Significance
Mechanisms of plant seed dormancy evolved to delay germination to a season favorable for seedling growth. Germination timing is an important adaptive early-life history trait which determines plant fitness in natural and agricultural ecosystems. The DELAY OF GERMINATION 1 (DOG1) gene provides natural genetic variation in dormancy, was the first dormancy-specific gene cloned, and encodes a protein of unknown function. We show here that DOG1 controls dormancy of different species by setting the optimal ambient temperature window for germination. This timing is achieved by temperature-dependent alteration of the gibberellin hormone metabolism, which in turn leads to altered expression of genes required for the biomechanical weakening of the coat encasing the embryo. The conserved DOG1-mediated coat-dormancy mechanism controls the timing of seed germination in a temperature-dependent manner.
Keywords: dormancy gene DOG1, gibberellin metabolism, germination temperature, cell-wall remodelling, Lepidium sativum
Abstract
Seed germination is an important life-cycle transition because it determines subsequent plant survival and reproductive success. To detect optimal spatiotemporal conditions for germination, seeds act as sophisticated environmental sensors integrating information such as ambient temperature. Here we show that the DELAY OF GERMINATION 1 (DOG1) gene, known for providing dormancy adaptation to distinct environments, determines the optimal temperature for seed germination. By reciprocal gene-swapping experiments between Brassicaceae species we show that the DOG1-mediated dormancy mechanism is conserved. Biomechanical analyses show that this mechanism regulates the material properties of the endosperm, a seed tissue layer acting as germination barrier to control coat dormancy. We found that DOG1 inhibits the expression of gibberellin (GA)-regulated genes encoding cell-wall remodeling proteins in a temperature-dependent manner. Furthermore we demonstrate that DOG1 causes temperature-dependent alterations in the seed GA metabolism. These alterations in hormone metabolism are brought about by the temperature-dependent differential expression of genes encoding key enzymes of the GA biosynthetic pathway. These effects of DOG1 lead to a temperature-dependent control of endosperm weakening and determine the optimal temperature for germination. The conserved DOG1-mediated coat-dormancy mechanism provides a highly adaptable temperature-sensing mechanism to control the timing of germination.
Seed dormancy is an important adaptive early-life history trait because it controls the distribution of germination in space (e.g., habitat selection) and time (e.g., in response to seasonal temperature changes). Ecophysiological work has shown that seed dormancy is a crucial fitness component with far-reaching consequences for the evolution of entire life histories (1–3). As an innate seed property, seed dormancy defines the environmental conditions in which a seed is able to germinate and ensures that the most vulnerable later phases of the plant life cycle occur during favorable seasonal and environmental conditions. Temperature during seed maturation defines the depth of primary dormancy established upon seed dispersal (4, 5). Soil temperature is the major environmental factor in the seasonal dormancy cycling of the soil seed bank in temperate regions (6, 7). Therefore, especially given a changing climate, it is important to understand the molecular mechanisms of temperature-related traits, including dormancy, and their role in the adaptation of populations to changing temperatures. The substantial influence of the environment on genetically controlled seed dormancy is mediated, at least in part, by the plant hormones abscisic acid (ABA) and gibberellins (GA) (8–10). Seed contents of and sensitivities to ABA and GA, as well as the properties of the embryo-encasing covering layers, are the physiological basis for the germination responses to distinct environments. Quantitative trait genes (QTGs), including DELAY OF GERMINATION1 (DOG1) (3, 10–12), provide the genetic basis for the observed natural variation in seed dormancy of Arabidopsis thaliana ecotypes. Arabidopsis thaliana DOG1 (AtDOG1) is a major dormancy QTG required for A. thaliana seed dormancy and is a decisive component for the environmental adaptation of populations (1, 2, 13–15). However, despite the central role of AtDOG1, neither its biochemical function nor its participation in a phylogenetically conserved dormancy mechanism has been elucidated.
Embryo-related developmental processes are mediated by tissue forces in animals and plants. They are determined by the interaction between the embryo and the encasing tissue layers and animal extracellular matrices or plant cell walls (16, 17). For example, in vertebrate embryos the elongation and straightening of the notochord depend on the interaction with the surrounding extracellular matrix sheath and its dynamic biomechanical properties (18). In the mature seed of most angiosperms the embryo is encased by two covering layers (“coats”): the living endosperm tissue and the dead testa (seed coat). Whether a plant undergoes a life-cycle transition by completing seed germination is controlled by the balance of opposing forces: Germination is promoted by the growth potential of the embryo growth zone (the embryonic radicle–lower hypocotyl axis, RAD) and is inhibited by the restraining tissue layers (i.e., coats) covering the RAD (17, 19, 20). In many angiosperms, including the Brassicaceae Lepidium sativum (garden cress) and A. thaliana, seed germination consists of two sequential steps: Shortly after imbibition, testa rupture (TR) takes place and is followed by endosperm rupture (ER) and radicle emergence, which is the visible completion of germination. Weakening of the micropylar endosperm (CAP) covering the RAD must occur before ER. Hormonal signaling and interaction between the RAD and CAP as the key seed compartments controls the expression of downstream genes encoding cell-wall–remodeling proteins (CWRPs) (21, 22). These proteins alter the biomechanical properties of cell walls, encouraging growth in RAD tissues and weakening in CAP tissues, to control the timing of germination. Little is known about the mechanisms by which QTGs such as DOG1 mediate the environmental and hormonal control of these processes.
AtDOG1 is a key regulator of seed dormancy because the A. thaliana dog1 mutant is completely nondormant and does not exhibit any obvious pleiotropic phenotypes apart from reduced seed longevity (11, 12). The time required for the release from seed dormancy during after-ripening storage is determined by AtDOG1 protein levels in dry seeds (5). These proteins accumulate during seed maturation, and their accumulation is controlled by temperature. In contrast to the situation during seed maturation, very little is known about the roles of AtDOG1 during seed germination. The AtDOG1 gene has been reported to belong to a small gene family, together with four other AtDOG1-like genes in A. thaliana, and encodes a protein of unknown function (11). Putative AtDOG1 orthologs are present in Lepidium species from varying environments of all continents, and these species exhibit considerable variation in dormancy (23, 24). Monocot DOG1-like genes with a low level of similarity to AtDOG1 have been found in cereals (25–27). Ectopic expression of some of these cereal DOG1-like genes in A. thaliana WT seeds delayed germination. It would be of interest to investigate whether DOG1 genes from different species provide a conserved dormancy mechanism with a common evolutionary origin, because most studies thus far have focused on A. thaliana.
The Brassicaceae originated as a tropical/subtropical family ca. 37 Mya in a warm and humid climate and subsequently evolved into a dry-adapted family (28). This diversification and radiation upon climate change also required the evolution of mechanisms that adapt seed responses to seasonal temperature cycling. Ancient whole-genome duplication (WGD) events leading to paleopolyploidy before climate changes play a crucial role in the genetic diversification, species radiation, and adaptation to new environments (28–30). The monophyletic Brassicaceae genus Lepidium (cress) contains a large number of polyploid species, suggesting a reticulate evolutionary history, and recent allopolyploidization is important for Lepidium speciation and range expansion (24, 28, 31). The cultivated spicy sprout crop L. sativum is characterized by nondormant seeds which do not have the after-ripening, cold stratification, or light requirements for germination that have been identified for the dormant seeds of A. thaliana. The larger seeds of L. sativum are an established Brassicaceae model system involving endosperm CAP weakening (21, 23, 32, 33) and thereby provide an interesting choice for studying the potential of candidate dormancy genes from other species: The lack of endogenous dormancy allows the biomechanical, transcriptional, and hormonal effects of transgenes to be studied immediately.
We show here by gene-swapping experiments between A. thaliana and L. sativum that DOG1 mediates a conserved GA-related coat-dormancy mechanism which determines the seed responses to ambient temperature and has CAP weakening as its major target. Our work provides an integrated view of the underlying molecular mechanisms by which a plant life-cycle transition is controlled in a temperature-dependent manner by the alteration of the biomechanical properties of key seed tissues regulating dormancy and germination.
Results
Two Seed-Expressed DOG1 Paralogs Are Present in the Diploid Species (2n = 24) L. sativum.
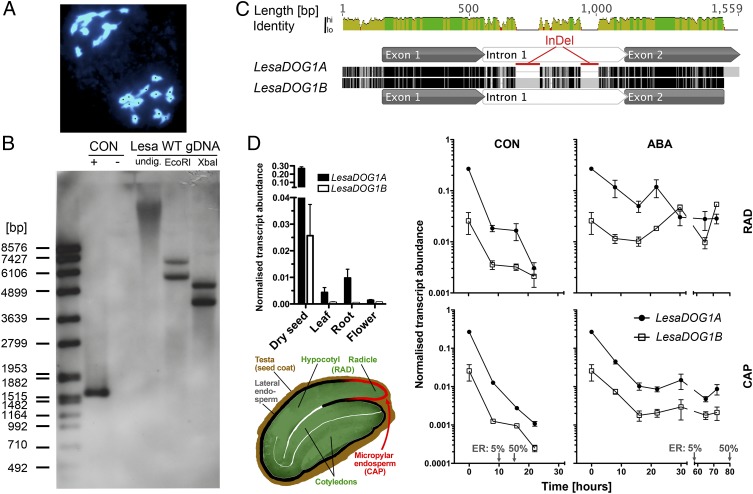
We have shown previously that L. sativum FR14 possesses the seed-expressed AtDOG1 gene homolog LesaDOG1 (23). Here we show by Southern blot analysis that L. sativum actually possesses two DOG1 genes (Fig. 1B), which we designated “LesaDOG1A” (described in ref. 23) and “LesaDOG1B.” We cloned a nearly full-length genomic DNA fragment from LesaDOG1B and its corresponding cDNA from dry seeds. LesaDOG1A and LesaDOG1B have conserved intron splice sites and show more than 93% sequence identity in their coding regions. Most differences between the two genes are located in the intronic regions, which have less than 50% identity and the presence of two large InDels (Fig. 1C). Similar variations in the DOG1 intron sequence also were found in other Lepidium species (24). The few single-nucleotide differences in the coding regions and the presence of large intronic indel blocks point to the recent origin of the gene duplication leading to these two paralogs. Such a duplication likely was caused by a recent Lepidium-specific polyploidization event (a postulated Le-Neo WGD) followed by diploidization. This possibility is supported by our finding that L. sativum is diploid (2n = 24) with regular meiosis with n = 12 (Fig. 1A and SI Appendix, Table S1) but with a much larger genome size than expected from closely related species. All the 202 L. sativum accessions investigated showed a nearly identical relative DNA amount of flow cytometry value (FC) = 2.77 ± 0.08, as compared with FC = 1 for Lepidium campestre with 2n = 16 (SI Appendix, Fig. S1 and Table S1).
Fig. 1.
Two seed-expressed DOG1 genes, LesaDOG1A and LesaDOG1B, in the diploid (2n = 24) species L. sativum. (A) DAPI-stained meiotic (metaphase II) chromosome spreads from flower bud tissue of L. sativum exhibiting 12 chromosomes, thus demonstrating regular meiosis. For better counting, chromosomes are labeled with black dots. (B) Southern blot analysis of the L. sativum FR14 genome indicates the presence of two DOG1 genes. Genomic DNA (Lesa WT gDNA) undigested (undig.) or digested with EcoRI or XbaI was hybridized with a LesaDOG1A probe. Hybridization controls were plasmids with (+) or without (−) LesaDOG1A full-length gDNA inserts. The left lane shows the DNA molecular mass ladder. CON, control. (C) Pairwise alignment of LesaDOG1A and LesaDOG1B (nearly full-length) gDNA sequences. Identical residues are colored black; mismatches are colored light gray; gaps are indicated by horizontal lines. Exon-intron annotations were derived by comparison with the respective cDNAs. Two large intronic InDels are marked in red. (D) Transcript abundances (Upper Left) of LesaDOG1A and LesaDOG1B in different plant tissues and during germination in the seed RAD and CAP tissues (as indicated by the schematic L. sativum seed drawing, Lower Left) determined by qRT-PCR analysis. n = 3; data are shown as mean ± SEM. (Right) Seeds were imbibed without (CON) or with the addition of 10 µM ABA. Gray arrows indicate time points of 5% and 50% ER. RAD comprises the radicle plus ca. one-third of the lower hypocotyl (the embryo growth zone); CAP is the micropylar endosperm tissue. Note that the dry seed samples (0 h) of the qRT-PCR analysis represent CAP+RAD tissue and not whole seeds.
We investigated the expression of the two LesaDOG1 paralogs and found both predominantly expressed in dry seeds, but the LesaDOG1A transcript was about 10 times more abundant than the LesaDOG1B transcript (Fig. 1D). Within the seed both genes were expressed in the RAD and CAP compartments, and their expression levels declined rapidly upon imbibition (Fig. 1D). ABA is known to inhibit the endosperm CAP weakening required for the completion of germination (21), and ABA also inhibited the decline in transcript abundance of both LesaDOG1 paralogs (Fig. 1D), suggesting that DOG1 has a key role in this process.
The L. sativum LesaDOG1A Gene Causes a Delayed-Germination Phenotype upon Overexpression in A. thaliana dog1-Mutant Seeds.
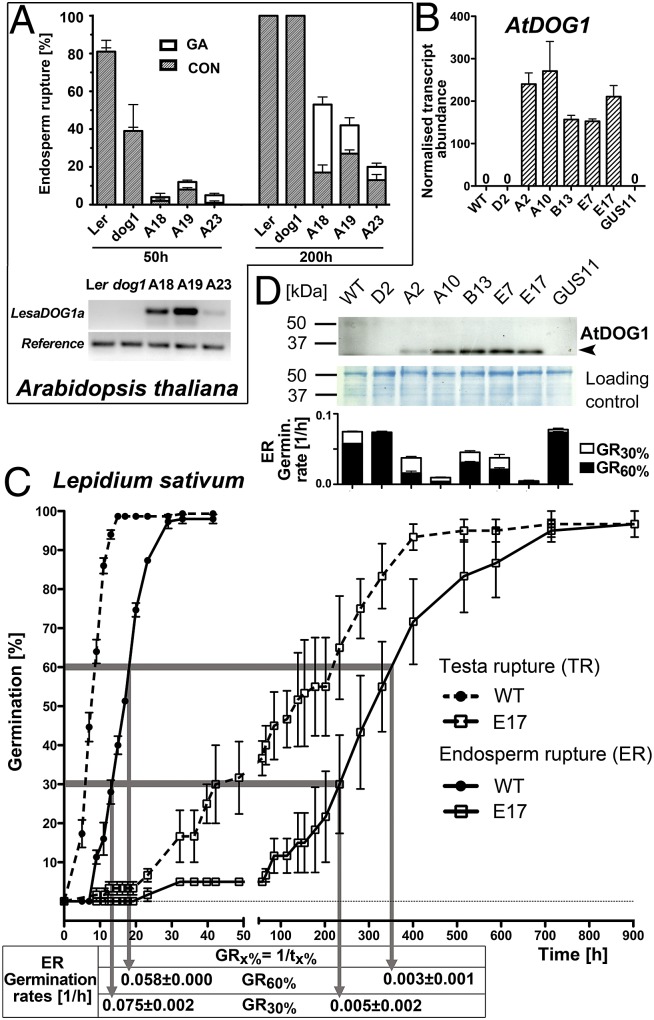
L. sativum produces nondormant seeds although both LesaDOG1 paralogs are expressed in seeds. Therefore, to investigate whether LesaDOG1A, as the more abundant paralog in seeds (Fig. 1D), encodes a functional DOG1 protein, we analyzed its ability to induce dormancy in A. thaliana. A transgene with the LesaDOG1A coding sequence driven by a CaMV 35S-promoter was introduced into the completely nondormant A. thaliana dog1-1 mutant. We compared the germination behavior of seeds from three independent homozygous transgenic A. thaliana dog1 lines overexpressing LesaDOG1A (At-OxLesaDOG1A-A18, -A19, -A23) with the Landsberg erecta (Ler) WT and the dog1 mutant. All At-OxLesaDOG1A lines showed a delayed-germination phenotype compared with WT and dog1-mutant plants and had markedly reduced germination capacity (Fig. 2A). Treatment with GA increased the germination percentages by releasing dormancy of the transgenic lines, especially at later times, but did not affect WT or dog1-mutant seeds (Fig. 2A). Taken together, these results demonstrate that LesaDOG1A overexpression confers GA-sensitive dormancy and delayed germination to A. thaliana dog1 seeds. Thus, LesaDOG1A and AtDOG1 seem to fulfill similar seed-related functions in A. thaliana.
Fig. 2.
Reciprocal DOG1 gene-swapping experiments between Brassicaceae reveal a conserved mechanism. Transgenic lines of the Arabidopsis thaliana dog1-mutant overexpressing LesaDOG1A (L. sativum) and transgenic lines of L. sativum FR14 overexpressing AtDOG1 (A. thaliana) exhibit a delayed-germination phenotype. (A) (Upper) ER of A. thaliana Ler WT, dog1 mutant, and independent homozygous transgenic dog1 lines overexpressing LesaDOG1A (At-OxLesaDOG1A-A18, -A19, -A23) during seed imbibition at 24 °C. n = 3; data are shown as mean ± SEM. The LesaDOG1A-overexpression lines showed a delayed-germination phenotype and a decreased ability to germinate, which did not increase beyond 200 h. Treatment with 10 µM GA4+7 (GA) partially released the dormancy of At-OxLesaDOG1A seeds. (Lower) Semiquantitative RT-PCR indicating expression of the LesaDOG1A transgene in dry A. thaliana seeds. At2G20000 was used as stable seed-expressed reference gene (45). (B) Independent homozygous transgenic L. sativum lines (A2, A10, B13, E7, E17) harboring a chimeric transgene with the CaMV 35S-promoter driving an A. thaliana Cvi DOG1 genomic fragment (Lesa-OxAtDOG1) strongly express AtDOG1 transcripts (qRT-PCR) in dry seeds. n = 4; data are shown as mean ± SEM. (C) Lesa-OxAtDOG1 lines show a strong delayed-germination phenotype evident from comparative TR and ER kinetics of WT and E17 seeds. The graph shows the calculation of the GR. n = 3; data are shown as mean ± SEM. (D) Lesa-OxAtDOG1 lines (A2, A10, B13, E7, E17) accumulate AtDOG1 protein in dry seeds and show lower ERs (and thus lower GRs) than WT and GUS11 transformation (control) lines.
Transgenic L. sativum Seeds Overexpressing AtDOG1 Have a Delayed-Germination Phenotype.
Proof that DOG1-homologous genes cause a delayed-germination phenotype has been obtained thus far only from work in A. thaliana, i.e., by transferring the garden cress LesaDOG1A gene into the nondormant A. thaliana dog1 mutant (this work) or by ectopically expressing putative cereal DOG1-like genes in the weakly dormant A. thaliana accession Columbia (26, 27). However, it is unknown if a dormancy mechanism involving DOG1 genes exists in other species or if the DOG1-signaling pathway is specific to Arabidopsis. We addressed the question about an evolutionary conserved DOG1-mediated pathway by overexpressing the AtDOG1 gene in nondormant L. sativum seeds to investigate whether the function of A. thaliana and L. sativum DOG1 genes is truly interchangeable.
To do so, we generated transgenic L. sativum lines overexpressing a genomic fragment of the A. thaliana Cape Verde Island (Cvi) DOG1 gene fused to a CaMV 35S-promoter. Overexpression has the advantage of maintaining a high DOG1 protein level that allows functional investigation during germination; this ability is especially important in determining the endogenous regulation of LesaDOG1A/B (Fig. 1D). We detected high levels of AtDOG1 transcript in dry seeds of most of the independent homozygous transgenic lines (Fig. 2B). The transgenic lines Lesa-OxAtDOG1-A2, -A10, -B13, -E7, and -E17 showed an extremely delayed-germination phenotype compared with WT (Fig. 2C and SI Appendix, Fig. S2A). Both TR and ER were delayed. For easier comparison of the TR and ER kinetics, we determined germination rates (GRs), which are the reciprocal values of the times needed for a seed population to complete a certain percentage of TR or ER (GRx% = 1/tx%). The L. sativum WT seed population reached 30% ER at 13.4 h (t30%) and thus has a GR30% value of 0.075/h (Fig. 2C). In contrast, the transgenic Lesa-OxAtDOG1-E17 line was far slower and reached 30% ER only at 213 h, resulting in a GR30% value of 0.005/h, ca. 16-fold lower (Fig. 2C). The various Lesa-OxAtDOG1 lines differed in the delay of germination, as reflected by their different GR values; only lines accumulating the transgenic AtDOG1 protein showed a lowered GR (Fig. 2D). We conclude that the delayed-germination phenotype is indeed caused by the transgenic overexpression of AtDOG1 in L. sativum seeds.
Overexpression of AtDOG1 in L. sativum Causes Coat-Imposed Seed Dormancy by Inhibiting Endosperm CAP Weakening Without Affecting the Embryo Growth Potential.
We investigated if the delayed-germination phenotype of the transgenic L. sativum Lesa-OxAtDOG1 seeds is indeed caused by the induction of physiological dormancy. To do so, we used several classical dormancy-breaking treatments and quantified their effect on germination behavior. We analyzed the influence of GA, the ABA-biosynthesis inhibitor fluridone, cold-stratification pretreatment, and after-ripening storage on the germination of freshly harvested mature seeds. Reflecting their nondormant state, the fast germination (high GR) of WT seeds was not affected appreciably by any of these treatments (Fig. 3A). In contrast, we found that the delayed germination (low GR) of Lesa-OxAtDOG1 seeds was accelerated drastically to a high GR by GA treatment, cold stratification, or after-ripening storage, indicating dormancy breaking (shown representatively for line E17 in Fig. 3A and SI Appendix, Fig. S2B). Interestingly, fluridone treatment did not affect the germination of E17 seeds, suggesting that de novo ABA synthesis is not involved in the AtDOG1-mediated dormancy of Lesa-OxAtDOG1 seeds. Taken together, these results demonstrate that AtDOG1 overexpression confers physiological dormancy to nondormant L. sativum seeds (Fig. 3A), as, conversely, LesaDOG1A overexpression does to A. thaliana dog1-mutant seeds (Fig. 2A).
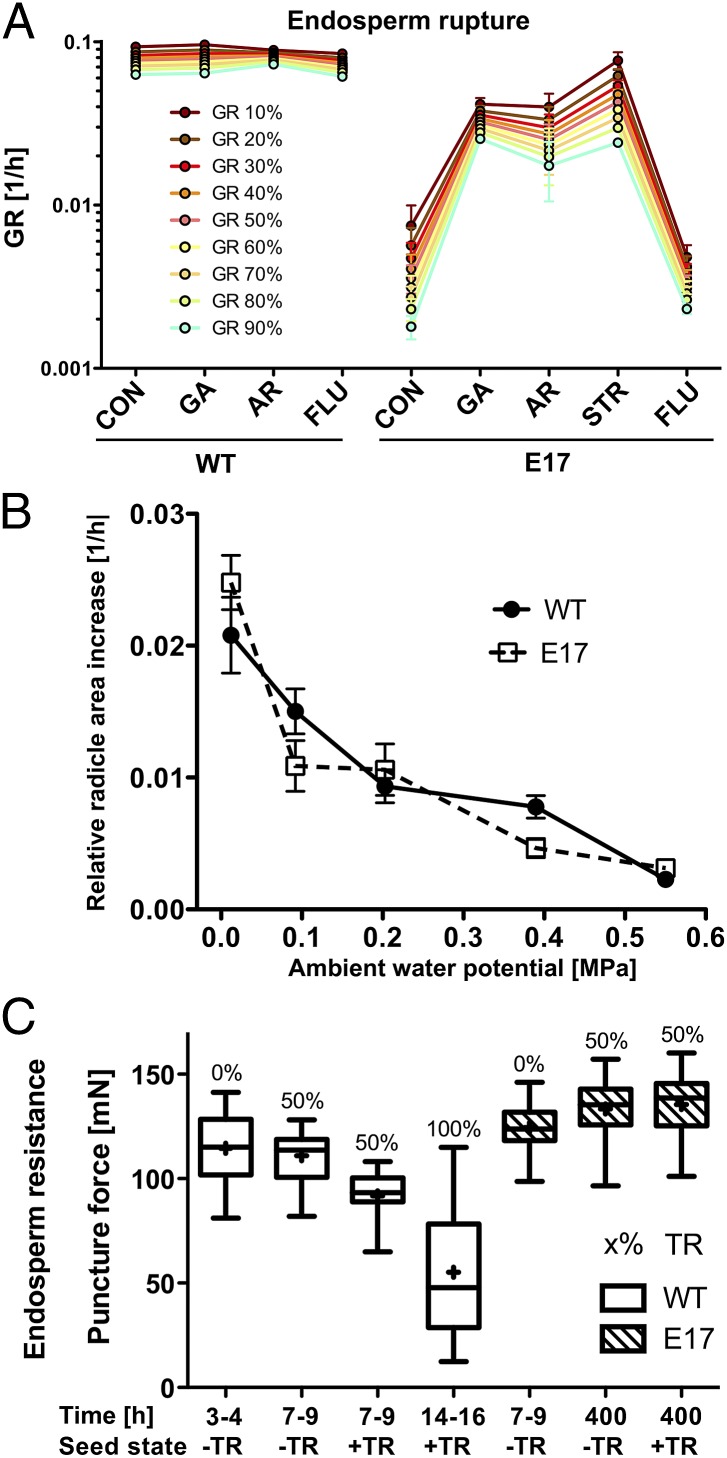
Fig. 3.
The delayed-germination phenotype of transgenic L. sativum is caused by inhibited endosperm CAP weakening. Overexpression of AtDOG1 in L. sativum caused delayed germination, which can be rescued by dormancy-breaking treatments and is caused not by altered embryo growth potential but by inhibited endosperm CAP weakening. (A) ER and resulting GRs of WT and Lesa-OxAtDOG1-E17 seeds at 24 °C without (CON) or with dormancy-breaking treatments: addition of 10 µM GA4+7 (GA), dry after-ripening storage for 9 mo (AR), addition of 10 µM fluridone (FLU), and cold-stratification pretreatment at 4 °C in the dark for 3 d (STR). n = 3; data are shown as mean ± SEM; for TR data, see SI Appendix, Fig. S2B. (B) Embryo growth potentials of WT and Lesa-OxAtDOG1-E17 seeds as measured by the increase in the radicle–hypocotyl axis area of excised embryos at different ambient water potentials at 24 °C. Note that there are no significant differences (P < 0.05) between the embryo GRs of WT and E17 at any water potential. Each data point represents the relative increase in average radicle area (±SEM) of at least 20 imbibed embryos during an incubation period of 27 h. (C) Endosperm weakening occurs during the germination of L. sativum WT seeds and is strongly inhibited in Lesa-OxAtDOG1-E17 seeds. Box plots show endosperm CAP puncture forces of imbibed seeds with (+TR) or without (−TR) testa rupture at 24 °C at the times indicated (n = 20). TR percentages of the seed population for the respective time points are indicated above the box plots.
Seed dormancy and germination are controlled by the balance between the resistance of the seed-covering layers (testa and endosperm) and the embryo growth potential (8, 19). The embryo growth potential determines embryo growth by water uptake and can be quantified using solutions that differ in water potential combined with image analysis (33). Interestingly, we found no significant difference in the growth potential of L. sativum WT and Lesa-OxAtDOG1-E17 embryos at any tested ambient water potential (Fig. 3B). Thus the overexpression of AtDOG1 does not alter the growth potential of isolated L. sativum embryos, although intact seeds germinate much more slowly. Initial scarification experiments (in which seed-covering layers are removed to lessen the resistance against the embryo growth potential) suggested that Lesa-OxAtDOG1 lines have coat dormancy, because the scarified seeds germinated faster. To test if overexpression of AtDOG1 alters the resistance of the seed-covering layers, we conducted puncture-force measurements (21). Analysis of E17 and WT testa plus endosperm tissues during very early imbibition showed that there is no significant difference in the initial resistance of these seed-covering layers and thus indicate the absence of general structural differences caused by AtDOG1 overexpression (SI Appendix, Fig. S3). However, puncture-force analysis of only the endosperm CAP tissue during the course of germination showed that endosperm CAP weakening was differentially affected in WT and Lesa-OxAtDOG1-E17 seeds (Fig. 3C). In WT seeds weakening was initiated in association with TR and progressed strongly thereafter, whereas in E17 seeds CAP weakening was strongly inhibited, and no weakening occurred at all before or after TR, even at very late time points during imbibition. We conclude that the delayed-germination phenotype of L. sativum seeds overexpressing AtDOG1 is caused not by a decreased embryo growth potential but rather by an AtDOG1-mediated severe inhibition of endosperm CAP weakening. Therefore AtDOG1 induced endosperm-mediated physiological coat dormancy in L. sativum Lesa-OxAtDOG1 seeds.
Germination Temperature Strongly Affects the Delayed-Germination Phenotype Caused by DOG1 Overexpression in L. sativum and A. thaliana.
We found that the delay of germination induced by AtDOG1 overexpression in L. sativum depended strongly on the seed imbibition temperature. Analysis of four different temperatures showed that 24 °C is optimal for the germination of L. sativum WT seeds, whereas 18 °C is optimal for Lesa-OxAtDOG1-E17 seeds (Fig. 4A). Germination of E17 seeds is drastically delayed at 24 °C (low GR) but is much faster and more similar to WT (high GR) at 18 °C. DOG1 overexpression in E17 seeds therefore generated a shift in the optimal germination temperature toward colder temperatures (Fig. 4 A and B).
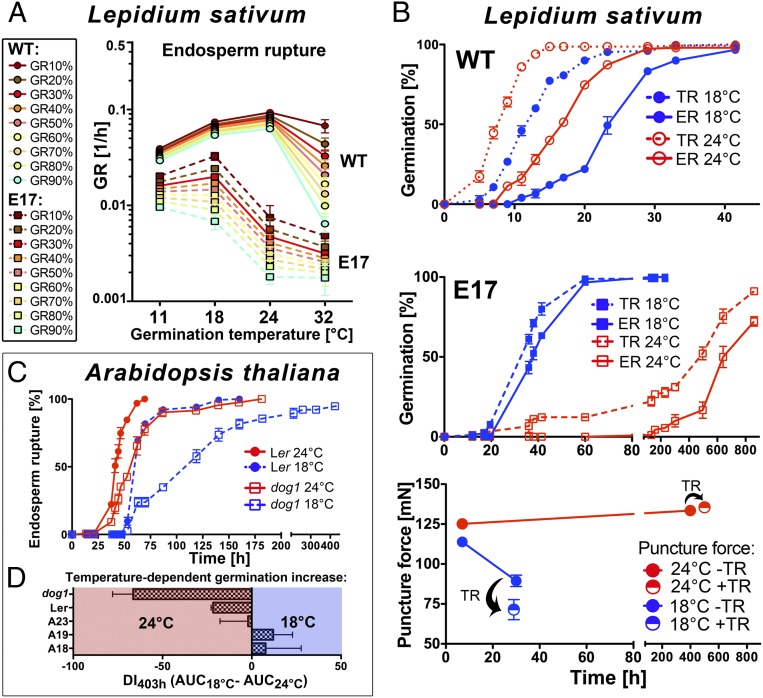
Fig. 4.
DOG1 influences the delay of germination in a temperature-dependent manner in L. sativum and A. thaliana. (A) GRs for ER of L. sativum WT and transgenic Lesa-OxAtDOG1-E17 seeds at different imbibition temperatures (TR data are shown in SI Appendix, Fig. S4A). (B) TR and ER in L. sativum WT and E17 seeds at 18 and 24 °C. Endosperm CAP resistance for E17 seeds quantified by puncture-force measurements are shown from seeds either with (+) or without (−) TR. n = 20; data are shown as mean ± SEM. (C) Temperature dependence of A. thaliana Ler and dog1-mutant seed germination. (D) Dormancy index (DI) calculated as the difference of the areas under ER curves at 18 °C and 24 °C between 0 and 403 h. DI is a measure of germination capacity. A positive DI indicates a positive effect of 18 °C on germination percentage, whereas a negative DI indicates a positive effect of 24 °C. n = 3; data are shown as mean ± SEM. Note that dog1 seeds germinate more slowly than Ler seeds at 18 °C (i.e., have a more negative DI). This effect is reversed in transgenic dog1 lines overexpressing LesaDOG1A (A18, A19, A23), in that either they germinate faster at 18 °C (positive DI) than at 24 °C, or no temperature effect is evident (DI = 0). For all germination kinetics, n = 3 plates; data are shown as mean ± SEM.
Puncture-force measurements of E17 endosperm CAPs at the two phenotypically very contrasting temperatures (18 °C and 24 °C) showed that endosperm CAP resistance of E17 seeds was differentially affected by the ambient temperature (Fig. 4B). No weakening of the E17 endosperm CAPs occurred at 24 °C, but at 18 °C CAP weakening commenced as germination proceeded in a pattern similar to that seen in WT seeds (Fig. 3C and Fig. 4B). Interestingly, the E17 endosperm CAPs weakened considerably when the testa ruptured at 18 °C, but at 24 °C no endosperm CAP weakening was detected even after TR (Fig. 4B). Because the embryo growth potentials of E17 and WT seeds at 24 °C are identical (Fig. 3B), the observed delayed-germination phenotype can be explained by temperature-dependent inhibition of endosperm CAP weakening caused by the overexpression of AtDOG1.
Between 18 °C and 24 °C there is a large shift in the germination response of Lesa-OxAtDOG1-E17 seeds (Fig. 4A): At 18 °C, 50% of seeds completed germination within 30 h, but at 24 °C 600 h were required to achieve the same percentage of germination (Fig. 4B). This temperature window is surprisingly narrow for such an immense difference. We investigated if this narrow temperature window also affected the A. thaliana dog1 mutant and the transgenic At-OxLesaDOG1A lines. Fig. 4C shows that A. thaliana Ler WT seeds germinated only slightly more slowly at 18 °C than at 24 °C, but the dog1-mutant seeds germinated much more slowly at 18 °C than at 24 °C. This delay in germination of the dog1-mutant seeds at the cooler temperature also was evident from their very negative dormancy index (DI) (Fig. 4D). In the transgenic A. thaliana lines (At-OxLesaDOG1A-A18, -A19, -A23), this dog1-specific temperature phenotype was completely reverted, as evident from their more positive DIs (Fig. 4D).
In conclusion, these results show that LesaDOG1A and AtDOG1 both affect the temperature responses of A. thaliana and L. sativum seeds. In both species a high DOG1 level seems to limit germination at warmer temperatures, whereas the absence or low levels of DOG1 permit germination at warmer temperatures. From the biomechanical analysis of Lesa-OxAtDOG1-E17 seeds, we conclude that overexpression of AtDOG1 in L. sativum defines the optimal temperature for endosperm CAP weakening, which then occurs at 18 °C rather than at 24 °C. We propose that temperature control of seed germination regulated by DOG1 depends on a conserved coat-dormancy mechanism within the Brassicaceae with endosperm CAP weakening as the target.
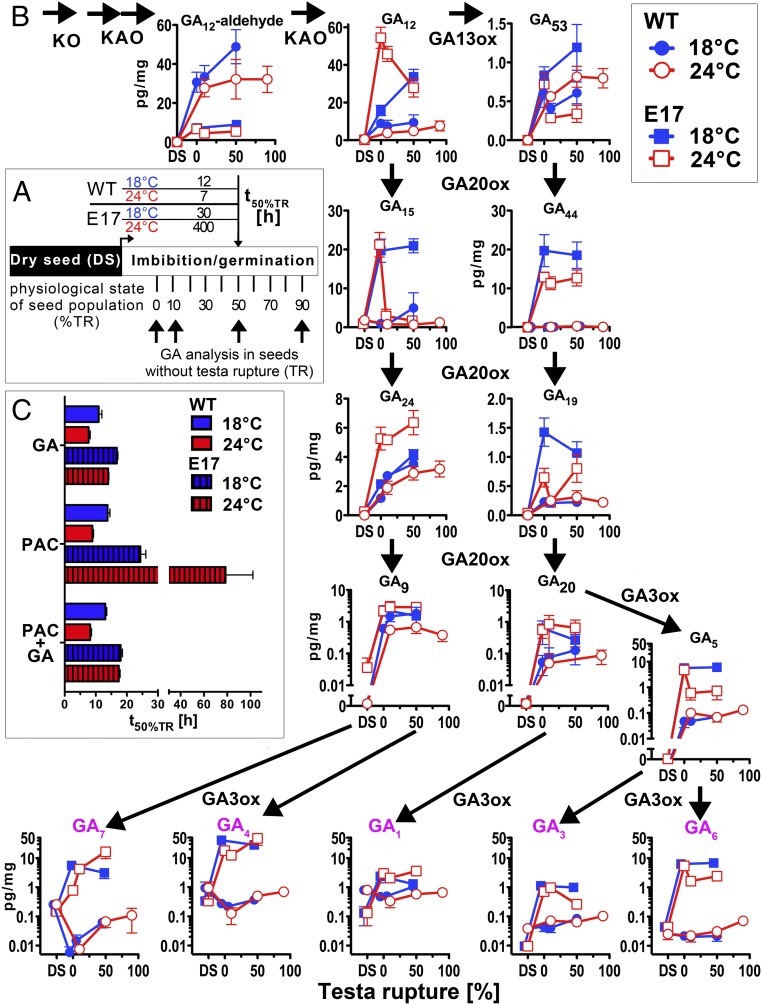
Germination Temperature Differentially Affects GA Contents in WT and Transgenic L. sativum Seeds Overexpressing AtDOG1.
ABA is known to maintain coat dormancy and inhibit endosperm weakening, whereas GA releases coat dormancy and promotes endosperm weakening (8, 10, 32). We found that during imbibition the ABA content of Lesa-OxAtDOG1-E17 seeds decreases, but there was no difference in this decrease at 24 °C as compared with 18 °C (SI Appendix, Fig. S4D). Therefore, absolute ABA content did not cause the remarkable differences in the temperature responses of E17 seeds, i.e., the strongly inhibited endosperm CAP weakening and delayed germination at the higher temperature (Fig. 4B and SI Appendix, Fig. S4D). We conclude that the ABA content does not cause the delayed and temperature-sensitive germination phenotype induced by AtDOG1 overexpression. This notion is in agreement with our finding that inhibiting ABA biosynthesis did not increase the delayed germination of E17 seeds (Fig. 3A).
To investigate the role of GAs, we quantified major GA metabolites in dry and imbibed L. sativum WT and Lesa-OxAtDOG1-E17 seeds at 18 and 24 °C (Fig. 5). We analyzed seed populations that had not yet undergone TR at physiologically and physically comparable times during germination (Fig. 5A). Thus we were able to identify the gradual changes that occur in seeds that are preparing to undergo the first visible committed step to the completion of germination. We found that GA metabolite contents in the imbibed state were strongly altered by AtDOG1 overexpression in combination with the ambient imbibition temperature (Fig. 5B and SI Appendix, Fig. S5). Surprisingly, bioactive GAs generally were far more abundant in E17 than in WT seeds. The total bioactive GA content (GA1, GA3, GA4, GA6, GA7) at 50% TR was 1.2 ± 0.6 pg/mg in WT seeds and 40.3 ± 16.1 pg/mg in E17 seeds at their respective optimal temperatures (24 °C for WT and 18 °C for E17). This ∼40-fold increase indicates that E17 seeds require a far higher GA content than WT seeds to reach the same germination progression under optimal conditions. Furthermore, the ambient temperature had opposing effects on the GA contents of WT and E17 seeds at the same physiological time point (50% TR). At 18 °C the bioactive GA content in E17 seeds was roughly double that at 24 °C, whereas in WT seeds it was almost halved. These temperature-dependent changes were in accordance with the observed germination phenotype; i.e., the temperatures for optimal germination were associated with higher bioactive GA content in both genotypes. However, the absolute bioactive GA content is far higher in E17 seeds than in WT seeds, even though E17 seeds germinate more slowly at any temperature (Fig. 4 A and B).
Fig. 5.
DOG1 controls the seed GA metabolism in a temperature-dependent manner. Overexpression of AtDOG1 in L. sativum seeds leads to an increase of GA metabolites and a strongly altered temperature regulation of GA metabolism during germination. (A) Experimental overview of the GA metabolite analysis shown in B. The physical time to reach TR differs depending on seed imbibition temperature and genotype. Shown is a representation of the time required to reach 50% TR (t50%TR) for L. sativum WT and the AtDOG1 overexpression line Lesa-OxAtDOG1-E17 imbibed at 18 °C and 24 °C. Arrows indicate physiological sampling time points for GA analysis. Only seeds without TR within the seed population were sampled. (B) The GA metabolite content of L. sativum WT and Lesa-OxAtDOG1-E17 seed populations reaches different physiological states during imbibition at 18 °C and 24 °C. Shown are main metabolites of the early 13-nonhydroxylated (Left) and 13-hydroxylated (Right) pathway. The GA biosynthetic enzymes catalyzing the respective steps are indicated. Bioactive GAs are indicated in purple. Results are presented as amount per dry weight (mean ± SEM); n = 5. DS, dry seed. Results for these and additionally quantified GA metabolites represented on a physical time scale are shown in SI Appendix, Fig. S5 and are shown as numeric values in SI Appendix, Table S2. (C) Effect of the GA biosynthesis inhibitor PAC (100 µM) on TR (t50%TR) of L. sativum WT and Lesa-OxAtDOG1-E17 seeds at 18 °C and 24 °C compared with germination on GA4+7 (GA, 10 µM) or on a combination of both (PAC+GA). n = 3; data are shown ± SEM.
To investigate if this observed higher GA accumulation is actually necessary for E17 seeds to germinate, we analyzed germination responses upon treatment with the GA biosynthesis inhibitor paclobutrazol (PAC) (Fig. 5C). Treatment with PAC strongly inhibited TR and subsequent completion of germination at 24 °C in E17 seeds, and this inhibition was rescued by combined application of PAC plus GA. Neither PAC nor GA affected the germination responses of WT seeds at any temperature. Interestingly, the PAC-induced inhibition of germination in E17 seeds was much weaker at 18 °C than at 24 °C.
These results show that DOG1 overexpression establishes a high GA requirement for germination. E17 seeds seem to react to this DOG1-mediated high GA threshold by producing larger amounts of GA, and the ability to produce these increased amounts is strongly temperature-dependent. In agreement with the observed temperature dependence of the PAC inhibition, the delayed-germination phenotype at 24 °C thus seems to be caused by lower GA biosynthesis at this temperature which does not compensate for the AtDOG1-induced GA requirement. This notion is in agreement with the findings that, for E17 seeds, the addition of GA stimulated germination at 24 °C (Fig. 3A), the observed endogenous GA levels were lower at 24 °C in most cases (Fig. 5B), and the inhibition of GA biosynthesis was very effective in delaying germination at 24 °C (Fig. 5C). DOG1 therefore seems to define the optimal germination temperature by mediating a germination block based on a high GA threshold for germination that can be reached only at temperatures that allow high GA biosynthesis.
Germination Temperature and DOG1 Presence Differentially Affect the Expression of GA Biosynthetic and Cell-Wall Remodeling Genes.
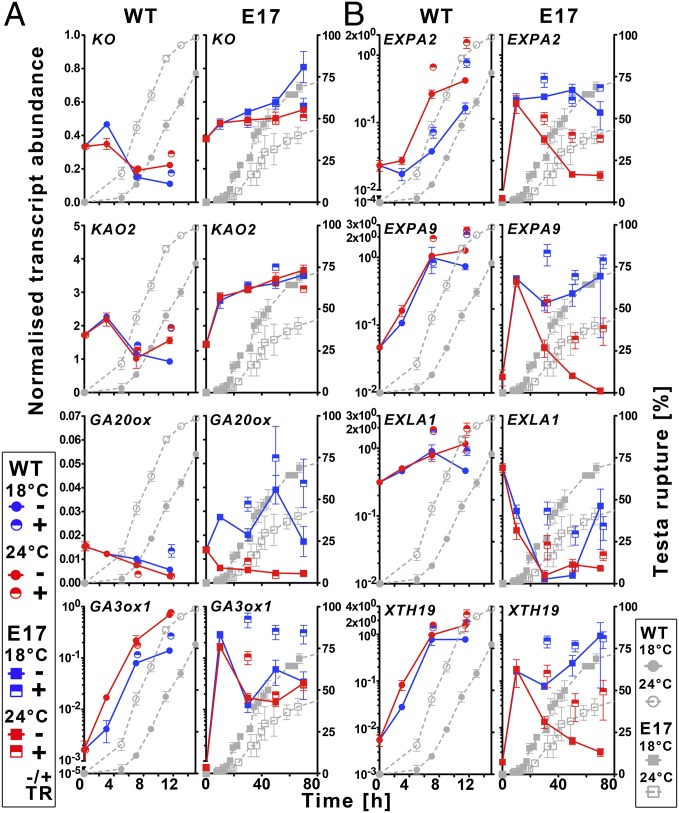
To elucidate the molecular basis of the drastically enhanced GA levels and their temperature regulation, we investigated the gene expression of key enzymes of the GA biosynthetic pathway in WT L. sativum and in the E17 line overexpressing AtDOG1. Biosynthesis of GA12, the common precursor for all GAs in plants, is catalyzed by ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) (34). The GA12 content was strongly increased in E17 seeds as compared with WT (Fig. 5B). In agreement with this finding, we found increased expression of KO and KAO2 during the germination of E17 seeds (Fig. 6A). Higher expression of these key enzymes thus may be the cause for the general elevation in E17 GA metabolite content. Further early reactions of GA biosynthesis are catalyzed by GA20-oxidases (GA20ox). Most interestingly, we found GA20ox expression to be strongly temperature-regulated in E17 but not in WT seeds (Fig. 6A and SI Appendix, Fig. S6). During germination GA20ox was up-regulated in E17 seeds at 18 °C but not at 24 °C, whereas it was down-regulated at both temperatures in WT seeds (Fig. 6A). Thus this specific expression pattern is highly associated with the accumulation patterns of the initial metabolites synthesized by GA20ox (GA15, GA44), which were high at 18 °C but low at 24 °C in E17 seeds (Fig. 5B). Bioactive GAs are synthesized by GA3-oxidases (GA3ox). Interestingly, we found that, in contrast to GA20ox, the GA3ox1 gene was up-regulated similarly during early germination of WT and E17 seeds at both temperatures (Fig. 6A). However, after this initial up-regulation, the transcript contents were down-regulated in E17 seeds at both temperatures but continued to be up-regulated in WT seeds. Surprisingly, later during germination (during TR), the expression of this gene in E17 seeds was strongly up-regulated at 18 °C but not at 24 °C. This pattern is intriguing, given that endosperm weakening of E17 seeds during TR occurs only at 18 °C, but not at 24 °C (Fig. 4B).
Fig. 6.
DOG1 influences the expression of GA biosynthesis and CAP-weakening genes in a temperature-dependent manner. Temperature-dependent gene-expression profiles of candidate GA biosynthesis and endosperm CAP-weakening genes during the germination of L. sativum E17 seeds overexpressing AtDOG1 compared with WT seeds. qRT-PCR analysis of transcript abundance of (A) GA biosynthesis genes (KO, KAO1, GA20ox, GA3ox1) and (B) candidate cell-wall remodeling genes (EXPA2, EXPA9, EXLA1, XTH19) at 18 °C and 24 °C in WT (Left) and E17 (Right) seeds without (−TR) or with (+TR) testa rupture were sampled at the indicated time points, which relate to the TR kinetics indicated by the gray curves. n = 3; data are shown as mean ± SEM. Results for other seed batches and lines are shown in SI Appendix, Fig. S6. Note that temperature affects transcript abundances and germination differently in WT and E17 seeds, but the overexpressed AtDOG1 transcript and protein amounts themselves were not temperature-regulated in E17 seeds during germination (SI Appendix, Fig. S7).
To gain insight into the underlying molecular mechanisms downstream of the strongly AtDOG1- and temperature-dependent CAP weakening, we analyzed the expression of GA-regulated candidate endosperm CAP-weakening genes (Fig. 6B). We investigated known genes encoding CWRPs of the expansin and xyloglucan endo-transglycosylases/hydrolase families (35). Transcript abundances of these genes (EXPA2, EXPA9, EXLA1, XTH19) increased steadily in WT seeds during germination at both 18 °C and 24 °C (Fig. 6B). In contrast, in E17 seeds EXPA2, EXPA9, and XTH19 were strongly temperature-regulated. After initially increasing in E17 seeds at both temperatures during the first hours of imbibition, transcripts declined dramatically at 24 °C but not at 18 °C (Fig. 6B). Therefore, the temperature- and AtDOG1-regulated patterns of EXPA2, EXPA9, and XTH19 transcript expression in WT and E17 seeds were highly associated with the accumulation patterns of bioactive GAs (Fig. 5B) and with the alteration of endosperm CAP weakening and the resulting germination phenotype (Fig. 4B). In contrast, EXLA1 was down-regulated independently of temperature in E17 seeds but was up-regulated in WT seeds.
We conclude that DOG1 established a high GA requirement for E17 seed germination by repressing the GA-induced expression of CWRP genes needed for endosperm weakening. A constantly high GA content is necessary to overcome this DOG1-induced repression of CWRP expression. In the presence of DOG1 this constantly high GA content is maintained only at colder and not at warmer temperatures. This result seems to be caused by the specific up-regulation of GA20ox expression only at the colder temperature. DOG1 therefore controls GA20ox expression and germination in a temperature-dependent manner.
Discussion
DOG1 Mediates a Conserved Physiological Coat-Dormancy Mechanism in the Brassicaceae L. sativum and A. thaliana.
We establish here that DOG1 genes mediate a common dormancy mechanism, i.e., that the function and role(s) of DOG1 are conserved. Environmentally and hormonally regulated DOG1 gene expression before seed dispersal (maturation) is important for the control of Brassicaceae seed germination (2, 4, 5, 11, 24), and it is known that in A. thaliana DOG1 provides adaptation to local environments (1, 14, 36). After seed dispersal, AtDOG1 transcript levels in the soil seed bank are central to sensing seasonal temperature patterns and differ characteristically during dormancy, cycling in summer and winter annual A. thaliana ecotypes (6, 7). We show here by reciprocal overexpression that AtDOG1 and LesaDOG1A confer dormancy to nondormant L. sativum WT and A. thaliana dog1-mutant seeds, respectively. We found that DOG1 genes induce primary physiological seed dormancy that can be released by after-ripening storage of dry seeds, by cold stratification of imbibed seeds, and by treatment of imbibed seeds with bioactive GAs.
From a mechanistic point of view, dormancy and germination are regulated by two opposing forces, the growth potential of the embryo counteracting the restraint imposed by the seed-covering layers (17, 19, 37). By imaging embryo growth (33) at different ambient water potentials, we demonstrate here that Lesa-OxAtDOG1 and WT embryos did not differ in growth potential. However, we found that the overexpression of AtDOG1 severely inhibits endosperm CAP weakening in imbibed Lesa-OxAtDOG1 seeds. Therefore, AtDOG1 confers endosperm-mediated coat dormancy to L. sativum seeds. Thus, we demonstrate that, to induce dormancy and delay germination, DOG1 targets not the embryo growth potential but the seed-covering layers.
We conclude that an evolutionarily conserved role of DOG1 confers physiological coat dormancy and delayed germination to Brassicaceae seeds. This DOG1-mediated dormancy pathway does not alter the embryo growth potential but instead has endosperm CAP weakening as its major target and enables temperature-dependent control of dormancy during imbibition by regulating CWRP gene expression and GA metabolism as outlined below.
DOG1 Determines the Temperature Window for Germination by Regulating the Expression of Endosperm CAP-Weakening Genes Through Temperature Control of the GA Metabolism.
We found for L. sativum that overexpression of AtDOG1 leads to a shift in the optimal germination temperature toward colder temperatures (from 24 °C to 18 °C). In agreement with this finding, the A. thaliana dog1-mutant germinated faster at the warmer temperature, and overexpression of LesaDOG1A abolished its preference for the warmer temperature as the germination optimum. These results show that the amount of functional DOG1 protein determines the seed’s optimal germination temperature. We conclude that increased levels of DOG1 shift the optimal germination temperature to colder temperatures and that reduced levels of DOG1 shift the optimal germination temperature to warmer temperatures. Endogenous AtDOG1 expression actually cycles in A. thaliana seeds in the soil seed bank throughout the year in association with soil temperature and dormancy state, and it has been proposed that AtDOG1 acts as a seed thermal sensor determining the depth of dormancy (6). In our L. sativum Lesa-OxAtDOG1 lines, overexpressed AtDOG1 does not cycle during imbibition (SI Appendix, Fig. S7), and the constantly high expression maintains the inhibition of germination at warmer temperatures. In agreement with this finding, germination of A. thaliana Cvi seeds known for high AtDOG1 expression also is inhibited at warmer temperatures (7). Thus, on the one hand, endogenous DOG1 expression is influenced by the environment, i.e., it is up-regulated at low temperature during maturation as well as in imbibed seeds of A. thaliana (2, 4–7) and L. sativum (SI Appendix, Fig. S8). On the other hand, the amount of DOG1 seems to determine the temperature-sensitivity window for germination. Higher DOG1 levels generate a narrow temperature window and restrict germination at higher temperatures, whereas lower DOG1 levels generate a wide temperature window and allow germination at higher temperatures. This regulation seems to be conserved in L. sativum and A. thaliana and points to a general underlying mechanism by which DOG1 regulates coat dormancy to define the temperature window for germination. As in embryo-related developmental processes in animals (16, 18), the embryo-encasing tissue layers of the seed are of key importance: For L. sativum we have shown directly that the temperature-dependent inhibition of germination is caused by the DOG1-mediated, temperature-specific inhibition of endosperm CAP weakening.
Our findings show that DOG1 is involved in the temperature-dependent control of GA metabolism. Strikingly, analysis of the GA metabolism demonstrated that imbibed dormant Lesa-OxAtDOG1 seeds have a strongly increased GA metabolite content, including the bioactive GAs. Furthermore, GA metabolites show a strong temperature-dependent accumulation in the presence of DOG1. In WT L. sativum seeds the KO and KAO2 genes encoding the enzymes for GA12 biosynthesis are expressed during early germination, and their expression decreases during late germination; this expression pattern is very similar to that in A. thaliana seeds (38). In contrast, these genes are constantly up-regulated when DOG1 is overexpressed in L. sativum E17 seeds. Interestingly, only KAO2—but not KAO1—expression is affected by DOG1 overexpression (SI Appendix, Fig. S9), indicating that different mechanisms are involved in the regulation of these two genes, as has been shown for duplicated KAO genes in pea and sunflower (34). KO expression is directly regulated by the bZIP transcription factor REPRESSION OF SHOOT GROWTH (RSG) (39), which also has been suggested to regulate KAO expression (40). Thus, it is tempting to speculate that DOG1 interacts with certain bZIP transcription factors to regulate GA biosynthesis. Because the drastic DOG1-mediated up-regulation of GA metabolite content is evident for GA12, but not for GA12-aldehyde, we propose that the DOG1-mediated induction of KAO2 plays a major role in the general increase in the GA metabolite content. Up-regulated DOG1-mediated KAO2 expression can explain the generally enhanced GA biosynthesis pathway but is not temperature-dependent and therefore does not explain the observed temperature-dependent differences in GA metabolite accumulation.
Most intriguingly, we found that GA20ox gene expression is temperature-regulated by DOG1 overexpression, and this regulation is in agreement with the temperature-dependent accumulation of GA metabolites catalyzed by GA20ox enzymes. In tobacco, gene expression of GA20ox, but not of KO or KAO, has been shown to be regulated by a GA negative feedback mechanism controlled by the RSG transcription factor (34, 39). In agreement with this finding, neither KO nor KAO2 expression is down-regulated by the high GA content in E17 seeds. However, GA20ox expression is down-regulated in E17 seeds imbibed at 24 °C, but not at 18 °C (Fig. 6A). This result suggests that DOG1 interferes with the negative feedback regulation of GA20ox in a temperature-dependent manner, potentially through interaction with transcription factors, such as RSG, that regulate GA20ox expression. In contrast to GA20ox, expression of GA3ox1, which also is feedback-regulated (41) but not by RSG (39), seems to be strongly down-regulated in E17 seeds during the course of germination when GA levels are high. Therefore we propose that DOG1 specifically interferes at the level of GA20ox expression to mediate the temperature-dependent regulation of bioactive GA accumulation.
The downstream mechanism for endosperm CAP weakening depends on the GA-induced expression of CWRP genes such as expansins and xyloglucans in the CAP (9, 35, 42–44). Although E17 seeds contain elevated GA contents compared with WT seeds at any imbibition temperature, the expression of candidate CWRP genes is reduced, especially at 24 °C. DOG1 therefore seems to repress GA-induced CWRP expression, especially at higher temperatures, and this repression subsequently inhibits endosperm weakening and germination. The elevated GA contents of E17 seeds at 18 °C caused by the GA20ox induction seem to overcome the DOG1-imposed repression of the expression of the CAP-weakening genes EXPA2, EXPA9, and XTH11. That DOG1 repressed EXLA1 at both temperatures in E17 seeds but not in WT seeds suggests that DOG1 might regulate the expression of diverse CWRP genes differently. Constantly high GA content seems to be required to overcome this repression and to release the DOG1-imposed coat dormancy. The elevated GA content in imbibed dormant Lesa-OxAtDOG1 seeds actually might be a feedback reaction of the seed to overcome the block to germination imposed by AtDOG1 overexpression. Nondormant L. sativum WT seeds do not require additional de novo GA biosynthesis for germination. In contrast, dormant Lesa-OxAtDOG1 seeds require de novo GA biosynthesis because they are highly responsive to the inhibition of GA biosynthesis. They are not GA-insensitive per se but have a high GA requirement which at 24 °C is not saturated by the elevated endogenous bioactive GA. This decreased GA sensitivity and increased sensitivity to the inhibition of GA biosynthesis in L. sativum seeds overexpressing AtDOG1 is in agreement with the finding that A. thaliana dog1-mutant seeds have increased GA sensitivity and decreased sensitivity to a GA biosynthesis inhibitor (5, 11). The A. thaliana dog1 mutant in a GA-deficient background needed 10-fold less added GA to reach the same germination progression as the corresponding control (11). Furthermore, ecophysiological work demonstrated that A. thaliana DOG1 alleles causing delayed germination in the field are associated with increased seed GA content (15). Interestingly, the enhanced content in GA metabolites were not evident in dry Lesa-OxAtDOG1 seeds but were induced during seed imbibition and underline a role of DOG1 in maintaining coat dormancy in the imbibed state.
Besides regulating the seed GA content in a temperature-dependent manner to control CWRP expression, DOG1 also may regulate GA-signaling pathways that are important for the GA induction of CWRP genes. Expression of the CWRP genes described above is known to be regulated via the GIBBERELLIN INSENSITIVE DWARF 1 (GID1)-type GA-signaling pathways in L. sativum and A. thaliana; both species possess the GA receptors GID1A, GID1B, and GID1C (35). In contrast to WT seeds, down-regulation of GID1A expression is evident in E17 seeds during early germination (SI Appendix, Fig. S9) and may be involved in causing lower CWRP expression in E17 seeds despite their elevated GA content. Furthermore, temperature-dependent up-regulation of GID1B in E17 seeds during late germination might provide the GA sensitivity needed to overcome the temperature-dependent repression of CWRP expression inhibiting germination.
Taken together, our findings show that DOG1 regulates Brassicaceae coat dormancy by repressing the GA-induced expression of CWRPs required for endosperm CAP weakening in imbibed seeds. DOG1 acts by modifying GA metabolism in a temperature-dependent manner to overcome the repression of CWRPs at certain temperatures. This mechanism confers temperature-responsive control of endosperm CAP weakening and thereby determines the optimal seed germination temperature.
Materials and Methods
Physiological Assays.
Analysis of seed germination (24) and embryo growth potential (33) was performed as described in SI Appendix, Supplementary Methods. Puncture-force measurements were conducted as described previously (32) with a modified custom-made biomechanics machine (load cell range, 0–1 N). Intact endosperm CAP tissue was dissected from the imbibed seeds and glued to a metal sample holder (hole size, 0.6 mm) using Loctite 454 glue (Henkel). A rounded metal pin was driven into the sample while force and displacement were recorded simultaneously. A probe with a diameter of 0.3 mm, a hemisphere-shaped tip, and a speed of 0.7 mm/min was used while force and displacement were recorded. The CAP puncture force (tissue resistance) was determined to be the maximal force from the displacement–force curves.
Molecular Methods.
LesaDOG1B genomic and cDNA sequences (GenBank ID KF501341) were cloned as described in refs.23 and 45. The A. thaliana DOG1 overexpression construct used for transformation of L. sativum FR14 was a pLEELA vector containing a double 35S CaMV promoter and the genomic ORF of A. thaliana Cvi DOG1, which was provided by Wim Soppe (Max Planck Institute for Plant Breeding Research, Cologne, Germany). The L. sativum LesaDOG1A overexpression construct used for transformation of A. thaliana dog1-1 was prepared by cloning the LesaDOG1A cDNA (23) ORF into the 35S CaMV promoter containing the pB2GW7 vector by using Gateway technology (Invitrogen). Southern blot analysis was performed using a digoxigenin-labeled probe covering 353 bp of exon 1 of LesaDOG1A, as described (24). Western blot analysis was performed as described (24) using a primary polyclonal antibody raised against AtDOG1 (5). RNA extraction, quantitative RT-PCR (qRT-PCR) analysis (45), GA and ABA metabolite quantification, and plant transformation were performed as described in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
We thank Wim Soppe and Kazumi Nakabayashi (Max Planck Institute for Plant Breeding Research) for kindly providing the AtDOG1 antibody and AtDOG1 overexpression construct and Safina Khan for expert technical assistance. This work and the positions of K.G., A.L., T.S., and A.V. were funded by Grants DFG LE720/6 and DFG LE720/7 from the Deutsche Forschungsgemeinschaft (to G.L.-M.). Further funding was provided by Grant GD522/08/H003 from the Czech Science Foundation, Grants LK21306 and LO1204 from the Ministry of Education and Sport of the Czech Republic, Grant 14-34792S by the Grant Agency of the Czech Republic (all to D.T. and M.S.), and Grant ED0007/01/01 from the Operational Program Research and Development for Innovations (to M.S.). T.U. received support from Grant CZ.1.07/2.3.00/30.0037.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KF501341).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403851111/-/DCSupplemental.
References
- 1.Kronholm I, Picó FX, Alonso-Blanco C, Goudet J, de Meaux J. Genetic basis of adaptation in Arabidopsis thaliana: Local adaptation at the seed dormancy QTL DOG1. Evolution. 2012;66(7):2287–2302. doi: 10.1111/j.1558-5646.2012.01590.x. [DOI] [PubMed] [Google Scholar]
- 2.Chiang GCK, et al. DOG1 expression is predicted by the seed-maturation environment and contributes to geographical variation in germination in Arabidopsis thaliana. Mol Ecol. 2011;20(16):3336–3349. doi: 10.1111/j.1365-294X.2011.05181.x. [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Blanco C, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21(7):1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kendall SL, et al. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23(7):2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakabayashi K, et al. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell. 2012;24(7):2826–2838. doi: 10.1105/tpc.112.100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Footitt S, Douterelo-Soler I, Clay H, Finch-Savage WE. Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc Natl Acad Sci USA. 2011;108(50):20236–20241. doi: 10.1073/pnas.1116325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Footitt S, Huang Z, Clay HA, Mead A, Finch-Savage WE. Temperature, light and nitrate sensing coordinate Arabidopsis seed dormancy cycling, resulting in winter and summer annual phenotypes. Plant J. 2013;74(6):1003–1015. doi: 10.1111/tpj.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171(3):501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 9.Holdsworth MJ, Bentsink L, Soppe WJJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179(1):33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 10.Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJJ. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35(10):1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 11.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103(45):17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentsink L, et al. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc Natl Acad Sci USA. 2010;107(9):4264–4269. doi: 10.1073/pnas.1000410107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton MW, et al. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat Genet. 2012;44(2):212–216. doi: 10.1038/ng.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silady RA, Effgen S, Koornneef M, Reymond M. Variation in seed dormancy quantitative trait loci in Arabidopsis thaliana originating from one site. PLoS ONE. 2011;6(6):e20886. doi: 10.1371/journal.pone.0020886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barua D, Butler C, Tisdale TE, Donohue K. Natural variation in germination responses of Arabidopsis to seasonal cues and their associated physiological mechanisms. Ann Bot (Lond) 2012;109(1):209–226. doi: 10.1093/aob/mcr264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heisenberg C-P, Bellaïche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153(5):948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Linkies A, Leubner-Metzger G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012;31(2):253–270. doi: 10.1007/s00299-011-1180-1. [DOI] [PubMed] [Google Scholar]
- 18.Keller R. Mechanisms of elongation in embryogenesis. Development. 2006;133(12):2291–2302. doi: 10.1242/dev.02406. [DOI] [PubMed] [Google Scholar]
- 19.Nonogaki H. Seed germination - the biochemical and molecular mechanisms. Breed Sci. 2006;56(2):93–105. [Google Scholar]
- 20.Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F. Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 2009;150(2):1006–1021. doi: 10.1104/pp.109.137901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linkies A, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;21(12):3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dekkers BJW, et al. Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol. 2013;163(1):205–215. doi: 10.1104/pp.113.223511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graeber K, et al. Cross-species approaches to seed dormancy and germination: Conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes. Plant Mol Biol. 2010;73(1-2):67–87. doi: 10.1007/s11103-009-9583-x. [DOI] [PubMed] [Google Scholar]
- 24.Graeber K, et al. Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium Delay of Germination1 genes. Plant Physiol. 2013;161(4):1903–1917. doi: 10.1104/pp.112.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugimoto K, et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc Natl Acad Sci USA. 2010;107(13):5792–5797. doi: 10.1073/pnas.0911965107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashikawa I, Abe F, Nakamura S. Ectopic expression of wheat and barley DOG1-like genes promotes seed dormancy in Arabidopsis. Plant Sci. 2010;179(5):536–542. doi: 10.1016/j.plantsci.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Ashikawa I, Abe F, Nakamura S. DOG1-like genes in cereals: Investigation of their function by means of ectopic expression in Arabidopsis. Plant Sci. 2013;208:1–9. doi: 10.1016/j.plantsci.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci. 2011;16(2):108–116. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Schranz ME, Mohammadin S, Edger PP. Ancient whole genome duplications, novelty and diversification: The WGD radiation lag-time model. Curr Opin Plant Biol. 2012;15(2):147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Haudry A, et al. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 2013;45(8):891–898. doi: 10.1038/ng.2684. [DOI] [PubMed] [Google Scholar]
- 31.Dierschke T, Mandáková T, Lysak MA, Mummenhoff K. A bicontinental origin of polyploid Australian/New Zealand Lepidium species (Brassicaceae)? Evidence from genomic in situ hybridization. Ann Bot (Lond) 2009;104(4):681–688. doi: 10.1093/aob/mcp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller K, Tintelnot S, Leubner-Metzger G. Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 2006;47(7):864–877. doi: 10.1093/pcp/pcj059. [DOI] [PubMed] [Google Scholar]
- 33.Voegele A, et al. Embryo growth, testa permeability, and endosperm weakening are major targets for the environmentally regulated inhibition of Lepidium sativum seed germination by myrigalone A. J Exp Bot. 2012;63(14):5337–5350. doi: 10.1093/jxb/ers197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444(1):11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 35.Voegele A, Linkies A, Müller K, Leubner-Metzger G. Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J Exp Bot. 2011;62(14):5131–5147. doi: 10.1093/jxb/err214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, et al. The earliest stages of adaptation in an experimental plant population: Strong selection on QTLs for seed dormancy. Mol Ecol. 2010;19(7):1335–1351. doi: 10.1111/j.1365-294X.2010.04557.x. [DOI] [PubMed] [Google Scholar]
- 37.Bewley JD. Breaking down the walls - a role for endo-β-mannanase in release from seed dormancy? Trends Plant Sci. 1997;2(12):464–469. [Google Scholar]
- 38.Ogawa M, et al. Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell. 2003;15(7):1591–1604. doi: 10.1105/tpc.011650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukazawa J, et al. bZIP transcription factor RSG controls the feedback regulation of NtGA20ox1 via intracellular localization and epigenetic mechanism. Plant Signal Behav. 2011;6(1):26–28. doi: 10.4161/psb.6.1.14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA. 2001;98(4):2065–2070. doi: 10.1073/pnas.041588998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh S, et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008;146(3):1368–1385. doi: 10.1104/pp.107.113738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen F, Bradford KJ. Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol. 2000;124(3):1265–1274. doi: 10.1104/pp.124.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrera E, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53(2):214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamauchi Y, et al. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16(2):367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graeber K, Linkies A, Wood ATA, Leubner-Metzger G. A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a Brassicaceae cross-species seed germination case study. Plant Cell. 2011;23(6):2045–2063. doi: 10.1105/tpc.111.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.