the displacement of the lid due to the proximity of neighboring protein molecules within the crystal lattice unlikely (Fig. S1C). The exposed cleft, with its catalytic residues and the oxyanion hole situated at one end, measures 16–18 Å in length and thus has the potential to accommodate a C16–C18 acyl chain from a lipid substrate within the adjacent hydrophobic ridge (Fig. 2E and Fig. S1D). Together, these findings provide evidence for an unprecedented heterotropic allosteric activation mechanism in which locally induced structural changes through Rab5c18–182(Q80L) binding are transmitted from the C-terminal domain of VipD19–564 to the N-terminal phospholipase domain, causing the displacement of the lid and exposure of the active site.
Fig. 2.
Allosteric activation of VipD through Rab5 binding. (A) Structural changes in VipD on Rab5 binding. Rab5c18–182 (colored in pink) is complexed to VipD19–564, which is colored from slate to red based on the root mean square deviation (RMSD) of C-α atom pairs when superimposed with the unbound form of VipD19–564 (PDB ID code 4AKF) shown in transparent gray. The black line represents the membrane plane. (B) Close-up of VipD–Rab5 interaction. The α17-α18 loop of VipD undergoes a Rab5-induced conformational rearrangement resulting in residue Phe442 of VipD being inserted into a hydrophobic pocket formed by Arg82, Tyr83, and Leu86 of Rab5. The displacement of the α16–17 loop favors the hydrophobic interaction between Leu432 and Ile433 of VipD with Ile54 of Rab5. Color code as in A. The remaining VipD structure has been omitted for clarity. (C) Close-up view of the catalytic site highlighting displacement of the lid (β10-α14 loop, light blue). (D and E) Surface representation of the unbound (D) and Rab5-bound (E) VipD molecule, respectively. Same view as in C.
VipD–Rab5 Interface.
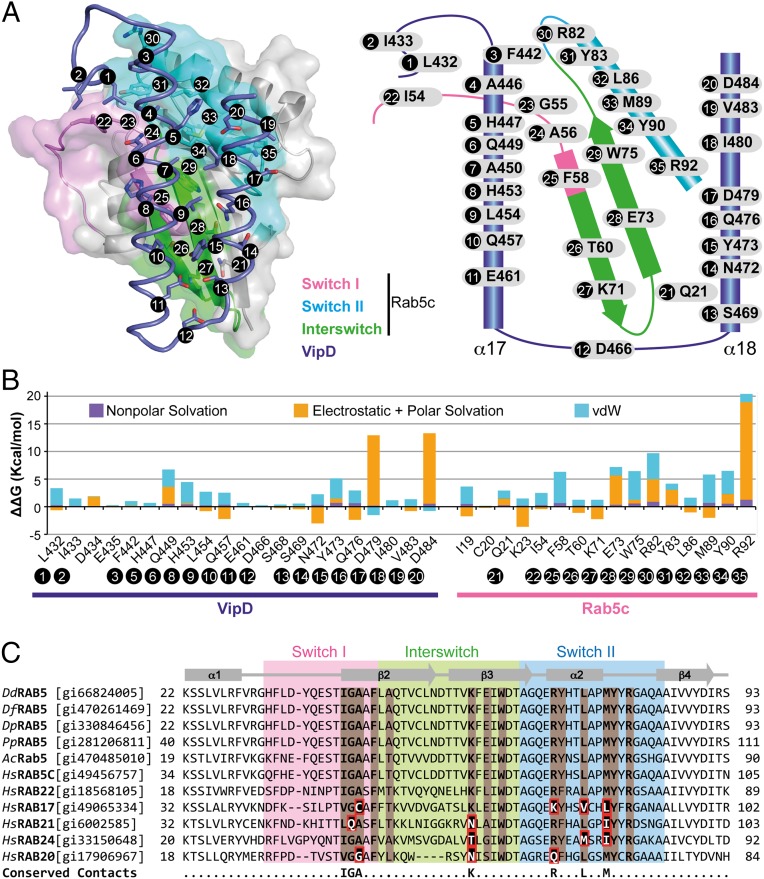
Our complex structure revealed a single interaction path between VipD19–564 and Rab5c18–182(Q80L) that occluded ∼722 Å2 of solvent-accessible surfaces. Although Rab5c18–182(Q80L) interacted with residues in the α16–α17 loop of VipD19–564 and residues in an helical hairpin formed by helices α17 and α18 (Fig. 3A), the VipD binding surface in Rab5c18–182(Q80L) included parts of the segment between α1 and β2 (the switch I region), the strands β2 and β3 (the interswitch region), and the β3–α2 segment (the switch II region) (Fig. 3A). The interface was composed of a core of hydrophobic contacts complemented by several polar interactions in the surrounding rim area (Fig. 3B). Specifically, the VipD binding epitope in Rab5c18–182(Q80L) included nonpolar residues in the switch I (Ile54, Gly55, Ala56, and Phe58), the interswitch (Trp75), and the switch II element (Tyr83, Leu86, Met89, and Tyr90), as well as polar/charged residues in the interswitch (Thr60, Lys71, and Glu73) and switch II element (Arg82 and Arg92) (Fig. 3 A and B). A comparison of the primary sequence of human Rab5 and Rab22 with Rab5 from several natural amoebean hosts found conserved residues at equivalent contact sites in the switch I (Ile54, Gly55, and Ala56), interswitch (Lys71), and switch II region (Arg82, Leu86, and Met89) that were variable in other Rab proteins (Fig. 3C), suggesting these residues are involved in the specific recognition by VipD. The corresponding epitope in VipD included several hydrophobic residues in helix α17 (Phe442, Ala446, Ala450, and Leu454) and in helix α18 (Tyr473, Ile480, and Val483) that wrapped around an elongated hydrophobic path in Rab5 formed by the conserved triad (Phe58, Trp75, and Tyr90) and Leu86. Surrounding this hydrophobic core were additional hydrogen bonds that enhanced the interaction.
Fig. 3.
Molecular interactions at the VipD–Rab5c interface. (A) (Left) Semitransparent surface of GppNHp-Rab5c18–182 in complex with the minimal Rab binding domain of VipD (slate ribbon model) highlighting the interfacial residues below 4.0-Å distance. (Right) Schematic diagram of interfacial residues in the VipD–Rab5c complex. (B) Detailed description of per-residue contribution from van der Waals (vdW) energy (blue), nonpolar solvation energy (purple), and the sum of electrostatic and polar solvation energy (orange) calculated by computational alanine scanning for interfacial residues in the VipD–Rab5c complex. Existing glycines and alanines are excluded in the calculation. (C) Sequence conservation between Rab5 GTPases from amoebean species and human homologs. Dd, Dictyostelium discoideum; Df, Dictyostelium fasciculatum; Dp, Dictyostelium purpureum; Pp, Polysphondylium pallidum; Ac, Acanthamoeba castellanii; Hs, Homo sapiens. Rab5c residues contacting VipD at a distance less than 4 Å are colored in light brown. Amino acid substitutions within the equivalently aligned interfacial residues of other Rabs are highlighted in a red box. Interfacial residues strictly conserved between Rab5 and Rab22, but variable in any of the other Rabs, are depicted in the bottom line of the alignment. Protein accession numbers are in brackets.
Like all GTPases, Rab5 exhibits structural changes within its switch regions dependent on its nucleotide-binding state (GDP vs. GTP), with the largest conformational variation in switch I (19). The structure of the VipD19–564–Rab5c18–182(Q80L) complex revealed that Leu432 and Ile433 of VipD19–564 interacted with Ile54 in switch I of Rab5c18–182(Q80L), therefore sensing its GTP-bound state (Fig. 2B). In fact, the conformation adopted by Rab5 in its GDP-bound state (19) resulted in a prominent steric clash between the switch I region and helix α17 of VipD19–564 (Fig. S1E), thus explaining why this activation state of Rab5 is only a poor ligand for VipD (13, 14).
Validation of the VipD–Rab5 Interface Through Mutational Analysis.
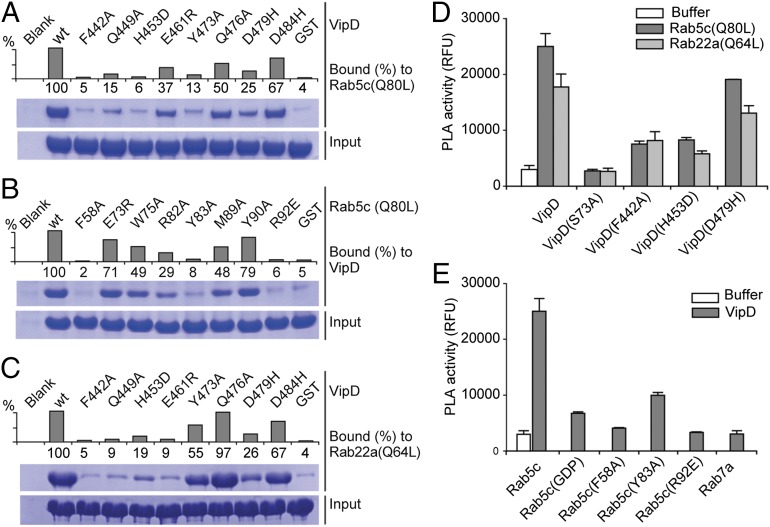
To experimentally validate the VipD–Rab5 binding interface seen in the crystal structure, we mutated several residues predicted to contribute to this protein–protein interaction and examined their role for complex formation in coprecipitation studies (Fig. 4A). Substitution of individual contact residues within VipD abrogated Rab5 binding either severely (F442A and H453D) or moderately (Q449A, E461R, Y473A, and D479H), whereas only a few of the tested substitutions in VipD were tolerated (Q476A and D484H). Similar results were observed for Rab5 interface mutants (Fig. 4B), with binding defects ranging from severe (F58A, Y83A, and R92E) to mild (E73R and Y90A). We also studied the mode of interaction between VipD and Rab22. As expected, substitution of individual contact residues of VipD required for Rab5c18–182(Q80L) binding (Fig. 4A) also resulted in a failure to stably associate with Rab22(Q64L) (Fig. 4C), suggesting that Rab22 occupies an epitope in VipD very similar to that of Rab5. None of the amino acid substitutions significantly altered the overall structure of the mutant proteins as evaluated by circular dichroism (CD) (Fig. S2), indicating that a reduction in binding was most likely not a consequence of protein misfolding.
Fig. 4.
Mutational analysis of interfacial recognition determinants. (A–C) Pulldown assays using the indicated VipD (A and C) or Rab5 (B) mutant proteins. The graphs are a densitometry-based quantification of the amount of query protein precipitated by the respective bait-coated beads. Input, total amount of query. (D and E) Fluorescence-based PLA activity assays using VipD (D) or Rab5c (E) variants. (D) The indicated VipD protein was incubated with Rab5(Q80L)18–182:GppNHp or Rab22(Q64L)16–181:GppNHp (molar ratio 1:2) or with buffer alone, and PLA1-dependent cleavage of the substrate Bis-BODIPYFL C11-PC was detected as an increase in fluorescence emission [relative fluorescence units (RFUs)]. (E) Same assay as in D using the indicated Rab protein variants.
Given that the PLA1 activity of VipD is triggered only in response to Rab5 binding, we analyzed how amino acid substitutions that attenuate VipD–Rab5 complex formation affect the PLA1 activity of VipD. Using a generic fluorogenic substrate (bis-BODIPY FL C11-PC), we found a tight correlation between loss of PLA1 activity and the inability of VipD mutant proteins (F442A, H453D, and D479H) to enter a stable complex with Rab5c18–182(Q80L) or Rab2216–181(Q64L) (Fig. 4D). Similar results were observed for Rab5c18–182(Q80L) mutant proteins (F58A, Y83A, and R92E) that had failed to stably associate with VipD and were hence unable to trigger its PLA1 activity (Fig. 4E). The observed crystallographic interaction between VipD and Rab5 thus corresponded to their molecular association in solution, and failure to form a stable VipD–Rab5 or –Rab22 complex caused the PLA1 domain to remain in its catalytically inactive state.
Disruption of the Interaction with Rab5 Precludes Endosomal Targeting of VipD.
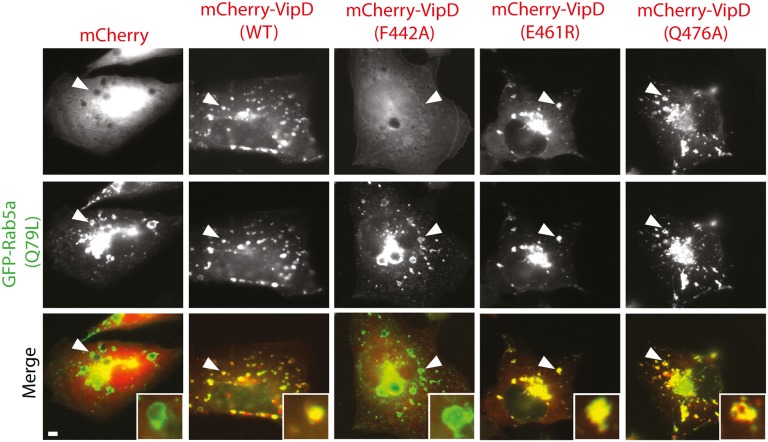
Within transiently transfected COS1 cells, fluorescently tagged VipD was enriched on Rab5-containing early endosomes, and this colocalization required the C-terminal Rab5 binding domain but not the N-terminal PLA1 domain (13, 14). A recent study reported that depletion of Rab5 (isoforms a-c) and Rab22a from HeLa cells by RNA interference (RNAi) did not affect VipD targeting to endosomes, claiming that endosomal localization of VipD would not simply depend on the interaction with Rab proteins (20). Given that RNAi rarely depletes the entire pool of a given cellular target and that VipD recruitment to endosomes could have been mediated not only by Rab5 and/or Rab22 but by additional yet unidentified Rab GTPases, we set out to reevaluate VipD’s endosomal targeting mechanism. For that, we analyzed the intracellular distribution pattern of four VipD mutant proteins that were either severely (F442A) or moderately (E461R and Q476A) attenuated for Rab5c binding in vitro (Fig. 4). Although WT VipD displayed robust colocalization with GFP-Rab5c18–182(Q80L)-positive endosomes, VipD(F442A) was entirely cytosolic (Fig. 5), consistent with this mutant’s inability to bind Rab5c. In contrast, VipD(E461R) and VipD(Q476A) showed no apparent difference in localization compared with WT VipD (Fig. 5). These findings strongly suggest that endosomal targeting of VipD is in fact dependent on the interaction with host cell Rab GTPases and that interference with the formation of these protein complexes results in the failure of VipD to properly localize to endosomes.
Fig. 5.
VipD localization to endosomes requires a functional Rab5 binding interface. Transiently transfected COS-1 cells producing Rab5a(Q79L) and the indicated mCherry-tagged VipD variants were analyzed by fluorescence microscopy to determine protein localization. The merged images (bottom row) show Rab5a(Q79L) in green and VipD variants in red. (Insets) Magnified view of endosomes marked by an arrowhead. Control, mCherry. (Scale bar, 2 μm.)
The N-Terminal Tail of VipD Is Crucial for PLA1 Activity.
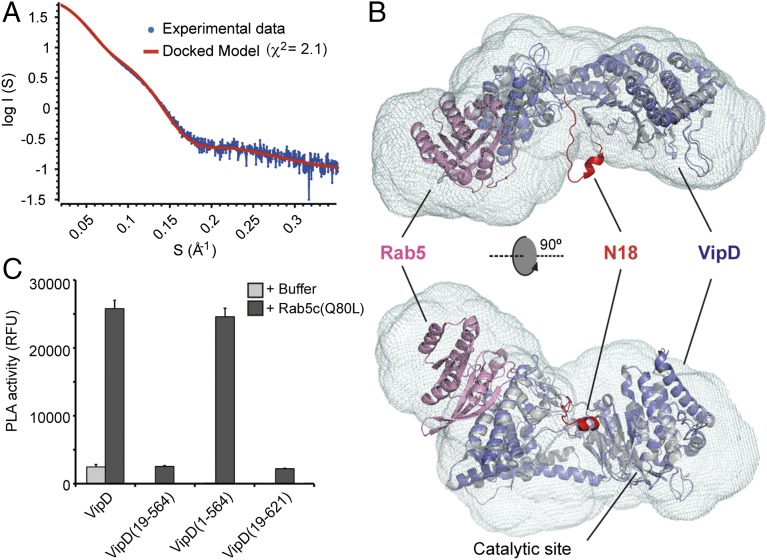
In the uncomplexed structure of VipD, the N-terminal tail (residues 1–18; N18) contained a small amphipathic helix (H1) that was involved in an intermolecular crystal contact (13). The structure of full-length VipD1–621 bound to Rab5c18–182(Q80L), on the other hand, contained no clear electron density for N18, suggesting that this region of VipD possessed high flexibility. Small angle X-ray scattering (SAXS) and gel filtration chromatography analysis suggest a heterodimeric VipD1–621–Rab5c18–182(Q80L) complex in solution, indicating that N18 was not involved in any oligomer formation (Fig. 6 A and B and Fig. S3). Given that the complexes of Rab5c18–182(Q80L) with either full-length VipD1–621 or truncated VipD19–564 exhibited nearly indistinguishable structures, we concluded that N18 was dispensable for the conformational changes induced by Rab5 binding. Consequently, we evaluated whether this short region was also dispensable for PLA1 activity of VipD. Unexpectedly, we found that, unlike VipD1–621, the truncated fragment VipD19–564 lacking N18 was strongly attenuated for PLA1 activity (Fig. 5C). To exclude the possibility that loss of PLA1 activity in VipD19–564 was caused by the lack of the C-terminal region (aa 565–621), we tested two additional constructs, VipD1–564 and VipD19–621, and detected robust PLA1 activity only in VipD1–564, indicating that N18 but not the C-terminal region critically contributed to the catalytic activity of this L. pneumophila effector (Fig. 5C). According to these observations, we propose that the flexible N18 with its amphipathic helix H1 and its close distance to the membrane plane may promote peripheral association of VipD with the lipid bilayer, possibly by orienting the catalytic site toward the membrane and/or assisting in substrate transfer.
Fig. 6.
The N-terminal 18 residues of VipD are essential for its PLA activity. (A) Fit of the optimized crystallographic VipD1–564-Rab5c18–182(Q80L) model (red line) to the experimental SAXS data of the complex (blue dots). (B) Fitting of the VipD19–564-Rab5c18–182(Q80L) crystallographic model (VipD in slate and Rab5 in pink) into the averaged ab initio envelope in two orthogonal views and superimposed with the unbound form of VipD (PDB 4AKF) in gray. Note the proximity of the N18 (residues 1–18 of VipD, PDB 4AKF) in red to the catalytic site. (C) Fluorescence-based PLA activity assays showing that the N18 segment of VipD is essential for its PLA1 activity.
Competitive Rab5 Binding Through Interface Expansion.
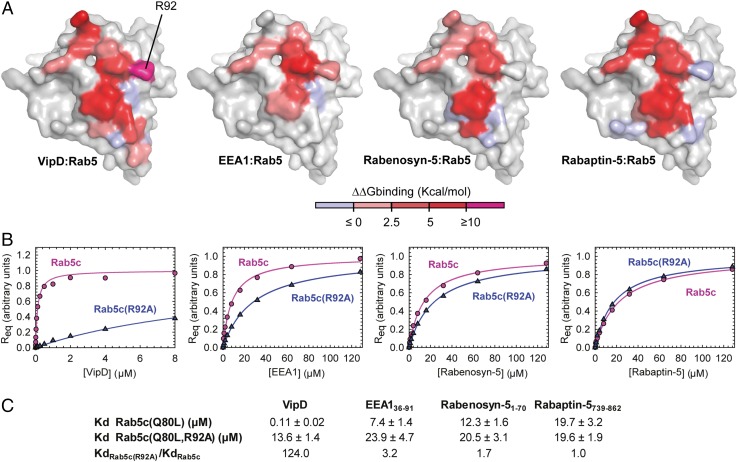
To localize to and stably associate with endosomal membranes, VipD needs to outcompete cellular ligands for Rab5 binding. EEA1, Rabaptin-5, and Rabenosyn-5 are each bound by Rab5 through a surface that includes the switch and interswitch region and that significantly overlaps with the epitope for VipD binding (19, 21, 22). To determine if and how the distribution of interaction energies differs within each of these complexes, we extended the computational alanine scan to the EEA1, Rabaptin-5, and Rabenosyn-5 epitopes and calculated the free binding energy for each of their residues (Fig. 7A and Figs. S4 and S5). All four analyzed protein interfaces share a number of nonpolar interacting residues in Rab5, namely the conserved triad (Phe58, Trp75, and Tyr90), with relatively similar energetic contributions to binding (Fig. 7A and Fig. S5B). The polar interactions surrounding this hydrophobic triad, however, determine their differential affinity, with the contact of Arg92Rab5 with Asp479VipD and Asp484VipD providing a particularly large energetic contribution to the interaction of VipD with Rab5 compared with the other cellular ligands (Fig. 7A and Fig. S5B). To verify the importance of this predicted hot-spot for VipD binding, we analyzed the affinities of either Rab5c18–182(Q80L) or Rab5c18–182(Q80L, R92A) toward VipD by surface plasmon resonance (SPR) spectroscopy. R92A mutation in Rab5 severely decreases the binding for VipD 124-fold while affecting the interaction with EEA1, Rabenosyn-5, and Rabaptin-5 to a much lesser extent (3.2-, 1.7- and 1.0-fold, respectively) (Fig. 7 B and C and Fig. S6). These findings pinpoint a binding hotspot for the superior affinity of VipD over the endogenous Rab5 ligands and confirm a good qualitative correlation between the computational analysis and the experimentally observed results.
Fig. 7.
Energy distribution between different effector binding epitopes in Rab5. (A) Space-filling model of Rab5c highlighting the epitopes for the interaction with VipD (Far Left), EEA1 C2H2 Zinc Finger (PDB ID code 3MJH) (Center Left), Rabenosyn-5 (rebuilt from PDB ID code 1Z0J) (Center Right), and Rabaptin-5 (PDB ID code 1TU3) (Far Right). Epitopes are colored as an intensity gradient according to the binding free energy change (ΔΔG) estimated as the difference between the binding ΔG of the WT and that of the alanine mutated complex. Existing glycines and alanines are excluded in the calculation. (B) Concentration dependence of the equilibrium surface plasmon resonance response for the binding of VipD WT, EEA1 C2H2 Zinc Finger (aa 36–91), Rabenosyn-5 (aa 1–70), and Rabaptin-5 (aa 739–862) to Rab5c18–182(Q80L) or Rab5c18–182(Q80L, R92A). Req represents the equilibrium SPR response normalized to the fitted maximum value for each dataset. (C) Table of the mean Kd values and SDs for at least two independent experiments showing the Kd ratio variation.
Discussion
VipD from L. pneumophila has long been predicted to function as a phospholipase during infection (11, 12), yet its catalytic activity had only recently been confirmed when it was shown that binding of host cell Rab GTPases (Rab5 and Rab22) is necessary for VipD to exhibit robust PLA1 activity (14). The crystallographic analysis described here provides an in-depth view of the Rab5-mediated activation mechanism. Above all, it uncovered a complex cascade of structural rearrangements within the C-terminal domain of VipD that result in the relocation of an active site-occluding lid and the exposure of the substrate binding pocket within the N-terminal PLA1 domain of VipD.
The structure of VipD in complex with Rab5c(Q80L) presented here is, to our knowledge, the first of a bacterial phospholipase bound to a host cell protein and the first of any translocated effector in complex with its allosteric activator molecule. Phospholipases constitute a common cellular tool to alter the lipid composition of membranes, and their activity must be carefully dosed and precisely directed toward the respective target membrane. There are more than 10,000 proteins (8,101 in Bacteria and 2,374 in Eukaryotes) containing potential patatin-like domains, most of them within a modular domain arrangement (23). Many members of the family of cytosolic phospholipases A2 (cPLA2), all of which share a patatin-like fold, contain a C2 domain crucial for membrane localization (24, 25). The patatin-like fold is also highly homologous to the group of calcium-independent phospholipases A2 (iPLA2), in which many members contain ankyrin repeats, a repetitive helix-turn-helix-loop structure considered to be a common platform for protein–protein interactions (24). Considering that Rab GTPases are key players in defining membrane identity and that many effectors from L. pneumophila have been acquired via horizontal gene transfer (26, 27), it is plausible that the scheme presented here for the concomitant localization and activation of VipD can be generalized across other microbial and eukaryotic phospholipases.
Human Rab5 interacted with VipD through a helix-turn-helix element that was similar to that used for Rabenosyn-5 binding (21), although the interface was slightly shifted toward the switch II region. Despite the observed overlapping contacts, the energy for VipD binding was not distributed uniformly across the interface but instead concentrated into a combination of hotspots that provide superior binding affinity and specificity (Fig. 7 and Fig. S6). A conserved hydrophobic triad in Rab5 (Phe58, Trp75, and Tyr90) supplied the core binding energy that was complemented by more specific polar and nonpolar contacts. Notably, most of these residues were highly conserved among Rab5 homologs from amoebean species, the natural host of L. pneumophila, or from the surrogate host Dictyostelium sp. (Fig. 3C). The ability to discriminate between GDP- and GTP-bound Rab5 and to compete with endogenous ligands evidences a remarkable adaptation for directing and retaining VipD on endosomal membranes. Interference with VipD–Rab5 complex formation, for example, by substituting Phe442 or His453 of VipD, strongly reduced the capability of these mutant proteins to interact with Rab5 (Fig. 4A), to exhibit PLA1 activity (Fig. 4D), and to efficiently localize to the endosomal compartment (Fig. 5), thus demonstrating that the function of VipD is intimately coupled to the presence of this host GTPase.
A hallmark feature of many phospholipases is to be minimally active on monomeric lipid substrates but undergo a substantial activation on binding to the surface of phospholipid membranes or micelles, a phenomenon known as interfacial activation (28–31). This behavior has been attributed to a flexible lid that at the lipid–water interface facilitates substrate diffusion to the catalytic site rather than being allosterically modulated through distant ligand binding (25). VipD does not display any interfacial activation despite having a short lid occluding the access to the catalytic site. Rather, when VipD is bound to Rab5, the lid is displaced through a chopstick-like activation mechanism in which the swing movement of two α-helices (α13 and α14) allosterically controls accessibility of the catalytic site. We cannot exclude the possibility that additional mechanisms contribute to the activity and/or specificity of the substrate catalysis by VipD. For example, the coil–helix transition of the β9–α13 loop to adjoin helix α13 relocates several charged residues closer to the catalytic groove, which might result in interactions with lipid head groups or other membrane components. Consistent with this notion, we discovered that the flexible N-terminal segment N18 of VipD is critical for the catalysis of a generic membrane-embedded substrate (Fig. 6). Deletion of N18 reduced the PLA1 activity of VipD19–564 but did not interfere with allosteric activation of the catalytic site. We hypothesize that N18 bearing a short amphipathic α-helix may facilitate the correct positioning of the PLA1 domain toward the lipid layer, promote substrate diffusion from the lipid–water interface into the catalytic site, or a combination of both effects as has been previously described for secretory PLA2 enzymes (32, 33). Interestingly, the N-terminal tail of VpdA (residues 11–54) and the N-terminal segment of P. aeruginosa ExoU (residues 57–96) showed structural similarity to the equivalent region of VipD despite lacking sequence homology (Fig. S7). Moreover, region 57–96 of ExoU, although not part of the phospholipase domain (34), was critical for cytotoxicity within transfected mammalian cells (35), suggesting that this segment contributes to the phospholipase activity of ExoU. We hypothesize that the equivalent region in VpdA may be equally important for the catalytic activity of this L. pneumophila effector and that appendixes, such as the N-terminal domains, may play important yet unresolved roles in membrane association and/or substrate transfer in other bacterial phospholipases.
In summary, our findings disclose an unexpected mode of long-range allosteric regulation of the PLA1 activity of VipD and explain how endosomal targeting is accomplished through competitive Rab5 binding. Our study also provides the basis for the development of novel therapeutic approaches that, rather than directly targeting the enzyme’s active site, specifically disturb the host factor-mediated activation process of VipD and related microbial phospholipases.
Materials and Methods
Plasmids and Cloning.
The DNA sequences encoding VipD, VipD19–564, VipD1–564, VipD19–621, Rab5c18–182(Q80L), Rab22a16–181(Q64L), EEA136–91, and Rabenosyn51–70 were cloned into the vector pGST-Parallel2 (36) using BamHI and XhoI restriction sites. Rabaptin5739–862 and Rab7a(Q67L) were cloned between the NcoI and XhoI restriction sites of pGST-Parallel2. PCR was performed using Phusion polymerase (Thermo). The PCR product was purified with QIAquick Gel Extraction Kit (NewEngland) and ligated into the digested pGST-Parallel2 vector using Quickligase (BioLab). The ligation mixture was used to transform Escherichia coli XL1 Blue competent cells, and transformants were then selected on Luria-Bertani (LB) plates containing 100 µg/mL ampicillin. The presence of the insert in the plasmid was tested by colony PCR. Quickchange mutagenesis was used to make directed mutations. The correct transformants were grown to isolate the plasmids that were sequenced on both strands. Plasmids and oligonucleotides used in this study are listed in Tables S1 and S2, respectively.
Protein Expression and Purification.
VipD was purified from E. coli BL21 (DE3) grown in LB medium and induced at an OD600 = 0.8 by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside. Cells were harvested after 16 h of growth at 18 °C. The cell pellet was resuspended in buffer A (50 mM Tris⋅HCl, pH 8.0, 300 mM NaCl, and 1 mM DTT) supplemented with 0.1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, and 1 mg/mL lysozyme and disrupted by sonication, and the lysate was cleared by centrifugation at 50,000 × g for 45 min. The supernatant was incubated for 2 h in batch with glutathione Sepharose beads (GE Healthcare) followed by extensive washing of the beads with buffer A in a gravity column. The N-terminal glutathione S-transferase (GST)-tag and linker were proteolytically removed by overnight incubation at 4 °C in the presence of tobacco etch virus (TEV) protease in 50 mM Tris⋅HCl, pH 8.0, 150 mM NaCl, and 1 mM DTT. The cleaved protein was eluted and further purified by ion exchange chromatography (HitrapQ; GE Healthcare) using a gradient of 50–1,000 mM NaCl, followed by size exclusion chromatography (Superdex200 16/60; GE Healthcare) in buffer B [25 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 5% (vol/vol) glycerol, and 1 mM DTT]. VipD mutants and truncated constructs were purified following the same procedure. The concentration of these proteins was calculated using the theoretical extinction coefficient.
Rab5c18–182(Q80L) was purified as described for VipD, with the difference that the HitrapQ column gradient was 25–1,000 mM, and the size exclusion chromatography was performed in a Superdex75 16/60 (GE Healthcare). Nucleotide exchange was achieved by incubation of the purified protein with a 20-fold excess of GppNHp in 50 mM Tris, pH 7.5, 150 mM NaCl, and 5 mM EDTA for 12 h at 4 °C. The exchange reaction was stopped by addition of MgCl2 (10 mM final concentration). Excess nucleotide was removed by gel filtration using a Superdex75 16/60 column in buffer C [25 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 5% (vol/vol) glycerol, 1 mM MgCl2, and 1 mM DTT]. Rab5c18–182, Rab5c18–182(Q80L) mutants, Rab22a16–181(Q64L), and Rab7a(Q67L) were purified as previously described. The concentration of the Rab proteins was determined by using Bradford’s procedure with BSA as standard.
For complex formation, VipD was incubated with GppNHp-bound Rab5c18–182(Q80L) in a 1:3 molar ratio for 2 h at 4 °C. The complex was further purified using a Superdex200 16/60 column equilibrated in buffer C and concentrated to 50 mg/mL using Amicon centrifugal concentrators (Millipore).
GST, GST-VipD, GST-Rabenosyn51–70, GST-EEA136–91, and GST-Rabaptin5739–862 were purified with the same protocol as described for VipD with the only difference that no TEV protease cleavage was performed during the purification.
Crystallization and Structure Determination.
Crystals were obtained by hanging-drop vapor diffusion at 18 °C by mixing 1 µL purified VipD19–564–Rab5c18–182(Q80L):GppNHp complex (50 mg/mL) and 1 µL precipitant solution (16% PEG6000, 0.1 M Tris⋅HCl, pH 8.0, and 0.2 M LiCl). Rod-shaped crystals grew within 2–3 d. Individual crystals were cryo-protected by a brief soak in well buffer supplemented with 25% (vol/vol) ethylene glycol and flash frozen in liquid nitrogen.
Diffraction data were collected at 100 K using radiation with a wavelength of 0.976 Å on beamline I04 at the Diamond Light Source (Didcot, UK). The data were integrated and scaled using XDS (37). The structure was solved by molecular replacement using the coordinates of VipD [Protein Data Bank (PDB) ID code 4AKF) and Rab5c (PDB ID code 1Z07) as a search model in Phaser (38). Subsequent rounds of refinement and interactive manual building were performed using Phenix (39) and Coot (40). For cross-validation, 5% of the original reflections was omitted from refinement and used to calculate the free R factor. The final model contained four complexes of VipD–Rab5c(Q80L). Only 10 residues (345–354) located in a connecting loop could not be modeled because of poor electron density in this region. Crystallographic data collection and model statistics are summarized in Table 1. The Ramachandran plot of the model calculated with the Rampage evaluation tool (41) shows 96.0% of the residues in the favored regions, whereas 3.9% fall in the allowed regions and 0.1% in disallowed regions. Graphics presented in this manuscript were generated using the program PyMOL (The PyMOL Molecular Graphics System; Schrödinger).
Pulldown Assays.
In vitro pulldown assays involving VipD–Rab5 interface mutants included GST-VipD, GST-Rab5c18–182(Q80L), VipD point mutants, and Rab5c18–182(Q80L) point mutants. The pulldown between VipD and Rab22 proteins included Rab22a16–181(Q64L) and VipD point mutants. No TEV protease cleavage was performed during the purification of GST-VipD, GST-Rab5c18–182(Q80L), or GST-Rab22a16–181(Q64L). For pulldowns involving VipD point mutants and GST-Rab5c18–182(Q80L), 13 µM VipD (WT or mutants) was mixed with 10 µM of GST-Rab5c18–182(Q80L) in binding buffer (50 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 1 mM DTT). Then 10 µL of equilibrated glutathione Sepharose beads were added to 70 µL of the protein mixture and incubated for 2 h at 4 °C with gentle agitation. Beads were washed several times with 0.5 mL of binding buffer and resuspended in sample buffer. The samples were subjected to 4–12% SDS/PAGE analysis, and gels were stained with Coomassie brilliant blue. The pulldown experiment between GST-Rab22a16–181(Q64L) and VipD point mutants was performed in a like manner. The binding between GST-VipD and Rab5c18–182(Q80L) point mutants was analyzed similarly using 20 µM GST-VipD and 30 µM Rab5c18–182(Q80L) (WT or mutants) in each individual reaction. Each pulldown was performed in triplicate with similar outcomes.
Phospholipase Assays.
The phospholipase activity of VipD and its mutants was assayed using bis-BODIPY FL C11-PC (Invitrogen), a glycerophosphocholine with BODIPY FL dye-labeled sn-1 and sn-2 acyl chains, respectively. This fluoregenic substrate is selfquenched and release of the fluorophores by acyl chain cleavage by either PLA1 or PLA2 results in increased fluorescence. To prepare fluorescence-labeled liposomes, we mixed 30 µL 10 mM dioleoylphosphatidylcholine (DOPC), 30 µL 10 mM dieloylphosphatidylglycerol (DOPG), and 30 µL of 1 mM bis-BODIPY FL C11-PC. All these compounds were dissolved in ethanol. The substrate was incorporated into liposomes by a slow injection of this ethanolic lipid mix into 5 mL assay buffer (50 mM Tris, pH 7.5, 150 mM NaCl, and 1 mM MgCl2) under continuous stirring. The mixture was pipetted into the side of the vortex using a narrow orifice gel-loading tip. Fifty microliters of each substrate solution was incubated with 50 µL bis-BODIPY FL C11-PC–labeled liposomes in 96-well plates for 2 h at 25 °C. The reaction mixtures contained VipD 2.5 µM and Rab 5 µM or the corresponding mutants in assay buffer. The fluorescence intensity was measured at 485-nm excitation and 530-nm emission in a multiwell reader (Biotek Synergy HT-1). All of the measurements were performed in triplicate. The assay buffer in the absence of enzyme was used as a blank.
Immunofluorescence Microscopy.
VipD localization was analyzed in COS-1 cells grown on coverslips in 24-well plates in 5% CO2 at 37 °C in DMEM media supplemented with 10% FBS. Semiconfluent monolayers were transiently transfected using Lipofectamine 2000 (Invitrogen) to produce fluorescently tagged (mCherry or GFP) proteins. Cells were fixed 10 h after transfection, and images were analyzed on a Zeiss Axio Observer.Z1 inverted light microscope using a Zeiss Plan-Apochromat 63×/ oil M27 objective and processed with Zeiss AxioVision 4.7.2 software.
SAXS.
Synchrotron SAXS data were collected on beamline BM29 at ESRF (Grenoble, France) with a 2D detector (Pilatus 1M) over an angular range qmin = 0.01 Å−1 to qmax = 0.5 Å−1. X-ray scattering patterns were recorded with the VipD1–564–Rab5c18–182(Q80L) complex at 2.2 and 6.4 mg/mL in 150 mM NaCl, 0.5 mM tris-(2-carboxyethyl)phosphine, and 25 mM Hepes, pH 7.5.
Data collection, processing, and initial analysis were performed using beamline software BsxCuBE. Further analyses were performed with the ATSAS suite. PRIMUS (42) was used for Rg determination with the Guinier method, and maximum distance (Dmax) was evaluated using GNOM (43), which was also used to calculate the distance distribution functions. Fitting of the model of the VipD–Rab5c structure to the SAXS data was calculated with CRYSOL (44) with a χ2 against raw data of 2.12 and 3.2 for samples at 2.2 and 6.4 mg/mL, respectively. To generate an ab initio model of the VipD–Rab5c complex, 20 runs of GASBOR (45) were performed using the merge of the two datasets (2 and 6.4 mg/mL) as raw data. Then, the most probable model was filtered with DAMSEL (46), and a 720 bead model was produced. Superposition of the bead model on the crystallographic VipD–Rab5c structure was carried out using the program SUPCOMB13 (47). The resulting bead model was converted to a mesh envelope and visualized using PYMOL (Schrödinger).
Molecular Dynamics Simulations.
A total of four molecular dynamics (MD) simulations were performed starting from the Rab5–VipD crystallographic structure and from three different Rab5 complexes previously described (Rab5–EEA1 C2H2 Zinc Finger, Rab5-Rabaptin5, and Rab5-Rabenosyn5). The initial coordinates of Rab5–EEA1 C2H2 Zinc Finger (PDB ID code 3MJH) and Rab5–Rabaptin5 (PDB ID code 1TU3) were taken from the Protein Data Bank, whereas the Rab5–Rabenosyn5 complex was rebuilt using the Rab22–Rabenosyn5 crystal structure (PDB ID code 1Z0J) as a template. In Rab5–VipD, Rab5–Rabaptin5, and Rab5–Rabenosyn5 structures, GppNHp molecules were replaced by GTP. In case of incomplete chains, acetyl and amide capping groups were added to the N-term and C-term residues flanking the mission regions to avoid improper charges on them. The protonation state of the ionizable residues was estimated at pH 6.5 using the server H++ (http://biophysics.cs.vt.edu/H++) (48–50). The parameter files for the GTP molecule and Zn2+ ion were prepared with the AMBER (51) module ANTECHAMBER, and the topology files for the protein complexes were generated using LEAP. Before running the molecular dynamics simulations, a short minimization and a five-step equilibration protocol were performed on the solvated structure, as previously described (52). On Rab5–VipD, Rab5–Rabaptin5, and Rab5–Rabenosyn5 complexes, unrestrained 10-ns MD simulations were performed in an isothermal-isobaric ensemble, setting pressure to 1 atm and temperature to 300 K. In the Rab5–EEA1 complex, 2.5-Å distance restraints between Zn2+ ion and each EEA1 zinc finger motif residue (Cys43, Cys46, His59, and His64) were applied during all of the equilibration and MD simulation step, to keep the same coordination as in the initial structure. The RMSD for the Cα atoms of each complex along the MD trajectory were calculated with the ptraj AMBER12 tool (51).
Computational Alanine Scanning.
We used the MMPBSA.py script in AMBER12 (51) to perform Computational Alanine Scanning calculations on 200 snapshots extracted from the last 2 ns of each complex MD trajectory (see above). All of the interface residues (defined as those located within 4-Å distance from the protein partner in the most representative structure along the last 2 ns of the trajectory) were mutated to alanine, and then the binding free energy change (ΔΔG) was estimated as the difference between the binding ΔG (MM-GBSA method) of the WT and that of the mutated complex. The contribution of conformational entropy was not included here, given the difficulty of computing it for a large protein– protein complex but that should not significantly affect the comparison of mutant and WT free energies.
Surface Plasmon Resonance Measurements.
The binding affinity of VipD, Rabenosyn51–70, EEA136–91, and Rabaptin5739–862 for Rab5c(Q80L) or Rab5c(Q80L,R92A) was calculated using SPR. SPR data were collected using a Biacore 3000 instrument (GE Healthcare) and a GST sensor chip. A research grade CM5 chip was first conditioned with three 5-µL injections of 100 mM glycine-NaOH, pH 12. Anti-GST antibody was covalently immobilized on the CM5 sensor chip injecting 45 µL at 30 µg/mL in 10 mM sodium acetate, pH 5.0, using the amine coupling kit [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and N-hydrosuccinimide] supplied by GE Healthcare. Nearly 30,000 resonance units (RUs) of the antibody were immobilized under these conditions in each flow cell, where 1 RU corresponds to immobilized protein concentration of ∼1 pg/mm2. The unreacted moieties on the surface were blocked with ethanolamine. The immobilization procedure was done at 5 µL/min with the running buffer containing 10 mM Hepes, pH 7.5, 150 mM NaCl, and 0.005% Tween20. Binding experiments were performed with the same buffer supplemented with 2 mM MgCl2. All of the proteins were dialyzed into this buffer. GST, GST-VipD, GST-Rabenosyn51–70, GST-EEA136–91, and GST-Rabaptin5739–862 were captured on the sensor chip with a 5-µL injection of 100 nM ligand at 5 µL/min on a reference and sample flow cell. Rab5c(Q80L) and Rab5c(Q80L, R92A) incubated with GppNHp were injected at different concentrations for a contact time of 2 min. Binding experiments were carried out at a flow rate of 20 µL/min at 25 °C. The anti-GST sensor chip was regenerated after each analyte injection with a 2-min injection of 10 mM glycine-HCl, pH 2.1. This regeneration procedure did not alter to any measurable extent the ability of the immobilized antibody to bind protein in subsequent cycles. Analysis of the data was performed using the BIAevaluation software supplied with the instrument. The steady-state binding response was determined by averaging the response over 5 s at the end of the injection and was corrected for background binding. The saturation binding values were fitted according to a one-site binding model. Each experiment was repeated in triplicate. Values of KD are reported as the means of independent experiments with corresponding SDs.
Supplementary Material
Acknowledgments
We thank A. Vidaurrazaga for technical assistance. We thank all the staff from the facilities where work was done for technical and human support. This work was supported by Carlos III Health Institute Grant PI11/00121 and Basque Government Grant PI2011-26 (to A.H.). This study made use of the Diamond Light Source (Oxfordshire, United Kingdom), partly funded by the BioStruct-X program (Proposal 2370), the European Synchrotron Radiation Facility (Grenoble, France) under the Block Allocation Group MX1283/MX1420, and ALBA synchrotron beamline BL13-XALOC under Proposal 2012100351.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The structural coordinates of the VipD19-564-Rab5c18-182(Q80L):GMP–PNP complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4KYI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405391111/-/DCSupplemental.
References
- 1.Christie PJ, Whitaker N, González-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843(8):1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fronzes R, et al. Structure of a type IV secretion system core complex. Science. 2009;323(5911):266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voth DE, Broederdorf LJ, Graham JG. Bacterial Type IV secretion systems: Versatile virulence machines. Future Microbiol. 2012;7(2):241–257. doi: 10.2217/fmb.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143(5):694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galán JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5(6):571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christen M, et al. Activation of a bacterial virulence protein by the GTPase RhoA. Sci Signal. 2009;2(95):ra71. doi: 10.1126/scisignal.2000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LaRock DL, Brzovic PS, Levin I, Blanc MP, Miller SI. A Salmonella typhimurium-translocated glycerophospholipid:cholesterol acyltransferase promotes virulence by binding to the RhoA protein switch regions. J Biol Chem. 2012;287(35):29654–29663. doi: 10.1074/jbc.M112.363598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohlson MB, et al. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe. 2008;4(5):434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson DM, et al. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol. 2011;82(6):1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: Actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA. 2000;97(17):9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci USA. 2005;102(13):4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VanRheenen SM, Luo ZQ, O’Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74(6):3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ku B, et al. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 2012;8(12):e1003082. doi: 10.1371/journal.ppat.1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaspar AH, Machner MP. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc Natl Acad Sci USA. 2014;111(12):4560–4565. doi: 10.1073/pnas.1316376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: A sum of parts. Curr Opin Microbiol. 2009;12(1):67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horwitz MA. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983;158(6):2108–2126. doi: 10.1084/jem.158.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: Aging gracefully. Biochem J. 2002;366(Pt 3):689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merithew E, et al. Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition. J Biol Chem. 2001;276(17):13982–13988. doi: 10.1074/jbc.M009771200. [DOI] [PubMed] [Google Scholar]
- 19.Zhu G, et al. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat Struct Mol Biol. 2004;11(10):975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
- 20.Zhu W, Hammad LA, Hsu F, Mao Y, Luo ZQ. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell Microbiol. 2013;15(11):1783–1795. doi: 10.1111/cmi.12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by rabenosyn-5. Nature. 2005;436(7049):415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra A, Eathiraj S, Corvera S, Lambright DG. Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1) Proc Natl Acad Sci USA. 2010;107(24):10866–10871. doi: 10.1073/pnas.1000843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, et al. Pfam: The protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111(10):6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dessen A, et al. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell. 1999;97(3):349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 26.de Felipe KS, et al. Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol. 2005;187(22):7716–7726. doi: 10.1128/JB.187.22.7716-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Quadan T, Price CT, Abu Kwaik Y. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 2012;20(6):299–306. doi: 10.1016/j.tim.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brzozowski AM, et al. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature. 1991;351(6326):491–494. doi: 10.1038/351491a0. [DOI] [PubMed] [Google Scholar]
- 29.van Tilbeurgh H, et al. Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography. Nature. 1993;362(6423):814–820. doi: 10.1038/362814a0. [DOI] [PubMed] [Google Scholar]
- 30.Roussel A, et al. Crystal structure of the open form of dog gastric lipase in complex with a phosphonate inhibitor. J Biol Chem. 2002;277(3):2266–2274. doi: 10.1074/jbc.M109484200. [DOI] [PubMed] [Google Scholar]
- 31.Aloulou A, et al. Exploring the specific features of interfacial enzymology based on lipase studies. Biochim Biophys Acta. 2006;1761(9):995–1013. doi: 10.1016/j.bbalip.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Qin S, Pande AH, Nemec KN, Tatulian SA. The N-terminal alpha-helix of pancreatic phospholipase A2 determines productive-mode orientation of the enzyme at the membrane surface. J Mol Biol. 2004;344(1):71–89. doi: 10.1016/j.jmb.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 33.Qin S, Pande AH, Nemec KN, He X, Tatulian SA. Evidence for the regulatory role of the N-terminal helix of secretory phospholipase A(2) from studies on native and chimeric proteins. J Biol Chem. 2005;280(44):36773–36783. doi: 10.1074/jbc.M506789200. [DOI] [PubMed] [Google Scholar]
- 34.Gendrin C, et al. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog. 2012;8(4):e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finck-Barbançon V, Frank DW. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol. 2001;183(14):4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15(1):34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovell SC, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 42.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36(5):1277–1282. [Google Scholar]
- 43.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst. 1992;25(4):495–503. [Google Scholar]
- 44.Svergun D, Barberato C, Koch MHJ. CRYSOL: A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28(6):768–773. [Google Scholar]
- 45.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36(3 Part 1):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Cryst. 2001;34(1):33–41. [Google Scholar]
- 48.Anandakrishnan R, Aguilar B, Onufriev AV. H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 2012;40(Web Server issue):W537–541. doi: 10.1093/nar/gks375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Myers J, Grothaus G, Narayanan S, Onufriev A. A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins. 2006;63(4):928–938. doi: 10.1002/prot.20922. [DOI] [PubMed] [Google Scholar]
- 50.Gordon JC, et al. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33(Web Server issue):W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Case DA, et al. 2012. AMBER 12 (University of California, San Francisco)
- 52.Meyer T, et al. MoDEL (Molecular Dynamics Extended Library): A database of atomistic molecular dynamics trajectories. Structure. 2010;18(11):1399–1409. doi: 10.1016/j.str.2010.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.