Significance
Congenital diaphragmatic hernia (CDH) is a common birth defect associated with high morbidity and mortality. Focusing on the coding sequence of 51 genes, discovered in human studies and in mouse models, we studied 275 CDH patients and identified multiple variants in CDH-causing genes. Information on gene expression in embryonic mouse diaphragms and protein interactions allowed us to prioritize additional compelling CDH-associated genes. We believe that an improved understanding of the genetics of CDH will be important to design new therapeutic strategies for patients with diaphragmatic defects.
Keywords: network analysis, diaphragm development, CDH genetics
Abstract
Congenital diaphragmatic hernia (CDH) is a common and severe birth defect. Despite its clinical significance, the genetic and developmental pathways underlying this disorder are incompletely understood. In this study, we report a catalog of variants detected by a whole exome sequencing study on 275 individuals with CDH. Predicted pathogenic variants in genes previously identified in either humans or mice with diaphragm defects are enriched in our CDH cohort compared with 120 size-matched random gene sets. This enrichment was absent in control populations. Variants in these critical genes can be found in up to 30.9% of individuals with CDH. In addition, we filtered variants by using genes derived from regions of recurrent copy number variations in CDH, expression profiles of the developing diaphragm, protein interaction networks expanded from the known CDH-causing genes, and prioritized genes with ultrarare and highly disruptive variants, in 11.3% of CDH patients. These strategies have identified several high priority genes and developmental pathways that likely contribute to the CDH phenotype. These data are valuable for comparison of candidate genes generated from whole exome sequencing of other CDH cohorts or multiplex kindreds and provide ideal candidates for further functional studies. Furthermore, we propose that these genes and pathways will enhance our understanding of the heterogeneous molecular etiology of CDH.
Congenital diaphragmatic hernia (CDH) is a common and severe congenital anomaly that occurs in ∼1:3,000 live births (1). In this defect, the diaphragm fails to form properly, causing displacement of the abdominal contents into the thoracic cavity. CDH is almost invariably accompanied by lung hypoplasia and pulmonary hypertension, severe clinical features that make caring for these infants exceptionally challenging (1). Although survival has improved in tertiary care centers, the average mortality rate worldwide remains at 50%, often because of complications associated with the intensive treatment modalities required for survival (2).
Several lines of evidence demonstrate the importance of genetic factors in CDH. First, a number of single-gene syndromic disorders and recurrent chromosomal abnormalities are associated with CDH (1, 3). Second, CDH has been reported in a number of multiplex kindreds (1). Third, more than 40 well-documented genetic mouse models with diaphragm defects have been described in the Mouse Genome Informatics Database (www.informatics.jax.org). The molecular etiology of CDH is likely to be highly heterogeneous and possibly polygenic. To date, in humans, several genes have been shown to play an important role in CDH, including some known to cause monogenic syndromes such as Donnai–Barrow, CHARGE, Cornelia de Lange, and Matthew–Wood syndromes, and Spondylocostal dysostosis (3). Other genes have been associated with isolated CDH (i.e., diaphragmatic defects and lung hypoplasia without co-occurring congenital anomalies in other systems) (4–6). However, disruption of these genes explains only a small fraction of CDH cases, leaving a large number of genes and pathways yet to be identified. Finally, knockout mice displaying herniation or abnormal diaphragm muscularization have pointed to CDH candidate genes, although their contribution in human CDH has not yet been determined.
Important clues to the genes and pathways involved in CDH are likely to come from knowledge of embryonic diaphragm formation. The mature diaphragm consists of posterior and antero-lateral muscular components and a central tendon. Studies in rodents suggest that the largest contributor to the mature diaphragm is the pleuroperitoneal fold (PPF), a transient tissue that eventually becomes fused with a mesodermal sheet between the heart and the liver, known as the septum transversum. Structural anomalies of the PPFs result in CDH in rodent models (7). Defects in migration, proliferation, or differentiation of premuscle cells, which originate from the cervical somites, result in abnormal diaphragm musculature (8).
Retinoic acid (RA) signaling is a pathway implicated in CDH by several developmental studies: vitamin A deprivation during rodent pregnancies, retinoid receptor null-mutant mice, and use of the CDH-inducing nitrofen compound that inhibits endogenous RA production all result in diaphragm defects (9); however, additional developmental pathways must also be important. A comprehensive transcriptome analysis of the developing diaphragm, which includes genes in the RA pathway, was made available recently by our laboratory (10) and can serve as a useful tool for evaluating CDH candidate genes.
Improved understanding of the genetic pathways that contribute to CDH is critical to improving survival and reducing complications for infants born with CDH. To this end, we report the results of whole-exome sequencing on 275 individuals with CDH, focusing on (i) rare variants in CDH-causing genes, identified in mice and/or humans, (ii) chromosomal hotspots of recurring deletion or duplications, (iii) candidates prioritized by embryonic diaphragm transcriptome and protein-interaction networks, and (iv) ultrarare and highly disruptive variants in these prioritized categories. Candidate gene analysis and the other strategies used point toward molecular pathways and a set of high priority targets for further studies with the ultimate aim of devising novel treatment paradigms.
Results
Variant Filtering.
Whole exome sequencing was performed on a cohort of 275 CDH patients, including both isolated and complex cases. Ninety-four percent of the patients had no family history of CDH, and all analyzed exomes were from unrelated individuals. Demographic and phenotypic details of this population can be found in Dataset S1. The average read depth for the targeted exome was 59×, with 81% covered at greater than 30×, and 96% covered at greater than 10×. Sequence alignment and variant calling revealed a total of 551,781 variants in 18,992 genes. Common variants, determined by a minor allele frequency (MAF) > 1% in any of the 1000 Genomes, National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) exomes, or Complete Genomics databases were excluded. This relatively generous cutoff of 1% was chosen to allow for the likelihood of low penetrance genes. After filtering out low quality and common variants, there were 219,889 in 18,458 genes remaining. Variants were then filtered based on predicted pathogenicity, including only those variants introducing stop codons, frameshifts, in-frame insertions and deletions, start site losses, splice site variants within two bases of an intron-exon boundary, and missense variants that were nonsynonymous and predicted to be pathogenic based on previous reports in the literature or by either the sorting intolerant from tolerant (SIFT) or PolyPhen-2 algorithms. This filtering resulted in a total of 34,973 variants in 12,551 genes, an average of 127 rare, predicted pathogenic variants per individual, which is consistent with the number reported for the 1000 Genomes Project (11).
Rare Variants in CDH-Causing Genes.
Given the heterogeneous nature of CDH genetics, we implemented a top-down strategy based on existing genetic evidence for CDH. Specifically, we began by searching for variants in 51 CDH-causing genes selected by stringent criteria, indicating involvement in CDH in humans (n = 11), and/or by the presence of a significant diaphragmatic phenotype (either hernia or thinning of the diaphragm muscle) in mouse models (n = 43) (Dataset S2). The targeted exome sequencing over these selected genes was high quality, with an average read depth of 66×. This analysis revealed a total of 154 rare, heterozygous, predicted pathogenic variants in 39 CDH-causing genes that were present in at least one of the 275 probands. Detailed information on each validated variant and proband phenotype is available in Dataset S3. The validation rate of selected variants was 84% by Sanger sequencing; false positives were primarily insertion/deletion calls (Dataset S4). The validated variants mapped to 34 genes, including six previously reported human CDH-causing genes and 28 CDH genes identified in mice. The number of variants detected in each of these genes and the evidence for their pathogenicity is depicted in Fig. 1A. Many were novel or ultrarare (MAF < 0.1%) (Fig. 1B). Additional algorithms supported the prediction of pathogenicity (Dataset S4). Functional domain information for protein variants is provided in Dataset S3.
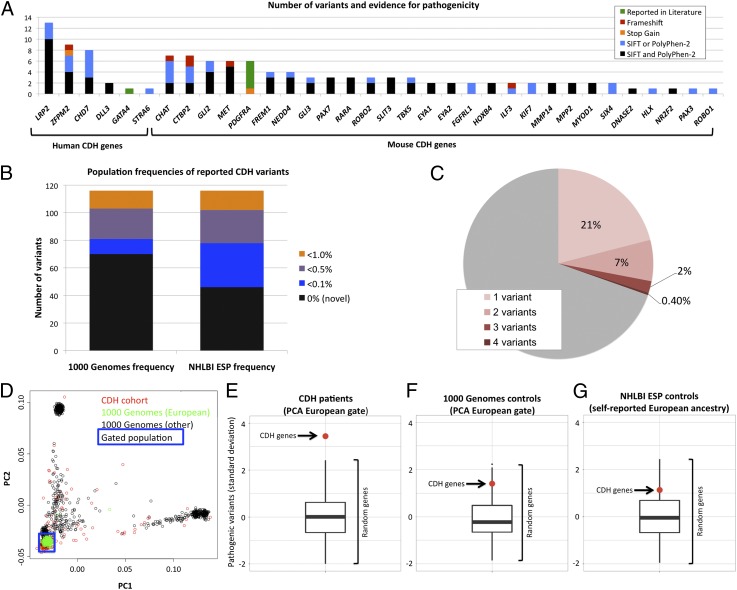
Fig. 1.
Overview of sequence variants in CDH patients. (A) Number of validated rare and predicted pathogenic variants identified in human (Left) or mouse (Right) CDH-causing genes. Colors indicate the evidence level for pathogenicity. (B) Reported population frequencies in 1000 Genomes and NHLBI ESP datasets for each of the 116 validated variants. (C) Individuals (%) with multiple (1–4, pink shades) validated rare and predicted pathogenic variants in CDH-causing genes. (D) PCA on CDH cohort exomes (red), 1000 Genomes European population (green), and the remainder of the 1000 Genomes population (black). Blue box indicates the gate used to determine which samples were included in the enrichment analysis. (E–G) Enrichment analyses for CDH patients and control cohorts, showing the frequency of pathogenic variants in CDH genes (red dot) compared with 120 sets of random size-matched genes (box plot: upper bar, maximum; lower bar, minimum; box, upper and lower quartiles; center line, median). y axis indicates number of SD from the mean of the random gene sets.
We concluded that 85 of 275 CDH probands (30.8%) had at least one predicted pathogenic variant in one of the 51 CDH-causing genes. Eighteen of 275 (6.5%) had two, 6 (2%) had three, and 1 (0.4%) had four variants in CDH-causing genes (Fig. 1C).
Variant Enrichment Analysis.
We determined whether the set of known CDH-causing genes in our cohort was enriched for predicted pathogenic variants compared with size-matched random gene sets. To control for different ancestries in our CDH cohort and control populations, we performed principal component analysis (PCA) on CDH cases and 1000 Genomes controls. Based on European ancestry assigned by common clustering, 195 CDH cases and 286 controls were selected (Fig. 1D).
We assessed the European subset of our CDH cohort for nonsense and predicted pathogenic missense variants (defined by a positive score by either SIFT or PolyPhen-2) in the 51 CDH-causing genes and compared them with 120 randomly generated gene sets that were size-matched based on the coding region of the longest predicted isoform. The burden of pathogenic variants in the CDH-causing genes was 3.5 SDs greater than the mean number of pathogenic variants found in the random gene sets (Fig. 1E) (P < 0.01, based on empirical distribution). The receptor LRP2 is encoded by a large and variable gene in normal populations. Therefore, we repeated the analysis excluding LRP2 and confirmed enrichment in the remaining genes (2.4 SD, P < 0.05).
As a control, we performed the same analysis in individuals from the 1000 Genomes Project matched by ancestry and showed that the number of predicted pathogenic variants in CDH-causing genes was not significantly different from the random gene sets (Fig. 1F). Similarly, no enrichment was detected in individuals with self-reported European ancestry in the NHLBI ESP group (n = 4,300) (Fig. 1G).
Inheritance Pattern.
Exome data were not available for unaffected family members. For this reason, the inheritance pattern of selected variants was determined by Sanger sequencing of parental DNA, available for ∼55% of the probands. For variants in the 51 CDH causing genes, two de novo variants were detected, whereas the remaining 63 variants were inherited from an unaffected parent (Dataset S3). The first de novo variant was in the zinc finger protein ZFPM2 (FOG2) (c.89A > G; p.E30G), also described in a focused report (12). The second was detected in the transcriptional coactivator EYA1 (c.164C > T; p.T55M), a previously reported pathogenic mutation causing Branchio-oto-renal syndrome 1 [BOR1; Online Mendelian Inheritance in Man (OMIM) no. 113650] (Dataset S3).
Chromosomal Hotspots.
We filtered our exome results for variants in genes mapped within “hotspots” for chromosomal deletions in patients with CDH, i.e., regions reported in the literature to be deleted in two or more individuals with CDH. The following seven chromosomal regions were given priority because they contained breakpoints precisely defined by molecular cytogenetics: 1q41-q42.12, 4p16.3, 6p25.2-p25.3, 8p23.1, 8q22.3-q23.1, 15q26.1-q26.3, and 16p11.2 (13–17). Fifteen genes in these critical regions have been proposed to play a role in CDH, seven of which were also included in the analysis above as CDH-causing genes. We found rare (MAF < 1%) and predicted pathogenic (by SIFT or PolyPhen-2) variants in all eight of the additional candidate genes from these regions (DISP1, FOXF2, FOXC1, NEIL2, MEF2A, TBX6, ARRDC4, and IGF1R), further substantiating their potential role in pathogenesis of CDH (Table 1).
Table 1.
SNVs in chromosomal hotspots
| Pt | Gene | Variant | Co-occurring |
| 12† | DISP1 | p.M1096T | — |
| 148 | DISP1 | p.M1096T | CTBP2 |
| 215† | DISP1 | p.M1096T | — |
| 246 | DISP1 | p.M1096T | — |
| 267 | DISP1 | p.M1096T | LRP2 |
| 108† | DISP1 | p.R1132Q | — |
| 124† | DISP1 | p.R1132Q | — |
| 62 | FOXF2 | p.V210M | — |
| 121 | FOXF2 | p.G425E | — |
| 226† | FOXC1 | p.A488_A489del | CHD7, NEDD4 |
| 105 | NEIL2 | p.R8fs | — |
| 57† | NEIL2 | p.S115C | — |
| 128† | NEIL2 | p.W142R | — |
| 77 | MEF2A | p.P279L | TBX6 |
| 77 | TBX6 | p.G162S | MEF2A |
| 260 | TBX6 | p.R272Q | MYOD1, TBX6 |
| 260 | TBX6 | p.G162S | MYOD1, TBX6 |
| 90† | ARRDC4 | p.Q139H | PBX3 |
| 186† | ARRDC4 | p.E217K | — |
| 127 | IGF1R | p.D433Y | — |
| 75† | IGF1R | p.V1021M | — |
| 103 | IGF1R | p.V1201I | — |
Pt, patient. Additional information available as Dataset S3.
complex CDH.
Diaphragm Development Genes and Protein Interaction Networks.
To identify novel CDH candidates, we reasoned that biologically significant genes are likely to be active in the diaphragm during a critical period of embryonic development and/or to interact with known CDH genes as part of a functional network. First, we focused on genes derived from a PPF expression array profiling, which we have previously shown to be significantly enriched for CDH-causing genes (10). Second, we generated a network of protein interaction partners with known CDH-causing genes, using the interactome-based affiliation scoring (IBAS) algorithm (18), which predicted 250 proteins as having statistically significant first- or second-order interactions (Fig. 2A). Four genes with rare and predicted pathogenic variants were identified from the overlap of the developmental expression and the protein interaction network genes (Fig. 2A). Variants in these genes were confirmed by Sanger sequencing and were shown to be inherited from an unaffected parent when both parental samples were available (n = 10/13) (Table 2).
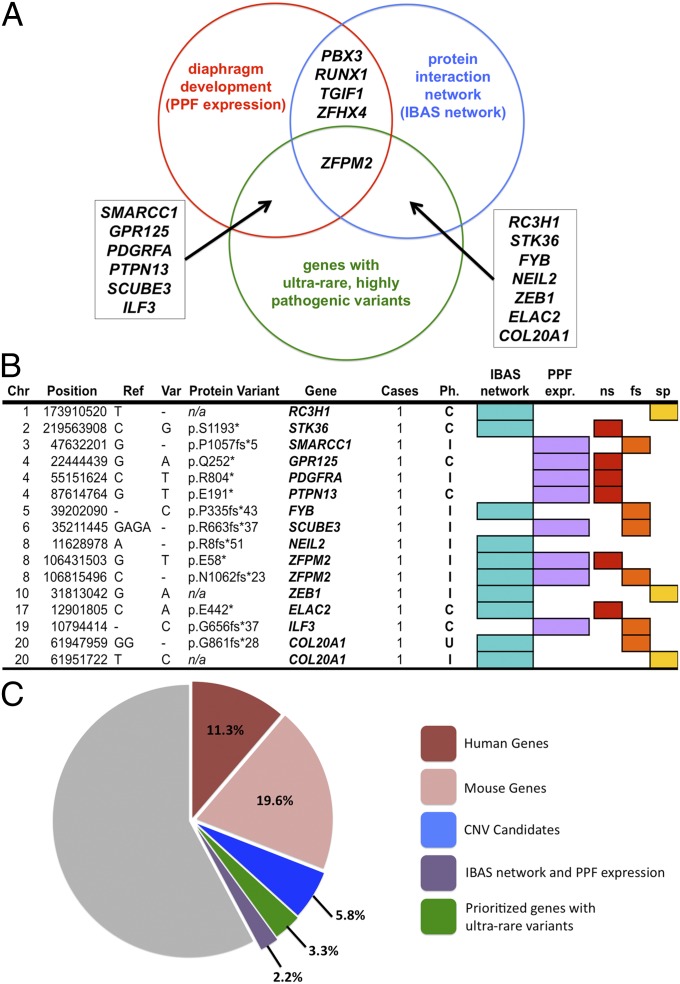
Fig. 2.
Candidate genes. (A) Candidate genes derived from the overlap of PPF transcriptomes, IBAS protein interaction networks, and genes with ultrarare and highly deleterious SNVs. (B) Ultrarare and highly deleterious SNVs in prioritized genes. C, complex CDH; Chr, chromosome; fs, frameshift; I, isolated CDH; n/a, not applicable; ns, nonsense; Ph., phenotype; Ref., reference allele; sp, splice site loss; Var, variant allele. (C) Exomes (%) with predicted pathogenic SNVs in human (red, 11.3%) or mouse (pink, 19.6%) genes, CNV candidates (blue, value = 5.8%), candidates selected by PPF transcriptomes and IBAS networks (green, value = 3.3%), and in genes with ultrarare and highly disruptive variants (2.2%).
Table 2.
SNVs filtered by IBAS protein networks and PPF transcriptomes
| Pt | Gene | Variant | Co-occurring |
| 34 | ZFHX4 | p.S2436W | CTBP2 |
| 61 | ZFHX4 | p.G3001V | — |
| 244 | ZFHX4 | p.E1776G | — |
| 11† | PBX3 | p.A136V | — |
| 32† | PBX3 | p.A136V | — |
| 49 | PBX3 | p.A136V | — |
| 172 | PBX3 | p.A136V | — |
| 239 | PBX3 | p.A136V | LRP2, ZFPM2 |
| 90† | PBX3 | p.L397P | ARRDC4 |
| 185† | TGIF1 | p.Q236L | — |
| 261 | TGIF1 | p.W30* | — |
| 25† | RUNX1 | p.D4G | — |
| 26 | RUNX1 | p.D4G | MYOD1 |
Ultrarare and Highly Disruptive Variants.
Because mutations causing birth defects are likely to affect reproductive fitness, we analyzed our exome data for ultrarare (MAF < 0.1%) and highly disruptive variants and identified ultrarare nonsense (n = 876), splice site (n = 555), and frameshift (n = 1,226) SNVs in 2,815 genes. Of these genes, 30% also had missense variants that were predicted pathogenic by SIFT or PolyPhen-2 in two or more additional patients (representing at least 1% of the entire CDH cohort), increasing the likelihood for their involvement in CDH. Fourteen also overlapped with either the IBAS network or the PPF expression genes. Three were known to be associated with CDH (PDGFRA, ZFPM2, ILF3) and one is mapped to a CNV hotspot (NEIL2), whereas the remaining 10 represent previously unreported, or novel, CDH candidates (Fig. 2 A and B). These ultrarare and highly disruptive variants were confirmed with Sanger sequencing.
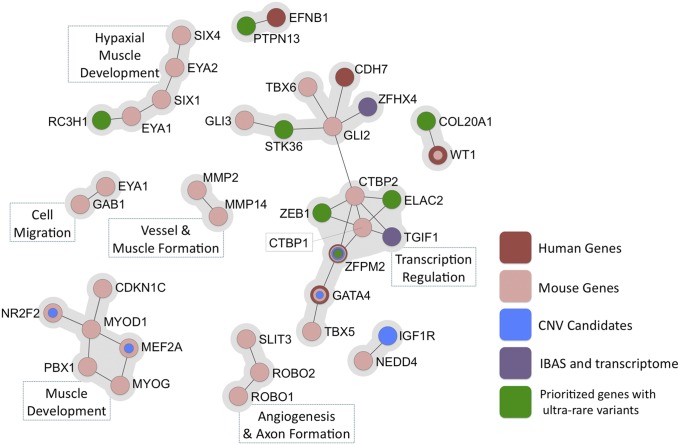
Using the approaches described in this manuscript, we were able to identify a potentially relevant variant in ∼42.2% of our patient cohort. These variants map to known CDH genes (30.9%), CNV candidate genes (5.8%), and candidates identified through the intersection of protein interaction networks and diaphragm expression profiles (3.3%), and genes with highly disruptive SNVs (2.2%) (Fig. 2C). Further, IBAS identified several coherent subnetworks of direct interactors predicted to be important in CDH, including muscle development, cell migration and signaling, and transcription factors with a specific role in cardiac development and blood vessel formation (Fig. 3).
Fig. 3.
Subnetworks of first-order (direct) interactors of CDH-causing genes, CNV candidates, and genes derived from the IBAS network and PPF expression profiles, and genes with ultrarare and highly disruptive variants. Corresponding functional annotations are listed.
Discussion
We report a large-scale exome study in patients with CDH, a common but incompletely understood birth defect. Data analysis, using a top-down approach based on existing genetic and developmental evidence for CDH, revealed multiple variants in reported CDH-causing genes, including 28 previously associated with abnormal diaphragm development in mice, but never implicated in human CDH. Additionally, we describe variants in eight CDH candidate genes mapped to CDH-associated chromosomal hotspots in patients. Finally, we used a bottom-up approach, including transcriptome and protein interaction data, and identified rare and predicted pathogenic variants in 14 novel CDH candidate genes.
CDH-Causing Genes Are Enriched for Damaging Variants.
We report a catalog of rare and predicted pathogenic variants within CDH-causing genes, which are present in 30.9% of our study cohort, further substantiating their role in diaphragm development. Patients showed a significant enrichment for predicted pathogenic variation within these CDH-causing genes compared with random gene sets, unlike a control cohort matched by ancestry. The enrichment analysis was designed to minimize the impact of population stratification, because the CDH-causing gene set is compared against 120 size-matched random gene sets in the same cohort, acting as internal controls.
Insights in the Genetics of CDH.
Our data are consistent with several proposed hypotheses about the genetic origins of CDH. First, we found multiple different genes affected across the CDH population, supporting the heterogeneous nature of the disease. Second, we identified rare and predicted pathogenic variants in two or more genes in multiple patients, raising the possibility of polygenic inheritance pattern in at least a subset of patients. However, this study does not allow us to assess the relative contribution of each variant within a given individual. Third, the majority of variants identified were inherited from an apparently unaffected parent, consistent with decreased penetrance for many of these putative causative genes. A model of decreased penetrance has been proposed for several other known CDH-causing genes, such as ZFPM2 (12, 13). Because this study did not include parental exomes, we are unable to determine the true contribution of de novo mutations to the pathogenesis of CDH. We did not identify any clear correlations between specific variants and patient phenotype, including isolated versus complex CDH, possibly due to limited sample size or incomplete clinical information. Further patient stratification will allow analyses of the effects of rare SNVs on disease severity.
SNVs in CDH-Causing Genes Associated with Noncanonical Phenotypes.
Rare and predicted pathogenic variants were identified in the DNA-binding protein CHD7, which is responsible for the CHARGE syndrome (OMIM no. 214800), in seven patients who did not match the classical clinical phenotype. CHD7 variants in our cohort differ from those reported in known CHARGE syndrome cases, suggesting that different variants in CHD7 may result in either the complex syndrome or an increased likelihood of developing CDH.
We also report multiple heterozygous rare and predicted pathogenic variants in LRP2, a gene that causes the autosomal recessive Donnai–Barrow syndrome (DBS; OMIM no. 222448), characterized by CDH, corpus callosum agenesis, and eye and kidney phenotypes (19). None of the 14 individuals with heterozygous LRP2 variants had other findings consistent with a diagnosis of DBS. Therefore, we suggest that variable levels of LRP2 activity may result in a spectrum of disorders, with complete loss of both alleles causing DBS, whereas other heterozygous variants in LRP2 may result in a milder phenotype manifested as an increased susceptibility to CDH.
Murine CDH-Causing Genes Are Associated with Human CDH.
The examination of murine CDH genes, not previously implicated in human CDH, identified multiple rare and predicted pathogenic variants in 28 genes, substantiating their importance in the human phenotype. Many of these genes play a role in a number of developmental pathways, notably retinoic acid (RARA) signaling, control of GLI transcription factor activity (GLI2, GLI3, KIF7), muscle development (MYOD, PAX3, PAX7), or cell migration and proliferation (MET, PDGFRA, SLIT/ROBOs). Many also cause cardiac anomalies in mouse knockouts (SLIT2, PDGFRA, TBX5, HOXB4), consistent with the frequent comorbidity between heart and diaphragm defects (20).
Two genes from this category are notable because of their association with recognizable human conditions, EYA1 and KIF7. We identified a de novo mutation in the EYA1 gene in one CDH patient. Mice lacking two alleles of Eya1 and one allele of Eya2 show diaphragm defects (21). Mutations in EYA1, responsible for 40% of BOR syndrome patients, have never been reported in CDH patients (22). In particular, this proband had CDH associated with bilateral hearing loss and unilateral microtia, a phenotype consistent with BOR1 syndrome. We also identified two rare and predicted pathogenic variants in the KIF7 gene, a cilia-associated protein that modulates GLI transcription factor activity (23). Mice harboring homozygous mutations in the Kif7 gene have CDH and lung hypoplasia as well as defects affecting the cardiac, skeletal, and central nervous systems (24). Homozygous KIF7 mutations have been associated with several severe, multisystem disorders in humans, without CDH (25, 26). Both patients in our cohort with KIF7 variants, however, have isolated CDH, suggesting that heterozygous variants in this gene may result in increased susceptibility to CDH.
Rare Sequence Variants in CDH Hotspot Regions.
Rare and predicted pathogenic variants are also present in candidate genes from recurrent regions of chromosomal copy number variation in CDH. These genes include the transcription factors MEF2A and NR2F2 (COUP-TFII) (15q26), which have been shown to cooperate with MYOD1 in the regulation of muscle differentiation (27–29), as well as GATA4, SOX7, and NEIL2 (8p23), which have been hypothesized to cooperate for proper diaphragm development (15, 30). The presence of rare and predicted pathogenic variants in these genes in the CDH population further supports their role in human diaphragm development.
Diaphragm Developmental Expression Profiles and Protein Interaction Networks Identify CDH Candidate Genes.
In the gene discovery phase of this study, we identified additional candidate genes by integrating exome sequencing data with (i) PPF gene expression profiles and (ii) IBAS protein interaction networks based on known CDH genes. We further prioritized these genes by focusing on ultrarare (MAF < 0.1%) variants present in multiple individuals, one of which had to be a highly disruptive mutation. This approach, designed to enrich for genes more likely to be functionally relevant, uncovered candidate genes, which should be given high priority in functional studies to assess their role in diaphragm development (Figs. 2A and 3).
Network Analyses.
Our network analyses also reveal several functional nodes that are likely to play key roles in the pathogenesis of CDH. RA signaling is a pathway known to be central to the pathogenesis of diaphragmatic defects (9), and many of the genes in which we describe rare and predicted pathogenic variants have been shown to interact directly or indirectly with the RA pathway. These candidates include TGIF1, PBX3, RUNX1, and ZFHX4 (31–34). Other affected pathways are muscle differentiation and Gli transcription activity (34, 35).
The genes delineated in this study harbor rare variants in 42.2% in CDH patients, thus establishing an important foundation on which future human and animal model CDH research can be built. Integrated sequencing, developmental, and bioinformatics strategies could direct future functional studies on CDH, could be applied to cohorts and consortia for CDH and other births defects, and could pave the way for potential therapies by providing molecular targets for drug discovery.
Methods
Patient Recruitment.
Probands and family members were enrolled in the “Gene Mutation and Rescue in Human Diaphragmatic Hernia” study. Informed consent, blood, and tissue samples were obtained according to Partners Human Research Committee and Boston Children’s Hospital clinical investigation standards (Protocol 2000P000372 and 05-07-105R, respectively). Whenever possible, consented individuals underwent a physical examination by a geneticist and review of medical records.
Whole-Exome Sequencing.
Samples were sequenced at the University of Washington, Seattle, by the NHLBI Resequencing and Genotyping Service (n = 92), at the Yale Center for Genome Analysis (n = 169), and at the Broad Institute (n = 14), on Illumina HiSEq. 2000 platforms. Sample libraries were captured on the SeqCap EZ Human Exome Library v2.0 (Roche NimbleGen).
Sequencing Data Analysis.
Raw sequencing data for each individual were aligned to the human reference genome (build hg19) by using Burrows–Wheeler Aligner (BWA 0.7.5a). The alignment files were converted from a sequence alignment map (SAM) format to a sorted, indexed, binary alignment map (BAM) file (SAMtools version 0.1.19; samtools.sourceforge.net) (36). To improve alignments and genotype calling, BAM files were realigned with The Genome Analysis Toolkit or GATK IndelRealigner (Broad Institute) (37, 38). Base quality scores were recalibrated, and duplicate reads were removed by the GATK base quality recalibration tool. SNP and insertion-deletion discovery and genotyping across all 275 samples were done by using GATK for those variants that have minimum probability of incorrect base call equal to 1 in 100,000 (Phred Quality Score equal or greater than 50). The resulting Variant Call Format (VCF; version 4.0) files were imported to the Ingenuity Variant Analysis platform for further filtering.
Principal Component Analysis.
Multidimentional scaling function, implemented in PLINK, was used to select ethnically matched individuals in the CDH and 1000 Genomes Project cohorts, using SNPs present in both groups, for enrichment analyses (39).
Enrichment Analysis.
The enrichment of sequence variants in CDH-causing genes was determined against 120 random in silico size-matched control gene sets [coding sequence ±10% as reported by ENSEMBL (www.ensembl.org)], using an approach that we call XRANGE (eXome RANdom Genesets Enrichment). Pathogenicity of missense variants was determined by using the Variant Effect Predictor version 73 [vepred] to run SIFT and Polyphen-2 (40, 41). The number of predicted pathogenic variants (by either of these algorithms) was counted for the CDH-causing genes and each random control gene set. All variants, regardless of population frequency, were included in this analysis to eliminate bias due to the fact that population frequencies are based on the 1000 Genomes and NHLBI ESP datasets that were used as controls for the analysis.
Network Analysis.
To uncover the proteins that interact with a list of known CDH-causing genes, we used IBAS (18), which is based on the updated human protein interaction network (InWeb) of ∼430,000 interactions among 23,000 human proteins (42). First, IBAS was trained with 51 known CDH-causing genes curated from the literature. Parameters were optimized to predict interacting phenotype-causing proteins by testing each one against 10,000 random proteins. Furthermore, using these parameters, all candidates (i.e., 23,000 proteins) covered by interaction data were scored and ranked, and permutation tests were used for determining the significance of the observed scores.
DNA sample preparation and variant confirmation were performed as described in SI Methods.
Supplementary Material
Acknowledgments
We thank The Association of Congenital Diaphragmatic Hernia Research, Awareness and Support (CHERUBS) for additional support; L. Kellndorfer, J. Kim, L. Luque-Bustamante, H. Al-Turkmani for technical assistance; and, for careful recruitment of patients, the surgeons at MassGeneral Hospital for Children and Boston Children’s Hospital: T. Buchmiller, C. C. Chen, D. Doody, S. J. Fishman, A. Goldstein, L. Holmes, T. Jaksic, R. Jennings, C. Kelleher, D. Lawlor, C.W. Lillehei, P. Masiakos, D. P. Mooney, K. Papadakis, R. Pieretti, M. Puder, D. P. Ryan, R. C. Shamberger, C. Smithers, J. Vacanti, and C. Weldon. We are grateful to the CDH support groups, CHERUBS and Breath of Hope, for providing families and caregivers information about our research study. Funding was provided by National Institute of Child Health and Human Development P01 HD068250-03 (to P.K.D.) and National Research Service Award 2T32GM007748-35 (to F.A.H.). Sequencing services were provided through Resequencing and Genotyping Service by the Northwest Genomics Center at the University of Washington, Department of Genome Sciences, under US Federal Government contract no. HHSN268201100037C from the NHLBI.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences have been deposited in the database of Genotypes and Phenotypes (dbGAP) (accession no. phs000783.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412509111/-/DCSupplemental.
References
- 1.Pober BR. Overview of epidemiology, genetics, birth defects, and chromosome abnormalities associated with CDH. Am J Med Genet C Semin Med Genet. 2007;145C(2):158–171. doi: 10.1002/ajmg.c.30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mohseni-Bod H, Bohn D. Pulmonary hypertension in congenital diaphragmatic hernia. Semin Pediatr Surg. 2007;16(2):126–133. doi: 10.1053/j.sempedsurg.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Pober BR. Genetic aspects of human congenital diaphragmatic hernia. Clin Genet. 2008;74(1):1–15. doi: 10.1111/j.1399-0004.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackerman KG, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1(1):58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu L, et al. Variants in GATA4 are a rare cause of familial and sporadic congenital diaphragmatic hernia. Hum Genet. 2013;132(3):285–292. doi: 10.1007/s00439-012-1249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu L, et al. University of Washington Center for Mendelian Genomics Whole exome sequencing identifies de novo mutations in GATA6 associated with congenital diaphragmatic hernia. J Med Genet. 2014;51(3):197–202. doi: 10.1136/jmedgenet-2013-101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greer JJ. Current concepts on the pathogenesis and etiology of congenital diaphragmatic hernia. Respir Physiol Neurobiol. 2013;189(2):232–240. doi: 10.1016/j.resp.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Merrell AJ, Kardon G. Development of the diaphragm — a skeletal muscle essential for mammalian respiration. FEBS J. 2013;280(17):4026–4035. doi: 10.1111/febs.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clugston RD, Zhang W, Alvarez S, de Lera AR, Greer JJ. Understanding abnormal retinoid signaling as a causative mechanism in congenital diaphragmatic hernia. Am J Respir Cell Mol Biol. 2010;42(3):276–285. doi: 10.1165/rcmb.2009-0076OC. [DOI] [PubMed] [Google Scholar]
- 10.Russell MK, et al. Congenital diaphragmatic hernia candidate genes derived from embryonic transcriptomes. Proc Natl Acad Sci USA. 2012;109(8):2978–2983. doi: 10.1073/pnas.1121621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longoni M, et al. Prevalence and penetrance of ZFPM2 mutations and deletions causing congenital diaphragmatic hernia. Clin Genet. 2014 doi: 10.1111/cge.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wat MJ, et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J Med Genet. 2011;48(5):299–307. doi: 10.1136/jmg.2011.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casaccia G, Mobili L, Braguglia A, Santoro F, Bagolan P. Distal 4p microdeletion in a case of Wolf-Hirschhorn syndrome with congenital diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol. 2006;76(3):210–213. doi: 10.1002/bdra.20235. [DOI] [PubMed] [Google Scholar]
- 15.Longoni M, et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A. 2012;158A(12):3148–3158. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaassens M, et al. Congenital diaphragmatic hernia and chromosome 15q26: Determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet. 2005;76(5):877–882. doi: 10.1086/429842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantarci S, et al. Characterization of the chromosome 1q41q42.12 region, and the candidate gene DISP1, in patients with CDH. Am J Med Genet A. 2010;152A(10):2493–2504. doi: 10.1002/ajmg.a.33618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miraoui H, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92(5):725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pober BR, Longoni M, Noonan KM. A review of Donnai-Barrow and facio-oculo-acoustico-renal (DB/FOAR) syndrome: Clinical features and differential diagnosis. Birth Defects Res A Clin Mol Teratol. 2009;85(1):76–81. doi: 10.1002/bdra.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Menon SC, et al. Clinical characteristics and outcomes of patients with cardiac defects and congenital diaphragmatic hernia. J Pediatr. 2013;162:114–119.e2. doi: 10.1016/j.jpeds.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 21.Grifone R, et al. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302(2):602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 22.Abdelhak S, et al. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15(2):157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 23.Liem KF, Jr, He M, Ocbina PJR, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci USA. 2009;106(32):13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coles GL, Ackerman KG. Kif7 is required for the patterning and differentiation of the diaphragm in a model of syndromic congenital diaphragmatic hernia. Proc Natl Acad Sci USA. 2013;110(21):E1898–E1905. doi: 10.1073/pnas.1222797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putoux A, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet. 2011;43(6):601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dafinger C, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121(7):2662–2667. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83(7):1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 28.Lee CT, et al. The nuclear orphan receptor COUP-TFII is required for limb and skeletal muscle development. Mol Cell Biol. 2004;24(24):10835–10843. doi: 10.1128/MCB.24.24.10835-10843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey P, Sartorelli V, Hamamori Y, Muscat GE. The orphan nuclear receptor, COUP-TF II, inhibits myogenesis by post-transcriptional regulation of MyoD function: COUP-TF II directly interacts with p300 and myoD. Nucleic Acids Res. 1998;26(23):5501–5510. doi: 10.1093/nar/26.23.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wat MJ, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21(18):4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartholin L, et al. TGIF inhibits retinoid signaling. Mol Cell Biol. 2006;26(3):990–1001. doi: 10.1128/MCB.26.3.990-1001.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin P, Cimildoro R, Kochhar DM, Soprano KJ, Soprano DR. PBX, MEIS, and IGF-I are potential mediators of retinoic acid-induced proximodistal limb reduction defects. Teratology. 2002;66(5):224–234. doi: 10.1002/tera.10082. [DOI] [PubMed] [Google Scholar]
- 33.Marcelo KL, et al. Hemogenic endothelial cell specification requires c-Kit, Notch signaling, and p27-mediated cell-cycle control. Dev Cell. 2013;27(5):504–515. doi: 10.1016/j.devcel.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemmi K, et al. A homeodomain-zinc finger protein, ZFHX4, is expressed in neuronal differentiation manner and suppressed in muscle differentiation manner. Biol Pharm Bull. 2006;29(9):1830–1835. doi: 10.1248/bpb.29.1830. [DOI] [PubMed] [Google Scholar]
- 35.Kim PC, Mo R, Hui Cc C. Murine models of VACTERL syndrome: Role of sonic hedgehog signaling pathway. J Pediatr Surg. 2001;36(2):381–384. doi: 10.1053/jpsu.2001.20722. [DOI] [PubMed] [Google Scholar]
- 36.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 41.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lage K, et al. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat Biotechnol. 2007;25(3):309–316. doi: 10.1038/nbt1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.