Significance
Factors influencing Wolbachia transfer into new species remain poorly understood. This is important as Wolbachia can influence speciation and is being developed as a novel arthropod-borne disease control approach. We show the native microbiota of Anopheles impede vertical transmission of Wolbachia. Antibiotic microbiome perturbation enables Wolbachia transmission in two Anopheles species. Mosquitoes with altered microbiomes do not exhibit blood meal-induced mortality associated with Wolbachia infection, suggesting that mosquitoes are killed by interactions between Wolbachia and other bacteria present in the mosquito. We identified Asaia as the bacterium responsible for inhibiting Wolbachia transmission, and partially responsible for blood meal-induced mortality. These results suggest that microbial interactions profoundly affect the host, and that microbiome incompatibility may influence distribution of Wolbachia in arthropods.
Keywords: holobiome, dysbiosis, competitive exclusion, microbe–microbe interactions, malaria
Abstract
Over evolutionary time, Wolbachia has been repeatedly transferred between host species contributing to the widespread distribution of the symbiont in arthropods. For novel infections to be maintained, Wolbachia must infect the female germ line after being acquired by horizontal transfer. Although mechanistic examples of horizontal transfer exist, there is a poor understanding of factors that lead to successful vertical maintenance of the acquired infection. Using Anopheles mosquitoes (which are naturally uninfected by Wolbachia) we demonstrate that the native mosquito microbiota is a major barrier to vertical transmission of a horizontally acquired Wolbachia infection. After injection into adult Anopheles gambiae, some strains of Wolbachia invade the germ line, but are poorly transmitted to the next generation. In Anopheles stephensi, Wolbachia infection elicited massive blood meal-induced mortality, preventing development of progeny. Manipulation of the mosquito microbiota by antibiotic treatment resulted in perfect maternal transmission at significantly elevated titers of the wAlbB Wolbachia strain in A. gambiae, and alleviated blood meal-induced mortality in A. stephensi enabling production of Wolbachia-infected offspring. Microbiome analysis using high-throughput sequencing identified that the bacterium Asaia was significantly reduced by antibiotic treatment in both mosquito species. Supplementation of an antibiotic-resistant mutant of Asaia to antibiotic-treated mosquitoes completely inhibited Wolbachia transmission and partly contributed to blood meal-induced mortality. These data suggest that the components of the native mosquito microbiota can impede Wolbachia transmission in Anopheles. Incompatibility between the microbiota and Wolbachia may in part explain why some hosts are uninfected by this endosymbiont in nature.
Bacteria in the genus Wolbachia are maternally transmitted Rickettsia-like endosymbionts that infect an estimated 40–69% of arthropod species (1, 2). In many cases, Wolbachia manipulate host reproduction to spread throughout arthropod populations (3). Incongruence between Wolbachia and host phylogenies indicate that horizontal transfer of the symbiont has been commonplace over evolutionary time (4, 5), enabling Wolbachia to invade new species. However, there is a poor understanding of barriers to horizontal transmission and why some species remain uninfected. An understanding of these factors is important from an evolutionary perspective given that Wolbachia influences speciation (6, 7), and from an applied perspective as Wolbachia is being transinfected into vector species for the control of arthropod-borne disease (8–10).
The ability to invade the host germ line is an important feature of Wolbachia biology that facilitates horizontal transmission, leading to the pervasive nature of this bacterium across invertebrate taxa. In order for Wolbachia to become established in a naïve host species, it must be acquired horizontally and successfully transmitted vertically (i.e., to offspring) to maintain the infection in the population. Multiple mechanisms of Wolbachia horizontal transmission have been proposed, including cohabitation, hemolymph transfer, predation, and parasitoid infection (11–15). After microinjection into Drosophila, Wolbachia infects the stem cell niches in the germ line (16, 17), and both Wolbachia-derived and host factors appear to influence tropism and bacterial density during oogenesis (17–20). Alternatively, somatic tissue may act as a reservoir for Wolbachia infection of the developing oocyte (20–23). Although pathways of horizontal transmission have been characterized in some species, identification of barriers to vertical transmission of the acquired Wolbachia infection remains elusive.
Microbial conflict or incompatibility within arthropods is a potential barrier to transmission of heritable symbionts. Studies in the tick Dermacentor variabilis demonstrate competitive exclusion between maternally inherited bacteria. Transovarial transmission of Rickettsia montanensis (formerly Rickettsia montana) and Rickettsia rhipicephali is inhibited by infection with the reciprocal species (24). Similarly, infection exclusion has been observed in D. variabilis between conspecific strains of Anaplasma marginale where one strain inhibits the infection of the other (25). Competitive inter- and intraspecific microbial interactions have also been observed with Wolbachia (26, 27).
Anopheles mosquitoes provide a unique system to examine microbial barriers to Wolbachia transmission. With few exceptions, Anophelines (which transmit the Plasmodium parasites that cause human malaria) are naturally uninfected with Wolbachia (28–31), suggesting the potential presence of innate barriers to infection in this genus. However, in vitro and in vivo studies indicate that Wolbachia are capable of infecting cultured Anopheles cells (32, 33), ex vivo cultured tissues (34), in vivo somatic tissue (35–37), and can stably infect the mosquito germ line (38). We investigated the ability of the native microbial community to influence vertical transmission of Wolbachia in Anopheles mosquitoes. We found that bacteria in the genus Asaia were responsible for inhibiting Wolbachia maternal transmission in this important mosquito genus.
Results
The wAlbB Wolbachia Strain Has a Greater Affinity for the Anopheles Germ Line Compared with wMelPop.
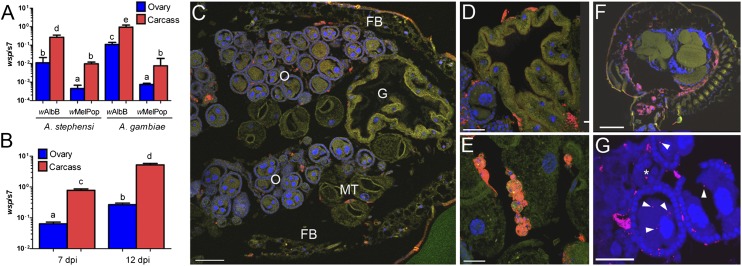
For Wolbachia to stably infect a novel insect species, the bacteria must colonize the germ line so they can be maternally transferred to progeny to perpetuate the infection. We investigated transfer of two Wolbachia strains (wMelPop from Drosophila melanogaster and wAlbB from Aedes albopictus) into Anopheles gambiae and Anopheles stephensi using adult intrathoracic microinjection. Although many transinfections have been accomplished using embryo microinjection (reviewed in ref. 9), this process may circumvent some barriers to vertical transmission (9). Injection of later life stages is more likely to resemble the process of horizontal transfer that occurs in nature (9, 11–15). After injection of the same number of Wolbachia bacteria into each mosquito, the wAlbB strain developed significantly higher germ-line titers compared with wMelPop, and invaded A. gambiae ovaries at significantly elevated titers compared with A. stephensi (Kruskal–Wallis, P < 0.05) (Fig. 1A). Similar results were observed for invasion of A. gambiae testes (Fig. S1). The carcass and ovarian density of wAlbB increased with time in A. gambiae (Kruskal–Wallis, P < 0.004), with ovarian density increase possibly due to replication within the ovary or more time to infect the germ line, or both (Fig. 1B). FISH was performed on mosquitoes to visualize Wolbachia in the germ line and other tissues. The wAlbB strain was observed to infect numerous tissues including hemocytes, gut, pericerebral fat body, and brain (Fig. 1 C–F), and was observed at low levels in the ovarian follicles of the mosquitoes, a location that is essential for maternal transmission (Fig. 1G). Subsequently, all further experiments were undertaken with the wAlbB strain.
Fig. 1.
Determining the optimal Wolbachia strain for vertical transmission in Anopheles mosquitoes. (A) Wolbachia density in microinjected mosquitoes assessed by qPCR at 7 dpi indicates the wAlbB strain infects Anopheles at significantly higher levels in both somatic and germ-line tissue. Approximately 9.3 × 106 bacterial cells were injected per mosquito. (B) The titer of wAlbB increases over time in A. gambiae somatic and germ-line tissue. Data for A and B were analyzed by Kruskal–Wallis test using the Dwass method for pairwise contrasts. The different letters (a–e) denote statistical significance [P < 0.05 in A and P < 0.004 in B]. Error bars in A and B represent SEM. (C) FISH performed on mosquitoes 17 dpi localizes the wAlbB infection in diverse tissues within the abdomen. O, ovaries; FB, fat body; G, gut; MT, malpighian tubule. Higher magnification of the (D) midgut, (E) hemocytes, (F) brain, and (G) ovarian follicles. The wAlbB strain infects the follicular epithelium of the oocyte and the nurse cells with the oocyte (white arrow heads in G). Wolbachia also infects the secondary follicles (asterisk in G). For ovarian follicles (G) mosquitoes were assessed 20 dpi. Red represents Wolbachia, blue the mosquito DNA, and green the mosquito tissue autofluorescence. (Scale bars: 300 μm in C and 60 μm in E–G.) FISH controls are available in Fig. S8.
Native Anopheles Microbiota Suppress Maternal Wolbachia Transmission.
To determine if microbial interactions influenced Wolbachia transmission in Anopheles, the microbiome of these mosquitoes was perturbed by antibiotic treatment. Wolbachia transmission was assessed in A. gambiae and A. stephensi mosquito lines reared continuously on a mixture of antibiotics (penicillin, streptomycin, gentamicin and kanamycin) and compared with a line reared on a conventional sugar diet. A. gambiae mosquitoes reared conventionally were capable of poorly transmitting the wAlbB strain of Wolbachia to their progeny, with only 10% of individuals being infected at very low titer. While fitness costs associated with Wolbachia infection in conventionally reared mosquitoes were evident in both species (affecting bloodfeeding and survival), they were extremely severe in A. stephensi and despite the high number of mosquitoes injected (n = 2680) no surviving adult offspring resulted from this line (Fig. 2). In stark contrast to conventionally reared mosquitoes, perfect vertical transmission (Fisher’s exact, P < 0.0001) at significantly higher titers (Mann–Whitney U test, P < 0.006) was seen in antibiotic-treated A. gambiae (Fig. 2), and offspring survived from antibiotic-treated A. stephensi, of which ∼90% were Wolbachia-infected (Fig. 2). Additionally, in A. stephensi both Wolbachia infection prevalence (Fisher’s exact test, P = 0.03) and titer (Mann–Whitney U test, P < 0.0001) could be significantly boosted by repeated injections over multiple generations into the antibiotic-treated line (Fig. S2).
Fig. 2.
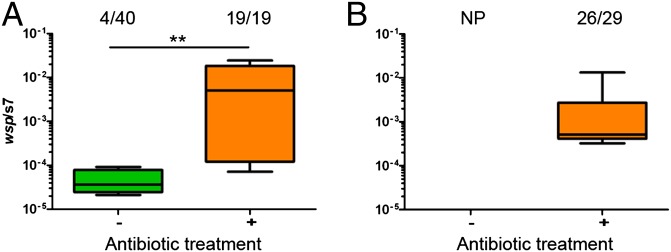
Antibiotic perturbation of the microbiome enables vertical transmission of Wolbachia in Anopheles mosquitoes. (A) In A. gambiae, Wolbachia is transmitted poorly to mosquitoes reared on conventional sugar (no antibiotics) but the infection frequency (Fisher’s exact test, P < 0.0001) and density (Mann–Whitney U test, **P < 0.006) increases significantly when mosquitoes are administered an antibiotic mixture. (B) In A. stephensi, no progeny (NP) were obtained from mosquitoes microinjected with Wolbachia reared on conventional sugar due to the severe fitness costs associated with infection. In mosquitoes administered an antibiotic mixture, Wolbachia was transmitted to 90% of offspring. Fractions represent the number of infected offspring over the total number. Green represents conventionally reared mosquitoes, whereas orange denotes antibiotic-treated mosquitos. Box and whiskers represent data quartiles and range, respectively.
Antibiotic Treatment Reduces Asaia Infection in Anopheles.
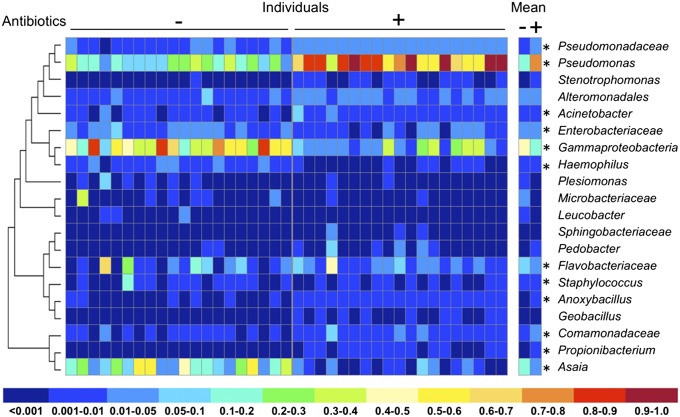
To determine the microbial composition of Anopheles lines reared on conventional sugar or antibiotics, we undertook high-throughput 16S amplicon sequencing for A. gambiae and A. stephensi (Fig. 3 and Figs. S3–S6). While there were species-specific alterations in microbiome composition due to antibiotic treatment (Fig. 3, Fig. S3, and Table S1), only operational taxonomic units (OTUs) belonging to the bacterial family enterobacteriaceae and the bacterial genus Asaia showed significantly reduced levels for both mosquito species [nonparametric t test with Monte Carlo simulation comparing OTU frequencies between treatments: A. gambiae (P < 0.01) and A. stephensi (P < 0.001) (Table S1)]. Asaia showed one of the largest changes in absolute number of reads for all significantly regulated taxon categories (Table S1). Reductions in Asaia titers were independently verified by quantitative PCR (qPCR) analysis (Fig. 4A).
Fig. 3.
Microbiome analysis of A. stephensi mosquitoes reared on conventional sugar (−) compared with those on an antibiotic mixture (+). OTUs were grouped by genus (where possible) or higher rank, and the relative abundance in individual samples calculated. The mean relative abundance per treatment is also shown. Asterisks denote presence of OTUs within that taxon grouping that significantly change in frequency (read count) between treatments (nonparametric t test with Monte Carlo simulation; see Table S1 for OTU-specific P values). A maximum likelihood phylogenetic tree was constructed based on the alignment of representative 75-nt OTU sequences for each taxonomic group. See Fig. S3 for a comparison of the relative abundance of bacterial taxa in A. gambiae.
Fig. 4.
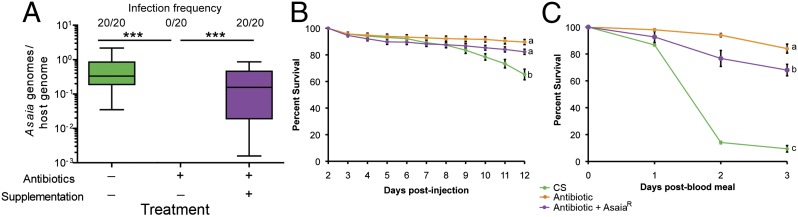
Asaia density and mosquito longevity in conventionally reared, antibiotic-treated, and Asaia-supplemented antibiotic-treated A. stephensi. (A) Levels of Asaia in control mosquitoes, antibiotic-treated mosquitoes, and antibiotic-treated mosquitoes supplemented with AsaiaR. Treating mosquitoes with an antibiotic mixture eliminates Asaia, whereas supplementation with AsaiaR in a sugar meal reestablishes the infection (ANOVA, ***P < 0.0001). Asaia levels in conventionally reared and AsaiaR supplemented mosquitoes are not significantly different (ANOVA, P = 0.06). Fractions represent the number of infected mosquitoes over the total number. Box and whiskers represent data quartiles and range, respectively. (B and C) Mosquito mortality trajectories after Wolbachia injection in mosquito lines pre- (B) and post-blood meal (C). Conventionally reared mosquitoes suffer slight fitness costs after injection and severe mortality after a blood meal, whereas antibiotic-treated mosquitoes do not suffer elevated mortality. Antibiotic-treated mosquitoes supplemented with AsaiaR exhibit a modest increase in mortality post-blood meal (ANOVA; letters a–c denote statistical significance P < 0.05). Green represents conventionally reared mosquitoes, orange the antibiotic-treated mosquitos, and purple the antibiotic-reared mosquitos supplemented with AsaiaR. Error bars in B and C represent SEM.
Microbiota–Wolbachia Interactions Kill Mosquitoes After a Blood Meal.
Asaia bacteria naturally infect the Anopheles germ line and are vertically transmitted (39, 40), and thus may conflict with other maternally transmitted bacteria. This, together with the observed significant and large decrease in frequency of this taxon upon antibiotic treatment in both mosquito species, led us to investigate Asaia as a candidate bacteria involved in the interference of Wolbachia transmission. To assess Wolbachia–Asaia interactions in Anopheles mosquitoes, an antibiotic-resistant mutant of Asaia (herein AsaiaR) was created and supplemented to mosquitoes reared on antibiotics to create a gnotobiotic line. The Asaia infection status of all lines was confirmed with qPCR; Asaia was not detected in the unsupplemented antibiotic-treated line and no significant difference was found between Asaia infection frequency (Fisher’s exact test, P = 1.0) or titer (ANOVA, P = 0.06) between conventionally reared and antibiotic-treated mosquitoes supplemented with AsaiaR (Fig. 4A). Given that we could not obtain offspring from the conventionally reared A. stephensi line, but could successfully raise progeny from the antibiotic-treated line post-Wolbachia injection (Fig. 2B), we assessed mortality trajectories of the three A. stephensi mosquito lines (conventionally reared, antibiotic treated, and antibiotic treated supplemented with AsaiaR) when infected with Wolbachia. Pre-blood meal, there was a modest but significant fitness cost of Wolbachia infection in conventionally reared A. stephensi compared with antibiotic-treated mosquitos or antibiotic-treated mosquitoes supplemented with AsaiaR (ANOVA, P < 0.0001; Fig. 4B). Post-blood meal, there was a dramatic cost to Wolbachia infection in conventionally reared mosquitoes, with massive mortality in mosquitoes observed 2 d post-blood meal. Antibiotic treatment completely alleviated this mortality. Antibiotic-treated mosquitoes supplemented with AsaiaR exhibited an intermediate mortality trajectory (ANOVA, P < 0.0001; Fig. 4C). These results indicate that interactions between Wolbachia and the native microbiota of A. stephensi induce blood meal mortality to which Asaia contributes in part, however the major microbial causative agent(s) of blood meal mortality have yet to be identified.
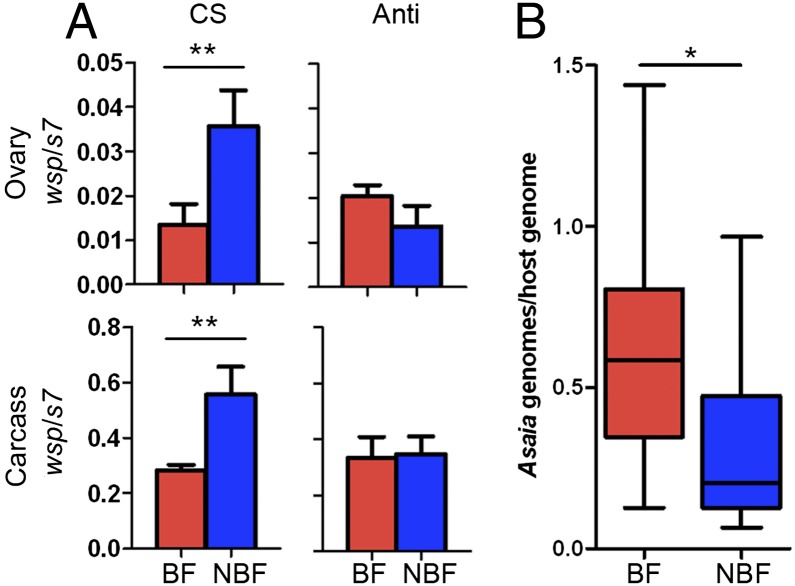
Microbiota Suppress Wolbachia Levels After a Blood Meal.
Blood feeding drastically influences microbial abundance and composition in mosquitoes (41–43). Given that we observed severe blood meal-associated fitness effects in Wolbachia-infected Anopheles, Wolbachia and Asaia levels were assessed in A. stephensi mosquitoes pre- and post-blood feeding. A blood meal significantly reduced Wolbachia titers in the ovary and carcass of mosquitoes that possessed their natural microbiota compared with non-blood fed individuals [Mann–Whitney U test, P < 0.001 (ovaries) and P < 0.01 (carcass); Fig. 5A]. However, reduction in Wolbachia titers was not observed in antibiotic-treated lines [Mann–Whitney U test, P = 0.18 (ovaries) and P = 0.93 (carcass); Fig. 5B]. In contrast to Wolbachia, Asaia levels were significantly elevated in mosquitoes post-blood meal (Mann–Whitney U test, P < 0.01; Fig. 5B). No evidence was found for antibiotic treatment influencing Wolbachia levels in the female germ line (Mann–Whitney U test, P = 0.1; Fig. S7).
Fig. 5.
Bacterial densities are modulated by blood feeding in A. stephensi. (A) By qPCR, Wolbachia levels in the ovary and the carcass significantly decrease after a blood meal in mosquitoes reared on conventional sugar (CS) [Mann–Whitney U test, **P = 0.0015 (ovary) and **P = 0.0178 (carcass)]. This reduction is abolished when mosquitoes are reared on an antibiotic mixture (Anti) [Mann–Whitney U test, P = 0.18 (ovary) and P = 0.93 (carcass)]. Error bars represent SEM. (B) In conventionally reared mosquitoes, blood feeding significantly increases Asaia levels compared with non-blood fed mosquitoes (Mann–Whitney U test, *P < 0.01). BF, blood fed (red); NBF, non-blood fed (blue). Box and whiskers represent data quartiles and range, respectively.
Asaia Inhibits Wolbachia Transmission in Anopheles.
To directly examine the effect of Asaia on Wolbachia transmission we compared transmission in antibiotic-reared A. stephensi lines with and without Asaia supplementation. In the antibiotic-treated line, Wolbachia was transmitted to the next generation with 83% of offspring infected. In contrast, Wolbachia transmission was completely abolished when Asaia was supplemented to female mosquitoes before injection (Mann–Whitney U test, P < 0.0001; Fig. 6).
Fig. 6.
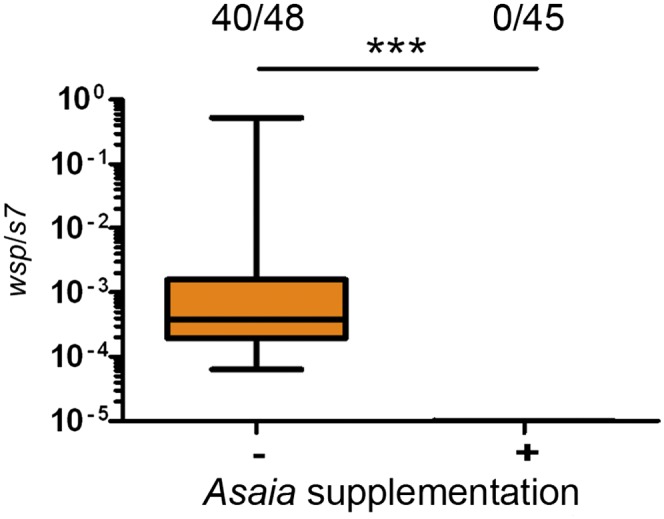
Asaia inhibits Wolbachia transmission in A. stephensi mosquitoes. Supplementing AsaiaR to antibiotic-treated A. stephensi abolishes Wolbachia transmission to offspring. Transmission is significantly different between nonsupplemented (orange) and supplemented mosquito lines that were reared on antibiotics (Fisher’s exact test, ***P < 0.0001). Fractions represent the number of infected offspring over the total number. Box and whiskers represent data quartiles and range, respectively.
Discussion
The microbiome affects the biology and physiology of the host across a broad range of eukaryotic taxa (44–46). In mammals, microbial interactions (particularly competitive exclusion) shape the composition of the microbiome (47). Similarly, the insect microbiome is important for host homeostasis, development, and immunity (48–50) and has recently been implicated to influence speciation (51). Although the microbiota of vector arthropods is known to be a potent modulator of pathogen vector competence (52, 53), little is known regarding bacterial interactions within these medically important insects. Our data suggest the native microbiota of Anopheles mosquitoes can impede transmission of the maternally inherited symbiont Wolbachia. Mosquitoes with an antibiotic-perturbed microbiome transmitted Wolbachia to their offspring and had reduced fitness costs associated with Wolbachia infection. In mosquitoes that possessed their native microbiota, decreases in Wolbachia transmission may be due to microbiome-induced reductions in Wolbachia titers post-blood meal, similar to what has been observed in naturally wFlu-infected Aedes fluviatilis mosquitoes (54).
Competitive exclusion between bacterial species can shape the composition of arthropod microbiomes (24–27). Using a combination of high-throughput sequencing and bacterial supplementation, we identified the bacterium Asaia as a specific agent inhibiting Wolbachia transmission in Anopheles. Recently, Wolbachia sequences were detected at low frequency in field populations of A. gambiae (55). High-throughput sequencing of the 16S rRNA gene amplified from the DNA extracted from the germ lines of 30 mating couples found one male that contained Wolbachia reads (55). Although lack of statistical power precludes definitive conclusions, examination of these data identified no Asaia sequence reads in the Wolbachia-infected individual, consistent with the hypothesis that Asaia acts competitively with Wolbachia infection in Anopheles. However, Asaia was also absent from other mosquitoes that were not infected with Wolbachia (55). A larger sample size is required to examine any correlation between Wolbachia and Asaia in potential natural A. gambiae infections. Our results provide a hypothesis to explain why the majority of Anopheles mosquitoes lack Wolbachia in nature, and more broadly, may in part explain the distribution of Wolbachia infections across taxa.
Strategies which exploit Asaia and Wolbachia are currently being devised for malaria control (38, 39). Further work is needed to determine if these approaches are complementary or incompatible, and whether the presence of Asaia in field populations will hinder the spread of Wolbachia into natural populations. Although a stable Wolbachia infection has been created in the Asian malaria mosquito A. stephensi (38), other important Anopheles vectors are yet to be transinfected with Wolbachia. Eliminating Asaia may facilitate the development of Wolbachia infections in these other vectors. The creation of Anopheles mosquito lines transinfected with a variety of Wolbachia strains will be worthwhile, given that some Anopheles–Wolbachia strain combinations may not be suitable for malaria control due to their propensity to increase certain Plasmodium parasites (36, 56, 57).
Materials and Methods
Wolbachia Culture and Extraction.
The wAlbB and wMelPop strains of Wolbachia were purified from infected A. gambiae Sua5B and Mos55 cells, respectively, according to previously published procedures (37). Purified Wolbachia were stained using the Live/Dead BacLight Bacterial Viability Kit (Invitrogen). Wolbachia were quantified using a hemocytometer under a fluorescence microscope (Olympus) to determine the density of viable cells.
Mosquito Rearing and Antibiotic Treatment.
A. gambiae (Keele strain) and A. stephensi (Liston strain) mosquitoes were reared on a conventional 10% (wt/vol) sugar diet and blood fed with an artificial feeder on expired human blood for reproduction. Antibiotic-treated lines of each mosquito species were reared with 10% sugar containing an antibiotic mixture of penicillin–streptomycin (10 units/mL, 10 µg/mL), gentamicin (15 µg/mL), and kanamycin (200 µg/mL) (58). Mosquito lines were continuously reared on the antibiotic mixture and all experiments in these lines were conducted after a minimum of three generations of antibiotic treatment. Wolbachia replication dynamics are not affected by kanamycin, penicillin or streptomycin, however Wolbachia is moderately susceptible to gentamicin (59). Therefore, gentamicin was removed from the antibiotic mixture before Wolbachia infection. Two to 5 d postemergence, mosquitoes were anesthetized on ice and injected with Wolbachia (∼107 bacteria per mosquito) according to a previously established methodology (37). Postinjection, mosquitoes were maintained at 28 °C with access to 10% sucrose (with or without antibiotics).
qPCR.
Ovaries were dissected from anesthetized mosquitoes under a dissecting microscope (Olympus) and DNA extracted either using QIAamp DNA Micro Kits (Qiagen) or DNeasy blood and tissue extraction kits (Qiagen) following the manufacturer’s protocols. qPCR was performed in triplicate on a Rotor Gene Q (Qiagen) using the Rotor Gene SYBR Green PCR Kit (Qiagen) according to Hughes et al. (35) and Wolbachia to host genome ratios were calculated using Qgene (60).
FISH.
Mosquitoes were fixed in acetone then embedded in paraffin wax and sectioned with a microtome. Slides were dewaxed with three successive xylene washes for 5 min, followed by 5-min washes with 100% ethanol then 95% ethanol before treatment with alcoholic bleach (6% H2O2 in 80% ethanol) for 3 d to minimize autofluorescence. FISH was completed as previously described (35). Images were captured with an LSM 510 META confocal microscope (Zeiss) and Olympus FV1000 Laser Scanning Confocal Microscope (Olympus). Images were processed using LSM image browsers (Zeiss), FV10-ASW Version 3.0 (Olympus), and Photoshop 7.0 (Adobe) software. Uninfected and no-probe controls were performed and are available in Fig. S8.
Microbiome Analysis.
The microbiomes of conventionally reared and antibiotic-treated Anopheles were characterized and compared using barcoded high-throughput amplicon sequencing of the bacterial 16S ribosomal gene. Whole conventionally reared A. gambiae (n = 5) and A. stephensi (n = 20), and antibiotic-treated A. gambiae (n = 5) and A. stephensi (n = 19) were surface sterilized by a wash in 20% bleach followed by three washes in 100% ethanol to reduce external contaminant bacteria. Total genomic DNA from whole mosquitoes was extracted following the Marriott extraction protocol (61), followed by washes and column elution for improved purity ratios (Powersoil DNA Elution kit; MO BIO). Initial preliminary sequencing was conducted on A. gambiae, where DNA samples underwent barcoded PCR of the 16S V3–V4 region followed by sequencing on the 454 Junior platform (Roche Inc). More in-depth sequencing to allow finer-scale changes to be detected was conducted on A. stephensi where DNA samples underwent barcoded PCR of the 16S V6 region followed by sequencing on the MiSeq platform (Illumina Inc.). For both species, primers and adaptors were subsequently removed from reads, the reads separated by sample barcode, and then the quality (minimum Q20) and length of the reads filtered. Sequences were clustered into OTUs at the 97% similarity level by reference-based picking with Quantitative Insights into Microbial Ecology (QIIME) 1.8.0 (62) implementation of UCLUST (63), against the 13_8 release of the Greengenes 16S database (64), with the remaining sequences clustered de novo. The QIIME default classifier UCLUST was used to assign taxonomy to OTUs. Only OTUs that contained more than five sequences were retained for subsequent analyses, reducing the likelihood of inclusion of OTUs formed through sequencing error. For beta diversity (between sample/treatment) measures, samples were standardized by randomly selecting a set number of sequences per sample. This value was chosen to equate that present in the sample with the lowest number of sequences (1,434 sequences for A. gambiae and 16,165 for A. stephensi), thus allowing inclusion of all samples but maximizing sequencing depth. For significance tests (nonparametric t test with Monte Carlo simulation) of OTU frequencies between treatments, OTUs were further filtered to remove low-frequency taxa. The filter threshold was taken as 1% of the mean sample sequence depth/read count (thresholds: 90 sequences for A. gambiae and 1,370 for A. stephensi). Microbial composition, rarefaction, diversity, and OTU frequency changes between treatments were analyzed using QIIME 1.8.0 pipelines. Asaia densities in all mosquito lines were corroborated by qPCR (n = 20) (39).
Wolbachia Transmission Experiments.
At 17–20 d post-Wolbachia injection, mosquitoes were provided with access to a blood meal and then given access to an oviposition site. For each experiment, both the treatment and control were injected with the same density of Wolbachia and blood fed at the same time postinjection. After mosquitoes oviposited, eggs were washed once with 10% bleach and three times with water, hatched in 1.5-L trays, and fed dried fish food. Emerging mosquitoes were collected and assayed by qPCR for Wolbachia. Transmission assays on antibiotic-treated mosquitoes were replicated at least twice. A kanamycin-resistant Asaia mutant was created by culturing an A. stephensi Asaia isolate on kanamycin (100 μg/mL) Gly media (25 g/L glycerol, 10 g/L yeast extract). The resistant Asaia was cultured in Gly media (kanamycin 100 μg/mL) for 2 d at 30 °C, supplemented to mosquito larvae (1 × 106), and provided to adults in the sugar. Mosquitoes were reared on 10% sugar containing an antibiotic mixture of penicillin–streptomycin (10 units/mL, 10 µg/mL) and kanamycin (200 µg/mL).
Mortality Assessment.
The mortality dynamics of mosquitoes pre- and post-blood meal were conducted as previously described (35, 36). wAlbB was injected into mosquitoes reared on conventional sugar, reared continuously on antibiotics, or reared continuously on antibiotics but supplemented with AsaiaR as described above. After Wolbachia injection, mosquitoes were placed into cups (60 per cup) with 6–10 replicate cups per treatment. Mortality of mosquitoes was monitored every 24 h for 12 d. After 12 d, mosquitoes were given a blood meal through a membrane feeder, and blood fed mosquitoes were sorted into new cups (30 mosquitoes per cup, 5 replicate cups per treatment), and mortality assayed for 3 d. Data were analyzed by ANOVA with Tukey’s correction for multiple comparisons.
Ovary Titer Analysis Pre- and Post-Blood Meal.
Wolbachia-injected A. stephensi mosquitoes were either given a blood meal as described above at 5 d postinjection (dpi) or continuously reared on 10% sucrose. At 8 dpi ovaries were dissected from both blood-fed and sugar-fed mosquitoes (n = 10). DNA was extracted from ovaries and carcasses and used as the template for qPCR as described above. To determine the effect of the microbiota on Wolbachia ovary titers after a blood meal, experiments were repeated with antibiotics supplemented in the 10% sucrose solution (penicillin–streptomycin and kanamycin at previously described concentrations). Data were analyzed by Mann–Whitney U test. All experiments were replicated twice.
Supplementary Material
Acknowledgments
We thank John Gibas, John Cantolina, and Missy Hazen for assistance with sectioning and confocal microscopy; Ping Xue for assistance with experiments; Dr. David Hughes for commenting on drafts of the manuscript; and especially Drs. Flaminia Catteruccia and Nicola Segata for kindly providing data on Asaia 16S sequence read frequency from ref. 55. We also thank the Microscopy and Cytometry Facility at the Huck Institutes of the Life Sciences and Penn State University. This research was supported by National Institutes of Health Grants R01AI067371, R21AI070178, and R21AI111175 (to J.L.R.); and also funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408888111/-/DCSupplemental.
References
- 1.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS ONE. 2012;7(6):e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol Lett. 2008;281(2):215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6(10):741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 4.O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89(7):2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: Reproductive parasites of arthropods. Proc Biol Sci. 1995;261(1360):55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 6.Miller WJ, Ehrman L, Schneider D. Infectious speciation revisited: Impact of symbiont-depletion on female fitness and mating behavior of Drosophila paulistorum. PLoS Pathog. 2010;6(12):e1001214. doi: 10.1371/journal.ppat.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telschow A, Flor M, Kobayashi Y, Hammerstein P, Werren JH. Wolbachia-induced unidirectional cytoplasmic incompatibility and speciation: Mainland-island model. PLoS ONE. 2007;2(8):e701. doi: 10.1371/journal.pone.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes GL, Rasgon JL. Transinfection: A method to investigate Wolbachia-host interactions and control arthropod-borne disease. Insect Mol Biol. 2014;23(2):141–151. doi: 10.1111/imb.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourtzis K, et al. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop. 2014;132(Suppl):S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 10.McGraw EA, O’Neill SL. Beyond insecticides: New thinking on an ancient problem. Nat Rev Microbiol. 2013;11(3):181–193. doi: 10.1038/nrmicro2968. [DOI] [PubMed] [Google Scholar]
- 11.Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc Biol Sci. 2004;271(1538):509–515. doi: 10.1098/rspb.2003.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaud T, Juchault P. Success and failure of horizontal transfers of feminizing Wolbachia endosymbionts in woodlice. J Evol Biol. 1995;8(2):249–255. [Google Scholar]
- 13.Vavre F, Fleury F, Lepetit D, Fouillet P, Boulétreau M. Phylogenetic evidence for horizontal transmission of Wolbachia in host-parasitoid associations. Mol Biol Evol. 1999;16(12):1711–1723. doi: 10.1093/oxfordjournals.molbev.a026084. [DOI] [PubMed] [Google Scholar]
- 14.Le Clec’h W, et al. Cannibalism and predation as paths for horizontal passage of Wolbachia between terrestrial isopods. PLoS ONE. 2013;8(4):e60232. doi: 10.1371/journal.pone.0060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath BD, Butcher RD, Whitfield WG, Hubbard SF. Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol. 1999;9(6):313–316. doi: 10.1016/s0960-9822(99)80139-0. [DOI] [PubMed] [Google Scholar]
- 16.Frydman HM, Li JM, Robson DN, Wieschaus E. Somatic stem cell niche tropism in Wolbachia. Nature. 2006;441(7092):509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- 17.Fast EM, et al. Wolbachia enhance Drosophila stem cell proliferation and target the germline stem cell niche. Science. 2011;334(6058):990–992. doi: 10.1126/science.1209609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serbus LR, et al. A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J Cell Sci. 2011;124(Pt 24):4299–4308. doi: 10.1242/jcs.092510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serbus LR, Sullivan W. A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog. 2007;3(12):e190. doi: 10.1371/journal.ppat.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toomey ME, Panaram K, Fast EM, Beatty C, Frydman HM. Evolutionarily conserved Wolbachia-encoded factors control pattern of stem-cell niche tropism in Drosophila ovaries and favor infection. Proc Natl Acad Sci USA. 2013;110(26):10788–10793. doi: 10.1073/pnas.1301524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacchi L, et al. Bacteriocyte-like cells harbour Wolbachia in the ovary of Drosophila melanogaster (Insecta, Diptera) and Zyginidia pullula (Insecta, Hemiptera) Tissue Cell. 2010;42(5):328–333. doi: 10.1016/j.tice.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Genty L-M, Bouchon D, Raimond M, Bertaux J. Wolbachia infect ovaries in the course of their maturation: Last minute passengers and priority travellers? PLoS ONE. 2014;9(4):e94577. doi: 10.1371/journal.pone.0094577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer K, Beatty WL, Jiang D, Weil GJ, Fischer PU. Tissue and stage-specific distribution of Wolbachia in Brugia malayi. PLoS Negl Trop Dis. 2011;5(5):e1174. doi: 10.1371/journal.pntd.0001174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol. 2002;39(6):809–813. doi: 10.1603/0022-2585-39.6.809. [DOI] [PubMed] [Google Scholar]
- 25.de la Fuente J, Blouin EF, Kocan KM. Infection exclusion of the rickettsial pathogen anaplasma marginale in the tick vector Dermacentor variabilis. Clin Diagn Lab Immunol. 2003;10(1):182–184. doi: 10.1128/CDLI.10.1.182-184.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto S, Anbutsu H, Fukatsu T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl Environ Microbiol. 2006;72(7):4805–4810. doi: 10.1128/AEM.00416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo N, Shimada M, Fukatsu T. Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol Lett. 2005;1(4):488–491. doi: 10.1098/rsbl.2005.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci I, Cancrini G, Gabrielli S, D’Amelio S, Favi G. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): Large polymerase chain reaction survey and new identifications. J Med Entomol. 2002;39(4):562–567. doi: 10.1603/0022-2585-39.4.562. [DOI] [PubMed] [Google Scholar]
- 29.Rasgon JL, Scott TW. An initial survey for Wolbachia (Rickettsiales: Rickettsiaceae) infections in selected California mosquitoes (Diptera: Culicidae) J Med Entomol. 2004;41(2):255–257. doi: 10.1603/0022-2585-41.2.255. [DOI] [PubMed] [Google Scholar]
- 30.Kittayapong P, Baisley KJ, Baimai V, O’Neill SL. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae) J Med Entomol. 2000;37(3):340–345. doi: 10.1093/jmedent/37.3.340. [DOI] [PubMed] [Google Scholar]
- 31.Osei-Poku J, Han C, Mbogo CM, Jiggins FM. Identification of Wolbachia strains in mosquito disease vectors. PLoS ONE. 2012;7(11):e49922. doi: 10.1371/journal.pone.0049922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes GL, et al. Wolbachia infections in Anopheles gambiae cells: Transcriptomic characterization of a novel host-symbiont interaction. PLoS Pathog. 2011;7(2):e1001296. doi: 10.1371/journal.ppat.1001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasgon JL, Ren X, Petridis M. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl Environ Microbiol. 2006;72(12):7718–7722. doi: 10.1128/AEM.01578-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes GL, Pike AD, Xue P, Rasgon JL. Invasion of Wolbachia into Anopheles and other insect germlines in an ex vivo organ culture system. PLoS ONE. 2012;7(4):e36277. doi: 10.1371/journal.pone.0036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7(5):e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes GL, Vega-Rodriguez J, Xue P, Rasgon JL. Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Appl Environ Microbiol. 2012;78(5):1491–1495. doi: 10.1128/AEM.06751-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin C, Ren X, Rasgon JL. The virulent Wolbachia strain wMelPop efficiently establishes somatic infections in the malaria vector Anopheles gambiae. Appl Environ Microbiol. 2009;75(10):3373–3376. doi: 10.1128/AEM.00207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bian G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340(6133):748–751. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 39.Favia G, et al. Bacteria of the genus Asaia stably associate with Anopheles stephensi, an Asian malarial mosquito vector. Proc Natl Acad Sci USA. 2007;104(21):9047–9051. doi: 10.1073/pnas.0610451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Damiani C, et al. Paternal transmission of symbiotic bacteria in malaria vectors. Curr Biol. 2008;18(23):R1087–R1088. doi: 10.1016/j.cub.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Gilbreath TM, 3rd, Kukutla P, Yan G, Xu J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE. 2011;6(9):e24767. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira JHM, et al. Blood meal-derived heme decreases ROS levels in the midgut of Aedes aegypti and allows proliferation of intestinal microbiota. PLoS Pathog. 2011;7(3):e1001320. doi: 10.1371/journal.ppat.1001320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327(5973):1644–1648. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 45.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 46.Broderick NA, Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes. 2012;3(4):307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13(11):790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Douglas AE. Lessons from studying insect symbioses. Cell Host Microbe. 2011;10(4):359–367. doi: 10.1016/j.chom.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6:146. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37(5):699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 51.Brucker RM, Bordenstein SR. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science. 2013;341(6146):667–669. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 52.Weiss B, Aksoy S. Microbiome influences on insect host vector competence. Trends Parasitol. 2011;27(11):514–522. doi: 10.1016/j.pt.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10(4):307–310. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baton LA, Pacidônio EC, Gonçalves DS, Moreira LA. wFlu: Characterization and evaluation of a native Wolbachia from the mosquito Aedes fluviatilis as a potential vector control agent. PLoS ONE. 2013;8(3):e59619. doi: 10.1371/journal.pone.0059619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baldini F, et al. Evidence of natural Wolbachia infections in field populations of Anopheles gambiae. Nat Commun. 2014;5:3985. doi: 10.1038/ncomms4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murdock CC, Blanford S, Hughes GL, Rasgon JL, Thomas MB. Temperature alters Plasmodium blocking by Wolbachia. Sci Rep. 2014;4:3932. doi: 10.1038/srep03932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hughes GL, Rivero A, Rasgon JL. Enhancement of vector-borne pathogen transmission by Wolbachia infection: An issue for malaria control? PLoS Pathog. 2014 doi: 10.1371/journal.ppat.1004182. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5(5):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fenollar F, Maurin M, Raoult D. Wolbachia pipientis growth kinetics and susceptibilities to 13 antibiotics determined by immunofluorescence staining and real-time PCR. Antimicrob Agents Chemother. 2003;47(5):1665–1671. doi: 10.1128/AAC.47.5.1665-1671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joehanes R, Nelson JC. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics. 2008;24(23):2788–2789. doi: 10.1093/bioinformatics/btn523. [DOI] [PubMed] [Google Scholar]
- 61.Post RJ, Flook PK, Millest AL. Methods for the preservation of insects for DNA studies. Biochem Syst Ecol. 1993;21(1):85–92. [Google Scholar]
- 62.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 64.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.