Natural products derived from fungi have a glorious pedigree in alleviating human suffering, with penicillins being the very first microbially derived compounds to be clinically used as antibiotics. Beyond the antibiotic activity of penicillins and contemporary cephalosporins, fungal natural products exhibit extensive utility in a myriad of clinical applications. Among these, fungal polyketides offer a vast array of bioactive molecules with prominent examples including the cholesterol-lowering lovastatin, immunosuppressant mycophenolic acid, and angiogenesis inhibitor cytochalasin E (1–3). Although polyketide natural products possess a comparatively higher “hit rate” for drug development (0.3%) compared with synthetic high-throughput screening libraries (<0.001%) (4), their complicated chemical structures that contain multiple stereocenters and numerous oxygen-containing substituents complicate synthetic diversity-oriented modifications (5). This structural complexity has precipitated the need to expand the chemical space naturally explored in microbes through the genetic reprogramming of secondary metabolism to yield designer molecules. Engineering of natural products via coexpression of heterologous gene combinations has been realized for bacterial polyketide antibiotics (6, 7). In PNAS, Xu et al. (8) extend this paradigm for a class of fungal polyketide natural products called the benzenediol lactones (BDLs) and generate novel “nonnatural” natural products while also providing means to generate previously described minor analogs in much improved yields.
BDLs are chemically interesting compounds possessing a dihydroxyphenyl ring fused to a macrolactone ring (Fig. 1). These fungal polyketide natural products exhibit numerous desirable bioactivities that include the estrogen agonist zearalenone, heat shock response modulators radicicol and monocillin II, and mammalian immune response regulator 10,11-dehydrocurvularin (1). BDLs are synthesized by the tandem action of two cooperative fungal iterative polyketide synthase (iPKS) assembly line enzymes comprising highly reducing (hrPKS) (9) and nonreducing (nrPKS) modules (10). The hrPKS acts first and constructs the lipid fragment that gets transferred to the nrPKS for further biosynthetic processing to install the aromatic fragment (Fig. 1). As more and more BDL structures and their associated biosynthetic gene clusters are revealed (11), a straightforward bioengineering approach to expand the chemical space naturally explored by BDL structures was envisaged by Xu et al. (8) to recombine unnatural pairs of BDL hrPKS and nrPKS elements. A major knowledge gap prior to this study by Xu et al. was whether noncognate hrPKS and nrPKS pairs could indeed interact with each other in a productive fashion. For the first time, the authors (8) provide experimental evidence that hrPKS and nrPKSs from different biosynthetic pathways can be combined to yield new chemical entities. Consequently, this work helps illuminate the underlying biochemical rules that dictate biosynthesis by these fascinating enzymatic machines.
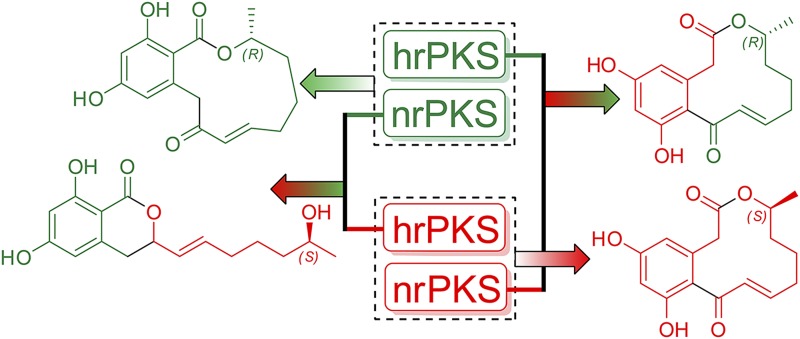
Fig. 1.
Combinatorial swapping of hrPKS and nrPKS enzymes that constitute a BDL biosynthetic iPKS leads to production of engineered BDLs. Physiological products of two BDL iPKSs are shown in green (trans-resorcylide) and red (10,11-dehydrocurvularin). On heterologous expression of different combinations of hrPKSs and nrPKSs, different nonnatural BDLs were synthesized (shown in mixed colors).
The Molnar group previously explored individual iPKS domain engineering and swapping approaches, such as for the product template (PT) (12) and thioesterase (TE) (13) domains between radicicol/monocillin II nrPKSs from Chaetomium chiversii (CcRad) (14) and 10,11-dehydrocurvularin nrPKS from Aspergillus terreus (AtCur) (15). Further building on the impressive body of work compiled by the Tang and Vederas laboratories on fungal iPKS systems (9, 16, 17), in the present study, Xu et al. take a significant step forward and explore how structural diversity for BDLs can be generated by pairwise combination of hrPKS and nrPKS modules derived from four different BDL biosynthetic gene clusters. In addition to CcRad and AtCur gene loci, they further expand their approach to include the newly discovered lasiodiplodin and resorcylide biosynthetic iPKSs from Lasiodiplodia theobromae (LtLas) and Acremonium zeae (AzRes), respectively (11), to provide 16 possible hrPKS–nrPKS combinations.
Most gratifyingly, the authors observe that polyketide intermediates can be shuttled between orthologous hrPKS–nrPKS pairs. Although this observation preserves the vectorial PKS hypothesis (18), it offers a significant step forward in the module-swapping type of surgical engineering of PKSs (4). This seminal finding makes accessible a vast chemical space to combinatorially assemble BDLs with simultaneous alterations at both the aromatic and lipid macrocycle cores. Based on the rich dataset obtained from 16 hrPKS–nrPKS combination experiments, the authors report that unnatural pairs of BDL biosynthetic iPKSs can often work synergistically. For instance, nrPKSs were found to be generally accommodating toward noncognate hrPKS intermediates that were equal or greater in chain length than their cognate hrPKS products. However, if these enzymatic compensations deviated too much against the natural pathway, then downstream processing events such as offloading and macrocyclization were impeded to yield byproducts rather than the cognate natural products.
These findings illustrate that even the cross-talk between highly related iPKS pairs can be rather fickle. They further illuminate significant shortcomings in our current understanding of polyketide biosynthesis about the intimate interplay between assembly line proteins and their covalently bound biosynthetic intermediates. Tremendous insight has been achieved with high-resolution structures of the complete fungal fatty acid synthase (19) and recently for bacterial PKS modules (20, 21) that begin to resolve the mechanistic decision-making process of these multifunctional enzymes. Such structural efforts in combination with prior (12, 13) and current (8) genetic reprogramming studies, such as those by Xu et al., are together contributing toward a clearer mechanistic understanding of PKS design rules that will undoubtedly facilitate how they can be tweaked to generate even greater chemical diversity.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
See companion article on page 12354.
References
- 1.Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: From genes to molecules. J Org Chem. 2012;77(22):9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiemann P, Keller NP. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41(2):301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 3.Schümann J, Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol. 2006;124(4):690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 4.Weissman KJ, Leadlay PF. Combinatorial biosynthesis of reduced polyketides. Nat Rev Microbiol. 2005;3(12):925–936. doi: 10.1038/nrmicro1287. [DOI] [PubMed] [Google Scholar]
- 5.Butler MS. The role of natural product chemistry in drug discovery. J Nat Prod. 2004;67(12):2141–2153. doi: 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- 6.McDaniel R, et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96(5):1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzella HG, et al. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol. 2005;23(9):1171–1176. doi: 10.1038/nbt1128. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, et al. Diversity-oriented combinatorial biosynthesis of benzenediol lactone scaffolds by subunit shuffling of fungal polyketide synthases. Proc Natl Acad Sci USA. 2014;111:12354–12359. doi: 10.1073/pnas.1406999111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma SM, et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326(5952):589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320(5873):243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Y, et al. Insights into the biosynthesis of 12-membered resorcylic acid lactones from heterologous production in Saccharomyces cerevisiae. ACS Chem Biol. 2014;9(5):1119–1127. doi: 10.1021/cb500043g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. Rational reprogramming of fungal polyketide first-ring cyclization. Proc Natl Acad Sci USA. 2013;110(14):5398–5403. doi: 10.1073/pnas.1301201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, et al. Thioesterase domains of fungal nonreducing polyketide synthases act as decision gates during combinatorial biosynthesis. J Am Chem Soc. 2013;135(29):10783–10791. doi: 10.1021/ja4041362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, et al. Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol. 2008;15(12):1328–1338. doi: 10.1016/j.chembiol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Xu Y, et al. Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat shock response-modulating anticancer fungal polyketide from Aspergillus terreus. Appl Environ Microbiol. 2013;79(6):2038–2047. doi: 10.1128/AEM.03334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Z, et al. Investigation of fungal iterative polyketide synthase functions using partially assembled intermediates. J Am Chem Soc. 2013;135(5):1735–1738. doi: 10.1021/ja4001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J Biol Chem. 2010;285(53):41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khosla C, Herschlag D, Cane DE, Walsh CT. Assembly line polyketide synthases: Mechanistic insights and unsolved problems. Biochemistry. 2014;53(18):2875–2883. doi: 10.1021/bi500290t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenni S, et al. Structure of fungal fatty acid synthase and implications for iterative substrate shuttling. Science. 2007;316(5822):254–261. doi: 10.1126/science.1138248. [DOI] [PubMed] [Google Scholar]
- 20.Whicher JR, et al. Structural rearrangements of a polyketide synthase module during its catalytic cycle. Nature. 2014;510(7506):560–564. doi: 10.1038/nature13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta S, et al. Structure of a modular polyketide synthase. Nature. 2014;510(7506):512–517. doi: 10.1038/nature13423. [DOI] [PMC free article] [PubMed] [Google Scholar]