Significance
The bacteriophage encoded λ Int protein is distinguished from other well-studied and widely exploited tyrosine recombinase family members as a heterobivalent DNA binding protein. With the help of accessory DNA bending proteins, Int bridges two different classes of DNA sites within the unique 400-kDa recombinogenic complexes of integrative and excisive recombination. The absence of any overarching investigations or structural models for these key complexes stems from the inability to determine which core-type and arm-type binding sites are joined to one another by Int-mediated bridges. We solved this long-standing problem using site-directed chemical cross-linking of Int in trapped Holliday junction intermediates and recombination reactions with chimeric recombinases, to identify the unique patterns of Int bridges for integrative and excisive recombination.
Keywords: site-specific recombination, regulation of directionality, recombinogenic architectures, molecular machines
Abstract
The site-specific recombinase encoded by bacteriophage λ [λ Integrase (Int)] is responsible for integrating and excising the viral chromosome into and out of the chromosome of its Escherichia coli host. In contrast to the other well-studied and highly exploited tyrosine recombinase family members, such as Cre and Flp, Int carries out a reaction that is highly directional, tightly regulated, and depends on an ensemble of accessory DNA bending proteins acting on 240 bp of DNA encoding 16 protein binding sites. This additional complexity enables two pathways, integrative and excisive recombination, whose opposite, and effectively irreversible, directions are dictated by different physiological and environmental signals. Int recombinase is a heterobivalent DNA binding protein that binds via its small amino-terminal domain to high affinity arm-type DNA sites and via its large, compound carboxyl-terminal domain to core-type DNA sites, where DNA cleavage and ligation are executed. Each of the four Int protomers, within a multiprotein 400-kDa recombinogenic complex, is thought to bind and, with the aid of DNA bending proteins, bridge one arm- and one core-type DNA site. Despite a wealth of genetic, biochemical, and functional information generated by many laboratories over the last 50 y, it has not been possible to decipher the patterns of Int bridges, an essential step in understanding the architectures responsible for regulated directionality of recombination. We used site-directed chemical cross-linking of Int in trapped Holliday junction recombination intermediates and recombination reactions with chimeric recombinases, to identify the unique and monogamous patterns of Int bridges for integrative and excisive recombination.
The tyrosine recombinase family, which includes the well-studied and highly exploited Cre, Flp, and λ Integrase (Int) recombinases, is responsible for such diverse functions as chromosome segregation, chromosome copy number control, gene expression, conjugative transposition, gene dissemination, and viral integration and excision [for reviews, see Mobile DNA II (1) and the in preparation Mobile DNA III]. The virally encoded λ Int recombinase is responsible for integrating and excising the λ chromosome into and out of the chromosome of its Escherichia coli host in response to a variety of physiological and environmental signals (2). Although all members of this family use the same isoenergetic chemistry and strand exchange mechanisms to execute DNA rearrangements, Int (in contrast to Cre and Flp) depends on an ensemble of accessory DNA bending proteins and carries out a recombination, between att site target DNAs, that is highly directional and tightly regulated (3–8).
Int is a heterobivalent DNA binding protein that binds to high-affinity “arm-type” DNA sites via its small amino-terminal domain (NTD), and to “core-type” DNA sites, where DNA cleavage and ligation takes place, via a central core binding domain (CB) and a C-terminal catalytic domain (CAT); the latter two domains are referred to here as the CTD. Each of the four Int protomers, within a multiprotein 400-kDa recombinogenic complex, is thought to bind and bridge one arm- and one core-type DNA site; the bridging interactions are facilitated by accessory DNA bending proteins IHF, Xis, and Fis. Differential occupancy of the 16 DNA protein binding sites (encoded by 240 bp of att site DNA) generates two overlapping ensembles that differentiate integrative from excisive recombination, as diagrammed in Fig. 1.
Fig. 1.
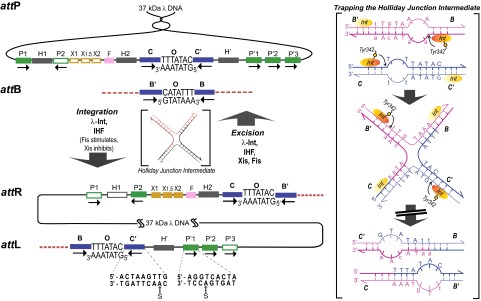
The overlapping ensembles of protein binding sites that comprise att site DNA and the DNA modifications used for cross-linking. Integrative recombination between supercoiled attP and linear attB requires the virally encoded Integrase (Int) (29) and the host-encoded accessory DNA bending protein IHF (30, 31) and gives rise to an integrated phage chromosome bounded by attL and attR. Excisive recombination between attL and attR to regenerate attP and attB additionally requires the phage-encoded Xis protein (which inhibits integrative recombination) (32) and is stimulated by the host-encoded Fis protein (33). Both reactions proceed through a Holliday junction intermediate that is first generated and then resolved by single-strand exchanges on the left and right side of the 7-bp overlap region, respectively. The two reactions proceed with the same order of sequential strand exchanges (not the reverse order) and use different subsets of protein binding sites in the P and P′ arms, as indicated by the filled boxes: Int arm-type P1, P2, P′1, P′2, and P′3 (green); IHF, H1, H2, and H′ (gray); Xis, X1, X1.5, and X2 (gold); and Fis (pink). The four core-type Int binding sites, C, C′, B, and B′ (blue boxes) are each bound in a C-clamp fashion by the CB and CAT domains, referred to here as the CTD. These core sites are where Int executes isoenergetic DNA strand cleavages and ligations via a high energy covalent 3′-phospho-tyrosine intermediate. The CTD of Int and the tetrameric Int complex surrounding the two overlap regions are functionally and structurally similar to the Cre, Flp, and XerC/D proteins. The bottom line shows the locations of the cystamine modifications (S) within the core- and arm-type consensus sequences (see main text and Tables S1 and S2). (Right) DNA sequence changes made in the 7-bp overlap regions to trap Holliday junction intermediates (lowercase letters). Following the first pair of Int cleavages (via the active site Tyr) on one side of the overlap regions (arranged here in antiparallel orientation) the “top” strands are swapped to form the HJ; this simultaneously converts the unpaired (bubble) bases to duplex DNA. On the other side, the sequence differences between the two overlap regions strongly disfavor the second (“bottom”) strand swap that would resolve the HJ, because this would generate unpaired bubbles in the product (11, 12). This diagram applies to both integrative and excisive recombination (even though the labels refer to integrative recombination).
Despite a wealth of genetic, biochemical, functional, and structural information, generated by many laboratories over the last 50 y, it has not been possible to determine which of the five arm-type DNA sites is paired via an Int bridge with which of the four core-type sites. This deficiency has been the major obstacle in understanding the architectures responsible for regulated directionality of integrative and excisive recombination. In this report, we address the need for a direct determination of the λ Int bridging patterns using site-directed chemical cross-linking of Int to trapped Holliday junction recombination intermediates, and we confirm the observed Int bridges in recombination reactions with chimeric recombinases.
Results
Trapping Recombination Intermediates.
λ Int is known to form off-pathway complexes and is also capable of promoting noncanonical recombination reactions under some conditions (9, 10). We therefore decided to study complexes that were trapped in the course of a canonical, bona fide recombination reaction (as opposed to assembling complexes from individual components). The most attractive candidate for this is the Holliday junction intermediate (HJ), the four-way DNA junction generated from two partner DNAs by the first pair of DNA strand exchanges and then resolved to two product DNAs by the second pair of DNA strand exchanges, i.e., the midpoint between reactants and products.
We efficiently trapped stable HJ complexes as described previously and diagrammed in Fig. 1 (11, 12) by recombining two att site partners with the following two features. On the left side of their respective overlap regions, each partner has an unpaired 2-bp heteroduplex bubble, such that strand exchange between them creates an HJ with fully base paired DNA. Reversal of this top-strand exchange would result in reformation of the unpaired heteroduplex bubbles and is thus energetically disfavored. On the right side of their respective overlap regions, the two partners differ in 2 bp of DNA duplex sequence. Therefore, the right hand strand exchange, which is required to resolve the HJ, would create a 2-bp heteroduplex in each recombinant product and is energetically disfavored. Thus, resolution of the fully duplex HJ in either direction is energetically disfavored and rarely occurs. We have characterized the stable nucleoprotein HJ complexes extensively in a wide variety of experiments, as described in SI Text and Fig. S1.
Chemical Cross-Linking of Int Bridges in the HJ Complex.
There are potentially four arm-core Int bridges in the integrative pathway and three in the excisive pathway, which requires only three arm-type sites. To map the Int bridges, we adapted the disulfide trapping technology developed in the Verdine laboratory (13, 14) to introduce disulfide cross-links at the protein–DNA interfaces between an Int NTD and its cognate arm-type site and between an Int CTD and its cognate core-type site. As described below, we used site-directed mutagenesis to introduce appropriately positioned Cys residues at the NTD and/or CTD DNA binding surfaces of an Int in which the two surface accessible Cys residues (C25S and C197S) had been mutated to generate an Int with two buried Cys residues (verified to be nonaccessible in the native protein) (SI Text). Additionally, we incorporated sulfhydryl groups with a two-carbon tether at N6 of appropriately positioned adenosine residues in the DNA of selected arm- and/or core-type sites.
Candidate amino acid and nucleotide residues that were juxtaposed at the protein–DNA interfaces were identified by inspecting the crystal structures of several Int–DNA complexes (15, 16). From these candidates, we constructed and characterized 14 different single Cys substitutions in the NTD or CTD domains and 12 oligonucleotides with appropriately positioned cystamine modifications in the P′1 arm or C′ core sites (Tables S1 and S2).
Based on the results of these analyses, we chose substitution N20C (hereafter called N-Cys) and position 4b of the arm-type consensus recognition sequence as the best-matched pair of targets for generating disulfide cross-links between the NTD and arm sites. We chose substitution G283C (hereafter called C-Cys) and position 8b of the core-type consensus recognition sequence as the best-matched pair of targets for generating disulfide cross-links between the CTD and core sites (Fig. 1 and Tables S1 and S2). We also constructed an Int in which the NTD-Cys and CTD-Cys were combined to give the double mutant, (N+C)-Cys. Each of the three Cys-mutant proteins is more than 70% as proficient as WT Int for recombination.
We generated an ensemble of HJ-trapping att sites with all possible combinations of a single cystamine-labeled arm-type site coupled with a single cystamine-labeled core-type site for disulfide cross-linking experiments, resulting in 12 att sites for excisive recombination (4 core sites and the 3 arm sites required for excisive recombination) and 12 att sites for integrative recombination (4 arm sites and 3 core sites, the C core site could not be labeled with cystamine at the required position in the bottom strand of the supercoiled attP substrate in our protocol; Materials and Methods and SI Text). We reasoned that an (N+C)-Cys Int, doubly cross-linked to both an arm- and a core-type site, would have an identifiably distinct electrophoretic mobility in SDS/PAGE compared with an N-Cys or C-Cys Int singly cross-linked to only one type of DNA site. The doubly cross-linked Int can only be generated when the single cystamine arm site and the single cystamine core site are bridged by the same Int. Indeed, the doubly cross-linked complex is identified as a band in an SDS gel that appears only in the lanes corresponding to reactions with (N+C)-Cys Int and not in the lanes from reactions where the reactive Cys resides only in the NTD or only in the CTD. The unique bands in the (N+C)-Cys Int lanes were shown to be HJs because they could be labeled with 32P from any one of the four core sites. As an additional control, we included a reaction containing a mixture of the single N-Cys and single C-Cys Ints, which cannot give rise to the doubly cross-linked, Int-bridged, product.
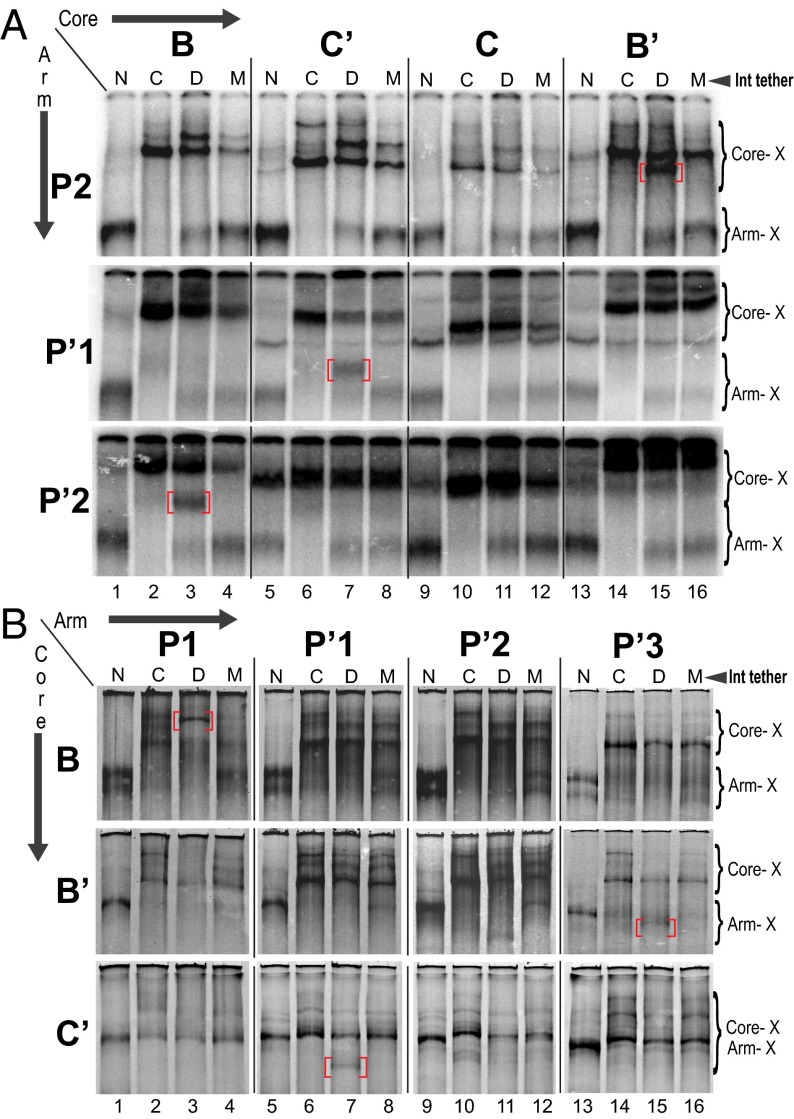
In Fig. 2, all four excisive recombination reactions contained identical DNA substrates, in which the single cystamine-modified core site was B and the single cystamine-modified arm site was P′2, chosen on the basis of the data in Fig. 3A. Each reaction contained different Int protomers: N-Cys, C-Cys, (N+C)-Cys, or a mixture of N-Cys and C-Cys. Aliquots of each reaction were analyzed by electrophoresis on SDS polyacrylamide gels of 4%, 6%, 8%, or 10%. All four gels show a single band that is unique to the (N+C)-Cys reaction (labeled lane D), consistent with results expected for an Int mediated bridge between the P′2 arm and the B core. This experiment highlights the well-known effect of shape on relative electrophoretic mobilities and anticipates our observation that the position of the double cross-link within the HJ influences its relative electrophoretic mobility (relative to singly and other doubly cross-linked complexes) in a way that depends on the gel composition. Because of this phenomenon, and the potential for comigration of bands, each of the experiments described below was analyzed on several different percentage gels (only one of which is shown in the experiments below). We also anticipated that the doubly cross-linked product would be generated in relatively low yield because many of the singly cross-linked products will derive from (abundant) non-HJ complexes, such as unrecombined substrate.
Fig. 2.
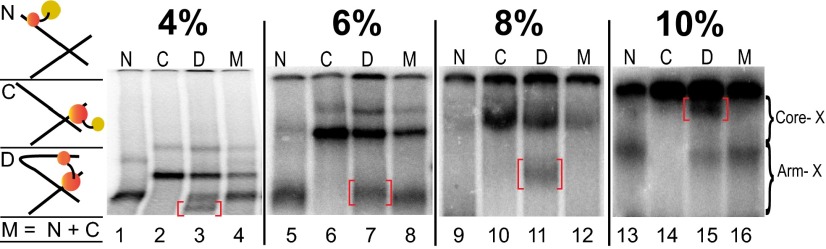
The effect of acrylamide composition on the relative electrophoretic mobility of an HJ with a doubly cross-linked Int bridge. Excisive recombination reactions were carried out between att site partners designed to trap HJ intermediates, as described in the main text. Each pair of recombining att sites contains a cystamine in the P′2 arm site and a cystamine in the B core site, two sites determined in other excisive recombination experiments (cf. Fig. 3, lanes 1–4) to be bridged by the same Int. A 32P-label was incorporated at the 5′ terminus of the P′ arm. Each of the four recombination reactions in a set contained a λ Int with a reactive Cys in the DNA binding pocket of either the NTD (N), the CTD (C), or both (D, for dual); the fourth reaction contained a mixture (M) of the N and C Ints. Following recombination, disulfide bond formation was promoted by adding hydrogen peroxide, which was then quenched by the addition of catalase. Reactions were terminated by the addition of SDS, fractionated by SDS/PAGE, and visualized on a phosphorimager. In each set of four reactions, the band unique to the dual labeled (bridging) Int is highlighted with a red bracket, and bands corresponding to complexes singly cross-linked at an arm or core site are labeled in the margin. See Materials and Methods and Results for details. (Left) Cartoon depicts the cross-linked species expected in lanes N, C, D, and M.
Fig. 3.
Mapping specific Int bridges by disulfide cross-linking. Excisive and integrative recombination reactions were carried out with the indicated Int proteins and processed as described in Fig. 2. Each pair of recombining att sites in a set of four reactions contains a single reactive cystamine in the indicated arm site and a single reactive cystamine in the indicated core site. A 32P-label was incorporated at the 5′ terminus of the P or P′ arm carrying the cystamine. The band unique to the bridging Int (D lanes) is highlighted with a red bracket, and bands corresponding to complexes singly cross-linked at an arm or core site are labeled in the margin. (A) Sets of four excisive recombination reactions between attL and attR, in which one of the three arm sites required for excision (y axis) and one of the four core sites (x axis) is labeled with cystamine. (B) Sets of four integrative recombination reactions between supercoiled attP and attB, in which one of the three tested core sites (y axis) and one of the four arm sites (x axis) is labeled with cystamine. The C core site could not be labeled with cystamine at the required position in the bottom strand of the supercoiled attP substrate (Materials and Methods and SI Text).
For the excisive recombination experiments shown in Fig. 3A, an att site, containing a single cystamine modification in one of the three arm-type sites required for excisive recombination (y axis), was recombined with att sites in which each of the four core sites (x axis) was modified with cystamine. This cycle was repeated with the cystamine label in each of the remaining arm sites. Each lane in these gels corresponds to a separate recombination reaction with its own 32P-labeled substrate; however, for each reaction condition (lane), the patterns of the bands and their relative intensities are very reproducible. The results show a unique band corresponding to an Int bridging B–P′2, C′–P′1, and B′–P2, leaving the C core site as the one that does not form an Int bridge with one of the three arm-type sites required for excisive recombination.
In a similar experiment for integrative recombination (Fig. 3B), there is a unique band corresponding to an Int bridging P1–B, P′1–C′, and P′3–B′. In lane 11, there is a weak unique band for the P′2 reactions, but its intensity relative to the reference band in the same lane (singly cross-linked CTD) was reproducibly four times weaker than the unique band in the P′3 reactions. We infer, and confirm in the genetic experiments described below, that P′2 forms an Int bridge with the C core site, which could not be labeled with cystamine at the required position in the bottom strand of the supercoiled attP substrate (Materials and Methods and SI Text).
Confirmation of the HJ Int Bridges in Complete Recombination Reactions.
Our strategy for genetically testing the Int bridges in a full recombination reaction depends on a previously constructed and characterized chimeric recombinase in which the amino-terminal domain of λ-integrase was fused with the Cre recombinase (17). Here we refer to this recombinase as Crn1. Substrates for Crn1, called lot sites, were generated by appending the appropriate P and P′ arms of the λ att sites to modified lox Cre target sites. The resulting four lot sites, lotP, lotB, lotL, and lotR (analogous to attP, B, L, and R), are recombined by Crn1 in a regulated directional reaction that depends on the λ arm-type sites and accessory proteins (IHF and Xis) in patterns identical to that of λ Int-mediated recombination (17).
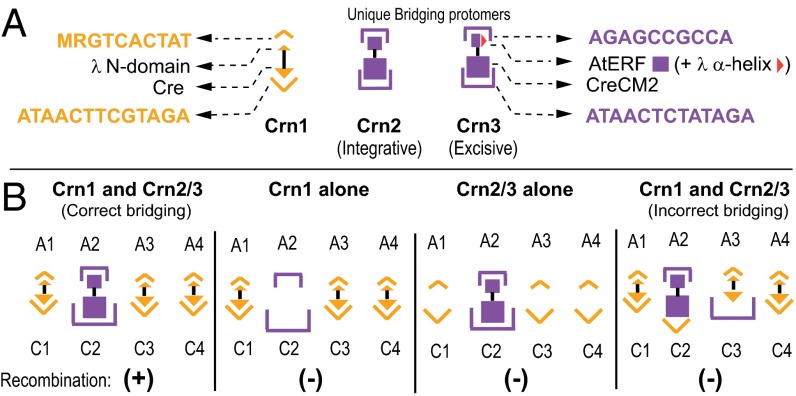
From Crn1, we created a recombinase, Crn2, with different arm- and core-type specificities. The arm-type DNA recognition domain of Crn1 (residues 16–55) was replaced with the G box DNA binding domain of the ethylene responsive factor from Arabidopsis thaliana (AtERF1) (residues 4–50) (Fig. S2 A and C). Like the Int NTD, this domain adopts a monomeric DNA binding fold consisting of a three-stranded, antiparallel β-sheet that is packed against a carboxyl-terminal α-helix (18, 19). The core-type DNA recognition specificity of Crn1 was changed by introducing the five amino acid changes of the well-characterized CreCM2 mutation, which has a DNA recognition specificity different from, and essentially nonoverlapping with, the WT Cre (20, 21). To enable Crn2 for excisive recombination, the carboxyl-terminal α-helix of the λ integrase NTD (residues 41–55 in Crn1), which has been shown to interact with Xis (22, 23), was used to replace the corresponding α-helix (residues 47–62) of Crn2 to generate Crn3 (Fig. 4A and Fig. S2). Crn3, which can be used in place of Crn2, was difficult to purify and was therefore only used for the excisive recombinations.
Fig. 4.
Confirmation of the Int bridges in complete recombination reactions. (A) Schematic of unique specificity bridging enzymes. Chimeric recombinase Crn1 (17) possesses the NTD of λ Integrase (small gold triangle), which binds to the DNA consensus sequence MRGTCACTAT, fused to the body of Cre (large gold triangle), which binds to the modified loxP (lot) site, ATAACTTCGTAGA. Crn2 possesses the NTD of AtERF (small purple box), which binds to the consensus GCC box sequence, AGAGCCGCCA, fused to the body of CreCM2 (large purple box), which binds to the lotM7 sequence, ATAACTCTATAGA. Crn3 is the same as Crn2, but the α-helix of AtERF has been replaced by a structurally analogous α-helix from λ Integrase (red triangle), which interacts with Xis (SI Text). The gold and purple color coding of the two DNA specificities is preserved in B. (B) Cartoon of the logic and possible outcomes of the experiments. In each of the excisive and integrative recombination reactions shown in Fig. 5, one of the arm-core pairs has the DNA sequences of the unique Crn2/3 specificity (purple), as indicated, whereas all of the other arm and core sites have the Crn1 specificity (gold). Recombinant product (+) is observed only when the complete and correct Int bridging pattern is achieved.
To make use of Crn1 and Crn2/3, we constructed hybrid lot sites in which one of the bridged arm-core pairs (identified by the chemical cross-linking experiments) has the arm and core sequences recognized by Crn2/3; we refer to this as the “unique bridge.” The remaining arm-core bridges have the arm and core sequences recognized by Crn1. If the chemically identified arm-core bridges of the HJs are functioning in the complete recombination pathway, Crn1 will not be able to carry out recombination on such substrates unless Crn2/3 is also present (Fig. 4B). The absence of cross-reactivity between Crn1 and Crn2/3 is shown in the proof-of-principle experiments (Fig. S3).
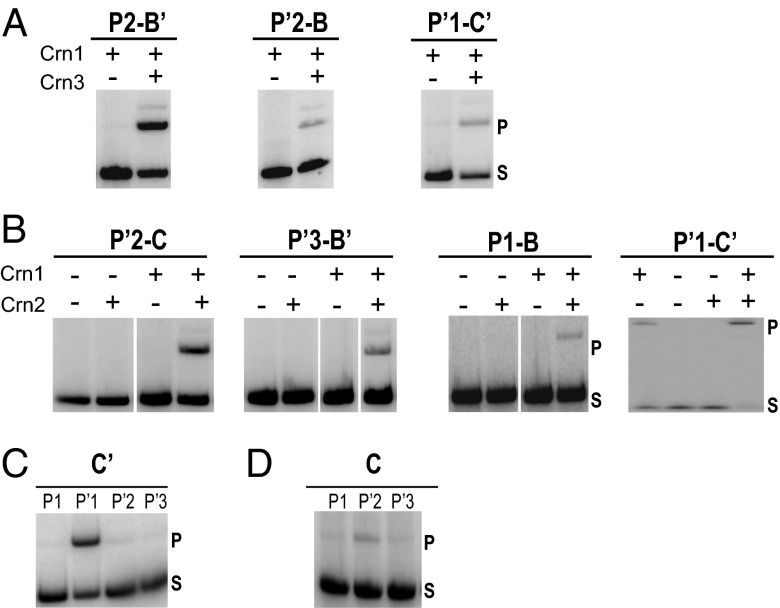
Recombination reactions in which one of the linear partners was 5′ end-labeled with 32P were analyzed by SDS/PAGE to assay for the appearance of the expected slower migrating product band. In each set of reactions, a different arm-core bridged pair has the indicated Crn2/3 specificity; all of the other arm and core sites in that set have Crn1 specificity. As predicted by the chemical cross-linking results, all of the recombination reactions required the presence of Crn2/3 in addition to Crn1 (Fig. 5).
Fig. 5.
Excisive and integrative recombination reactions carried out with a mixture of two chimeric recombinases and att site substrates containing the indicated unique arm and unique core specificities. In each of the excisive (A) and integrative (B) recombination reactions, one of the arm-core pairs identified by chemical cross-linking has the DNA sequences of the unique Crn2/3 specificity (purple in Fig. 4), as indicated, whereas all of the other arm and core sites have the Crn1 specificity (yellow in Fig. 4). In C and D, all of the integrative reactions contained both Crn1 and Crn2. The unique core specificity was fixed either at C′ (C) or at C (D), and the unique arm specificity was moved to a different arm site in each of the recombination reactions, as indicated. All of the att sites in D had Crn2 specificity at the previously characterized P′1–C′ bridged pair, so there are only three recombination reactions. Integrative recombination reactions contained supercoiled attP, linearized 32P-labeled attB, and IHF. Excisive recombination reactions contained linear attL, linear 32P-labeled attR, and accessory proteins IHF and Xis. All integrative and excisive recombinations require IHF or Xis, as shown previously (17). Recombinase concentrations were optimized for each set of experiments and typically were in the range of 50–100 nM; the gel lanes shown in A and B have been selected from a large titration gel to present the most informative concentrations. Each DNA substrate was at 6 nM. The reactions were terminated by the addition of SDS; electrophoresed on SDS/PAGE gels; and visualized on a phosphorimager (Materials and Methods and SI Text).
For the unique P′1–C′ integrative bridge, there was a small amount of recombination with Crn1 alone (Fig. 5B), but this was ∼10-fold lower than the recombination seen with both Crn1 and Crn2. This low level of background Crn1 activity is abolished by the addition of a catalytically inactive core binding domain, Cre-CM2(K201A) (24), with specificity for the Crn2 core sites. This competitor, which lacks an NTD, completely suppresses the small amount of nonspecific Crn1 binding without reducing the overall efficiency of the bona fide bridging Ints in recombination reactions (see the proof-of-principle experiments described in Fig. S3).
The integrative P′1–C′ bridge was also analyzed by constructing a set of additional hybrid att sites, in which the core-type Crn2 specificity was fixed at C′ and the arm-type Crn specificity was moved from P′1 to each of the other arm sites in turn. As predicted, the most efficient recombination was observed only when the Crn2 specificities of P′1 and C′ were paired with each other (Fig. 5C, Left).
A similar experiment was used to provide additional confirmation of the integrative P′2–C bridge, inferred from the chemical cross-linking experiments. Another set of hybrid att sites was constructed in which the core-type Crn2 specificity was fixed at C and the arm-type Crn specificity was moved from P′2 to each of the other arm sites in turn. In this set of experiments, all of the att sites also had Crn2 specificity at the previously characterized P′1–C′ bridged pair, so there are only three recombination reactions. As predicted, the most efficient recombination was observed only when the Crn2 specificities of C and P′2 were paired with each other (Fig. 5C, Right). The patterns of Int bridges consistent with both the genetic and cross-linking experiments are schematically summarized for both integration and excision in Fig. 6.
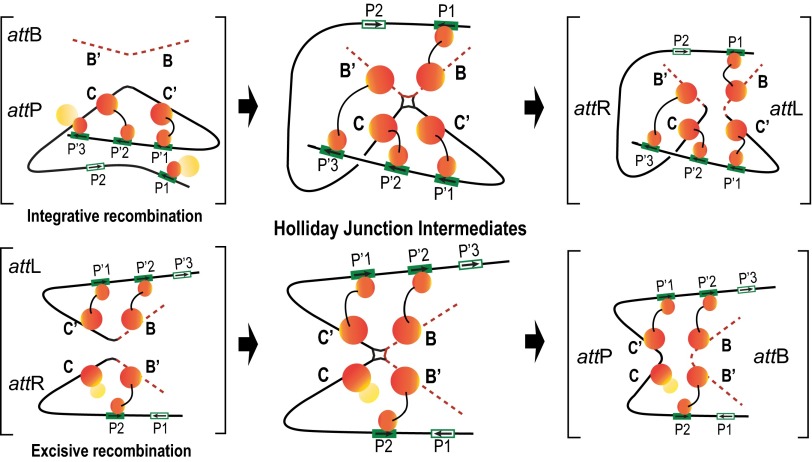
Fig. 6.
Schematic summary of the Int bridges in integrative and excisive recombination. The middle panel diagrams the Int bridges of the Holliday junction recombination intermediates determined in this study. In the integrative complex, all four core sites and four of the five arm sites enjoy an Int bridge, whereas the excisive complex engages three of the four core sites and three of the five arm sites. The flanking panels (brackets) depict extrapolations from the HJ complexes to the respective att site recombination partners (substrates) and recombinants (products) based on the deduction that Int bridges are not broken and reformed during recombination (Discussion).
Discussion
The patterns of Int bridges described here for the integrative and excisive recombinogenic complexes (Fig. 6) differ significantly from previous speculations on this subject. One of the earliest attempts to discern these patterns used a combination of synthetic lethality and protein gel shift experiments with attL and attR (25). Unfortunately, the synthetic lethality results were not definitive, and the DNA binding experiments depended on the assumption, we now know to be incorrect, that the assembled protein–DNA complexes were on the recombination pathway. The most recent attempt to identify the Int bridges depended on an extrapolation from the X-ray crystal structure of an Int tetramer bound, via the CTDs, to a chemically synthesized core site Holliday junction and also bound, via the NTDs, to a pair of short oligonucleotides encoding P′1–P′2 arm sites (16). Subsequent experiments (26), and the results reported here and in the companion paper (27), have shown that the simplified, symmetric complex required for crystallization is not an accurate mimic of the native recombinogenic complexes, which are in fact highly asymmetric.
The chemical cross-linking experiments identify the specific Int bridges comprising the two HJ recombination intermediates, i.e., they provide a snapshot of one intermediate in each of the reactions. Conversely, the genetic experiments afford an integrated view over the course of each entire recombination reaction; they report on the Int bridging requirements from the beginning to the end of the reaction. Therefore, the ability of a single arm site and a single core site (as a unique Int-bridged pair) to satisfy the requirements for recombination indicates that in each of the pathways every Int protomer is involved in only one type of bridge, and it does not have to reform new bridges over the course of the reaction.
The two kinds of experiments reported here are also complementary in another way. The chemical cross-linking benefits from flexibility and micromovements, which are conducive to sampling an optimal orientation for the precise short-range contact. In contrast, the genetic experiments benefit from a more stable and rigid environment that can better compensate for the imperfect protein–DNA interfaces inherent in the design of these experiments. Thus, where the bridging patterns, and the models described in ref. 27, predict a more rigidly held Int bridge, such as P′1–C′ in integration, the genetic signal is relatively strong and the cross-linking signal is relatively weak. Conversely, where the bridging pattern predicts a more loosely held Int bridge, such as P1–B in integration, the genetic signal is relatively weak and the cross-linking signal is relatively strong. Indeed, the loosely held P1–B bridge figures prominently in the architectures described in the companion paper (27).
The monogamous relationship of each arm-core bridged pair throughout the course of the recombination reaction makes it possible to extrapolate from the patterns observed in the HJ recombination intermediate to those predicted for the presynaptic recombination partners and the post-HJ recombination products. Inspection of Fig. 6 reveals that for excisive recombination, the presynaptic partners have only intramolecular bridges, suggesting that Int bridging is not a driving force in synapsis of attL and attR. In contrast, integrative recombination has two intermolecular bridges that are responsible for the capture of attB by supercoiled attP, as predicted by Richet and Nash (28).
Our results do not address the question of whether DNA cleavage of the top strands precedes synapsis but it may be relevant that, in both integrative and excisive recombination, one of two protomers responsible for the first strand exchange is not held tightly by an Int bridge. In excisive recombination, the Int bound at C does not bridge to an arm site and in integrative recombination the Int bound at B is bridged to P1, which resides on a weakly constrained P arm that is postulated to move during synapsis [see above discussion and companion article (27)].
Before these results, there was an appealing class of models in which regulated directionality of recombination depended on some degree of Int bridge remodeling during the course of the reaction. The results reported here argue strongly against such models. They additionally suggest that one of the contributors to directionality could be the progression from intra- to intermolecular Int bridges. Most importantly, these results lay the foundation for additional experiments aimed at determining the overall architectures of the recombinogenic complexes responsible for regulated directionality in integrative and excisive recombination (27).
Materials and Methods
Proteins, Oligonucleotides, and Recombination Reactions.
All proteins and oligonucleotides were made as described in SI Text, as are standard recombination buffer and reactions conditions.
Chemical cross-linking.
To incorporate a disulfide bond tether at the N6 position of dA, a commercially synthesized oligonucleotide containing O6-phenyl-2′-deoxyinosine (O6-phenyl-dI) at the desired position (Operon, HPLC purified) was treated with cystamine, as previously described (13, 14).
Genetic test of int bridges.
The chimeric recombination substrates were constructed from the lot sites previously described (17). The sequences for the mixed specificity core sites are shown in Fig. S2C. IHF binding at the H1 site in the P-arm inhibits excisive recombination and therefore was mutated in the original lotR substrates and also the lotR derivatives described here.
Supplementary Material
Acknowledgments
We thank Christine Lank, Lindsay Steele, and Joanne Nelson for technical assistance and Joan Boyles for administrative support. We thank Jeffrey Mumm for a gift of Cy3-labeled Int. We thank former A.L. laboratory members, especially Mihaela Matovina, Theron Hamilton, and Jeffrey Mumm, for contributions to the intellectual and experimental foundations of this work, which was supported by National Institutes of Health Grants GM062723 and GM033928 (to A.L.) and GM108751 (to G.D.V.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413007111/-/DCSupplemental.
References
- 1.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 2.Campbell AM. Episomes. In: Caspari EW, Thoday JM, editors. Advances in Genetics. New York: Academic Press; 1962. pp. 101–145. [Google Scholar]
- 3.Azaro MA, Landy A. Int and the λ Int family. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148. [Google Scholar]
- 4.Grainge I, Jayaram M. The integrase family of recombinase: Organization and function of the active site. Mol Microbiol. 1999;33(3):449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 5.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 93–117. [Google Scholar]
- 6.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 7.Nash HA. 1996. Site-specific recombination: Integration, excision, resolution, and inversion of defined DNA segments. Escherichia coli and Salmonella, eds Neidhardt FC, et al. (ASM Press, Washington, DC), 2nd Ed, Vol 2, pp 2363–2376.
- 8.Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: Examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29(11):2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segall AM, Nash HA. Architectural flexibility in lambda site-specific recombination: Three alternate conformations channel the attL site into three distinct pathways. Genes Cells. 1996;1(5):453–463. doi: 10.1046/j.1365-2443.1996.d01-254.x. [DOI] [PubMed] [Google Scholar]
- 10.Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25(19):4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matovina M, Seah N, Hamilton T, Warren D, Landy A. Stoichiometric incorporation of base substitutions at specific sites in supercoiled DNA and supercoiled recombination intermediates. Nucleic Acids Res. 2010;38(18):e175. doi: 10.1093/nar/gkq674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun KQ. 1990. A study of DNA-DNA interactions during bacteriophage lambda integrative recombination. PhD doctoral thesis in biochemistry (Univ of Illinois at Urbana-Champaign, Champaign, IL)
- 13.Huang H, Harrison SC, Verdine GL. Trapping of a catalytic HIV reverse transcriptase*template:primer complex through a disulfide bond. Chem Biol. 2000;7(5):355–364. doi: 10.1016/s1074-5521(00)00113-7. [DOI] [PubMed] [Google Scholar]
- 14.Verdine GL, Norman DP. Covalent trapping of protein-DNA complexes. Annu Rev Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- 15.Aihara H, Kwon HJ, Nunes-Düby SE, Landy A, Ellenberger T. A conformational switch controls the DNA cleavage activity of lambda integrase. Mol Cell. 2003;12(1):187–198. doi: 10.1016/s1097-2765(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 16.Biswas T, et al. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435(7045):1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warren D, Laxmikanthan G, Landy A. A chimeric Cre recombinase with regulated directionality. Proc Natl Acad Sci USA. 2008;105(47):18278–18283. doi: 10.1073/pnas.0809949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen MD, Yamasaki K, Ohme-Takagi M, Tateno M, Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17(18):5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wojciak JM, Sarkar D, Landy A, Clubb RT. Arm-site binding by lambda -integrase: Solution structure and functional characterization of its amino-terminal domain. Proc Natl Acad Sci USA. 2002;99(6):3434–3439. doi: 10.1073/pnas.052017999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci USA. 2002;99(7):4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saraf-Levy T, et al. Site-specific recombination of asymmetric lox sites mediated by a heterotetrameric Cre recombinase complex. Bioorg Med Chem. 2006;14(9):3081–3089. doi: 10.1016/j.bmc.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Swalla BM, Cho EH, Gumport RI, Gardner JF. The molecular basis of co-operative DNA binding between lambda integrase and excisionase. Mol Microbiol. 2003;50(1):89–99. doi: 10.1046/j.1365-2958.2003.03687.x. [DOI] [PubMed] [Google Scholar]
- 23.Warren D, et al. Identification of the lambda integrase surface that interacts with Xis reveals a residue that is also critical for Int dimer formation. Proc Natl Acad Sci USA. 2003;100(14):8176–8181. doi: 10.1073/pnas.1033041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J Biol Chem. 2007;282(33):24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Landy A. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science. 1992;256(5054):198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazelbaker D, Azaro MA, Landy A. A biotin interference assay highlights two different asymmetric interaction profiles for lambda integrase arm-type binding sites in integrative versus excisive recombination. J Biol Chem. 2008;283(18):12402–12414. doi: 10.1074/jbc.M800544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seah N, et al. Nucleoprotein architectures regulating the directionality of viral integration and excision. Proc Natl Acad Sci USA. 2014;111:12372–12377. doi: 10.1073/pnas.1413019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richet E, Abcarian P, Nash HA. Synapsis of attachment sites during lambda integrative recombination involves capture of a naked DNA by a protein-DNA complex. Cell. 1988;52(1):9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 29.Nash HA. Purification of bacteriophage λ Int protein. Nature. 1974;247(5442):543–545. doi: 10.1038/247543a0. [DOI] [PubMed] [Google Scholar]
- 30.Craig NL, Nash HA. E. coli integration host factor binds to specific sites in DNA. Cell. 1984;39(3 Pt 2):707–716. doi: 10.1016/0092-8674(84)90478-1. [DOI] [PubMed] [Google Scholar]
- 31.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell. 1996;87(7):1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 32.Abbani MA, et al. Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly. Proc Natl Acad Sci USA. 2007;104(7):2109–2114. doi: 10.1073/pnas.0607820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papagiannis CV, et al. Fis targets assembly of the Xis nucleoprotein filament to promote excisive recombination by phage lambda. J Mol Biol. 2007;367(2):328–343. doi: 10.1016/j.jmb.2006.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.