Abstract
We developed a cell-culture/biosensor platform consisting of aptamer-modified Au electrodes integrated with reconfigurable microfluidics for monitoring of transforming growth factor-beta 1 (TGF-β1), an important inflammatory and pro-fibrotic cytokine. Aptamers were thiolated, labeled with redox reporters, and self-assembled on gold surfaces. The biosensor was determined to be specific for TGF-β1 with an experimental detection limit of 1 ng/mL and linear range extending to 250 ng/mL. Upon determining figures of merit, aptasensor was miniaturized and integrated with human hepatic stellate cells inside microfluidic devices. Reconfigurable microfluidics were developed to ensure that seeding of “sticky” stromal cells did not foul the electrode and compromise sensor performance. This microsystem with integrated aptasensors was used to monitor TGF-β1 release from activated stellate cells over the course of 20 h. The electrochemical response went down upon infusing anti-TGF-β1 antibodies into the microfluidic devices containing activated stellate cells. To further validate aptasensor responses, stellate cells were stained for markers of activation (e.g., alpha smooth muscle actin) and were also tested for presence of TGF-β1 using enzyme linked immunosorbent assay (ELISA). Given the importance of TGF-β1 as a fibrogenic signal, a microsystem with integrated biosensors for local and continuous detection of TGF-β1 may prove to be an important tool to study fibrosis of the liver and other organs.
The liver is at the center of body’s metabolism, and its injury by toxicants or infections is the main cause of several diseases such as cirrhosis, fatty liver, hepatitis, jaundice, and liver cancer.1,2 Liver fibrosis is an inflammatory condition that is present during liver injury, cancer, or infection.3 Transforming growth factor-beta 1 (TGF-β1) is an important factor associated with fibrosis of the liver and other organs.4 In the liver, TGF-β1 is secreted by the activated hepatic stellate (stromal) cells, causing stellate cells to begin aberrant production of extracellular matrix proteins and leading to loss of differentiated hepatic phenotype.5−7
Given that TGF-β1 is a key molecular trigger of fibrosis and liver injury, it is important to know how fast it appears and what its dynamics are over the course of injury or insult. Immunoassays traditionally used for detection of signaling molecules such as TGF-β are limiting when it comes to determining secretion dynamics. We wanted to leverage aptamer-based biosensors for continuous monitoring of TGF-β1 secreted by liver cells. These aptasensors are based on the concept of structure switching pioneered by Plaxco and co-workers.8,9 Our lab has been interested in placing aptasensors at the site of cells for local, sensitive, and continuous detection of secreted molecules.10−12 Our focus has previously been on detecting inflammatory cytokines secreted from immune cells.11,12 In this paper, we wanted to develop an aptasensor for monitoring activation and TGF-β1 release from hepatic stellate cells. The aptamer was based on the DNA sequence described in the literature.13 Unlike our previous work with anchorage independent immune cells, stellate cells are quite adhesive, capable of attaching to and fouling the electrode surfaces. To remedy this, a reconfigurable microfluidic device was developed to allow for lowering of a microstructured poly(dimethylsiloxane) (PDMS) membrane to protect the electrodes during stellate cells seeding and for raising during cell detection experiments.
The miniaturized aptasensor was constructed by immobilizing methylene blue (MB)-tagged TGF-β1 aptamer13 on top of Au electrodes placed inside of microfluidic devices. Stellate cells were cultivated inside microfluidic devices next to sensing electrodes. The aptamer immobilized electrodes were protected with PDMS microcups in order to avoid nonspecific attachment of cells during seeding. The cells were then activated by infusion of platelet-derived growth factor (PDGF). Onset and progression of TGF-β1 release was monitored using square wave voltammetry (SWV) over the course of 20 h. This TGF-β1 sensor provides highly specific and sensitive detection. The PDGF activation of stellate cells was verified with immunostaining and enzyme linked immunosorbent assay (ELISA).
Materials and Methods
Chemicals and Reagents
Chromium (CR-4S) and gold etchants (Au-5) were purchased from Cyantek Corporation (Fremont, CA). Positive photoresist (S1813) and its developer solution (MF-319) were bought from Shipley (Marlborough, MA). Poly(dimethylsiloxane) (PDMS) and silicone elastomer curing agent were purchased from Dow Corning (Midland, MI). Amine functionalized thiolated transforming growth factor (TGF)-β1 DNA aptamer (MW 23 689.9) was purchased from Integrated DNA Technologies, USA. Recombinant human TGF-β1, platelet-derived growth factor (PDGF), 6-mercapto-1-hexanol (MCH), triton-X 100, bovine serum albumin (BSA), tris(2-carboxyethyl)phosphine hydrochloride (TCEP), sodium bicarbonate (NaHCO3), collagen (Type I), and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) were purchased from Sigma-Aldrich, USA. Methylene blue (MB), carboxylic acid, and succinimidyl ester (MB-NHS) (MW 598.12) was received from Biosearch Technologies, Inc. (Novato, CA). Dulbecco’s modified Eagle’s medium (DMEM), sodium pyruvate, fetal bovine serum (FBS), and penicillin/streptomycin (PS) were purchased from Invitrogen (Carlsbad, CA, USA). Paraformaldehyde was purchased from Electron Microscopy Sciences, USA. Anti-α-smooth muscle actin (α-SMA) and goat antirabbit IgG conjugated with Alexa-488 were obtained from Abcam and Invitrogen, respectively. 4,6-Diamidino-2-phenylindole (DAPI) was from Molecular Probes, Invitrogen. All other chemicals were used without further purification. The ELISA kit was obtained from R&D Systems, USA.
In the present study, we utilized
a TGF-β1-binding aptamer (IDT Technologies, San Diego, CA) having
a loop structure with amino functionality at the 5′ and thiol
functionality at the 3′ end.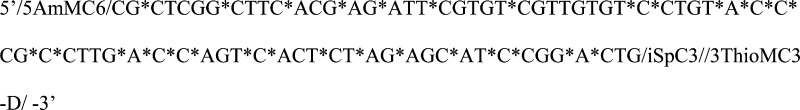 The backbone
of single-stranded DNA aptamers was modified
to have phosphorothioates (represented by “∗”
in the aptamer sequence) on 5′ of both A and C. This modification
is believed to provide enhanced nuclease resistance as well as higher
affinity than that of a phosphate counterpart.13 The aptamer stock solution was made in 40 mM HEPES buffer
containing 100 mM NaCl, 5 mM KCl, and 5 mM MgCl2.
The backbone
of single-stranded DNA aptamers was modified
to have phosphorothioates (represented by “∗”
in the aptamer sequence) on 5′ of both A and C. This modification
is believed to provide enhanced nuclease resistance as well as higher
affinity than that of a phosphate counterpart.13 The aptamer stock solution was made in 40 mM HEPES buffer
containing 100 mM NaCl, 5 mM KCl, and 5 mM MgCl2.
Functionalization and Immobilization of Aptamers
To fabricate an electrochemical biosensor, the TGF-β1 aptamer was tagged with MB, an electroactive redox label. NHS-labeled MB was conjugated to the 5′-end of amino-modified TGF-β1 aptamer through the succinimide ester coupling reported previously.9,14 Prior to modification of the Au electrodes, aptamer stock solution was reduced in 10 mM TCEP for 1 h to cleave disulfide bonds. This solution was then diluted in HEPES buffer to achieve the desired aptamer concentration (1 μM, the optimal concentration for TGF-β1 binding). For immobilization of aptamer onto the microfabricated Au electrodes, 1 μM MB-tagged TGF-β1 aptamer solution was infused into the working channels of the microfluidic device followed by incubation for about 18 h at 4 °C in the dark. Following incubation, the channels were rinsed with copious amounts of deionized water and HEPES and then incubated with 1 mM MCH for 15 min to displace nonspecifically adsorbed aptamer molecules and to passivate the electrode surface.
Design and Fabrication of Microfluidic Device with Integrated Electrodes
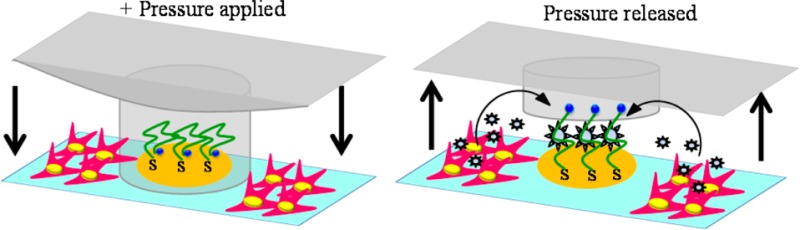
In order to avoid nonspecific attachment of the stellate cell to the sensing electrodes during cell seeding, we designed a reconfigurable microfluidic device of the kind shown in Scheme 1A. This device consisted of two PDMS layers assembled onto a glass slide with micropatterned Au electrodes. The micropatterned Au arrays of eight electrodes of diameter 300 μM were prepared using standard photolithography and a metal-etching process.15,16 One PDMS layer contained a linear channel with length × width × height dimensions of 15 mm × 1.7 mm × 0.075 mm. This was the control layer used for actuation of the working microfluidic layer. The latter consisted of eight microchannels, each with length × width × height dimensions of 6 mm × 1.2 mm × 0.075 mm. Each microchannel contained four microcups with inner and outer diameters of 500 and 700 μm, respectively, and the height of 0.060 mm. The device assembly steps included integration of the control PDMS layer (thickness ∼5 mm) on another thin PDMS layer (thickness ∼1 mm) containing eight microchannels with microcups followed by assembling the joint layers of PDMS with microfabricated Au electrodes. The layers of the device were bound together and to the substrate by O2 plasma treatment. Alignment of each layer during the device assembly was checked optically under the microscope. The device was left for 15 min at room temperature and then filled with deionized water to remove the bubbles. The tubing was attached in the control layer, and glass cylinders (diameter 8 mm) were placed at the inlets of the device using 10:1 ratio of PDMS and curing agent followed by baking for 20 min at 70 °C. This device was utilized to culture stellate cells next to the aptasensing electrodes preventing nonspecific attachment of cells on the sensor by lowering and raising the PDMS microcups (Scheme 1B).
Scheme 1. (A) Microfluidic Device Was Composed of Three Layers: Glass Slide with Micropatterned Au Electrodes, PDMS Layer with Fluid Channels and Microcups, and Another PDMS Layer for Controlling of Microcups. (B) Diagram Showing Actuation of Microcups to Protect Electrodes during Collagen Coating and Cell Seeding into the Channel.
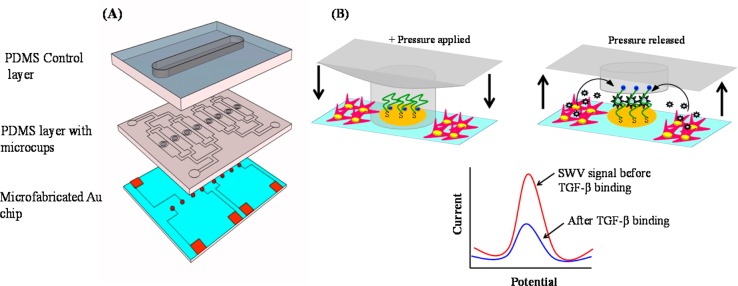
Once cells are activated and microcups are raised, the electrodes may be used for detection of TGF-β1. Redox labeled aptmer molecules immobilized on the electrode interact with TGF-β1, leading to a change in redox current. The redox current is decreased when TGF-β1 binds.
Cultivation of Stellate Cells inside Sensing Microfluidic Devices
A human hepatic stellate cell line (LX2) was maintained in DMEM supplemented with 0.5% FBS, 200 units/mL penicillin, and 200 μg/mL streptomycin at 37 °C in a humidified 5% CO2 atmosphere. Cells were cultured in a tissue culture flask (growth area of 75 cm2) until 90% confluence and then passaged. For cell seeding experiments, aptamer functionalized working Au electrodes were first protected with PDMS microcups by applying positive pressure through the control layer and HEPES buffer was flowed into the microchannels. Subsequently, 0.1 mg/mL solution of collagen (type I) was infused into the channels and kept at room temperature for about 1 h to adsorb collagen on the glass surface around aptasensing electrodes. The channels were again washed with buffer and sterilized under UV. Stellate cells, resuspended at a concentration of 1.6 × 106 cells/mL in DMEM described above, were infused into a microfluidic device and incubated at 37 °C.
Characterization of TGF-β1 Aptasensor Using Electrochemistry and Surface Plasmon Resonance
In order to obtain the optimal TGF-β1 aptasensor, we characterized aptamer immobilization and target binding via a surface plasmon resonance (SPR) flow through system (Biosensing Instruments, USA). In all the SPR experiments, the system was purged with the running buffer solution (HEPES) to set and stabilize an initial baseline. Subsequently, TGF-β1 aptamer was loaded at the flow rate of 40 μL/min in the SPR flow system and allowed to interact with the Au surface. At the completion of binding, the surface was washed with running buffer and the final baseline was recorded. The SPR angle (m°) difference (represented by the y-axis in Figure S1, Supporting Information) between the final and initial baselines in the binding curve corresponds to the change in refractive index due to bound TGF-β1aptamer. The aptamer modified SPR surface was challenged with a series of recombinant TGF-β1 concentrations to find the binding affinity of TGF-β1 aptamer with recombinant TGF-β1.
Additional initial optimization studies to find out the response and specificity of TGF-β1 aptamer were carried out by performing square wave voltammetry (SWV) using a potentiostat (CHI instruments, model 842B) at a frequency of 60 Hz over the potential range from 0 to −0.50 V. Au-coated Si substrates were incubated with TGF-β1 aptamer (1 μM) and were then placed into a homemade electrochemical cell creating an electrode area of 1.13 cm2 (Figure S2, Supporting Information). Pt wire counter electrode and Ag/AgCl reference electrode were immersed into the electrolyte solution to complete the three-electrode cell. The aptasensors were also challenged with nonspecific proteins such as IgG, BSA, IL-2, and IFN-γ as well as the analyte of interest TGF-β1. Responses to other isoforms of TGF-β were also studied.
Electrochemical Sensing of TGF-β1 Release from Stellate Cells within the Microfluidic Device
The aptamer functionalized microfluidic device was calibrated by infusing recombinant TGF-β1 ranging in concentration from 0.5 to 300 ng/mL in cell culture media (DMEM supplemented with 0.5% FBS and 1% PS). For cell experiments, miniaturized aptasensing electrodes were protected by lowering down the PDMS microcups (Scheme 1B) via application of positive pressure created in the control layer (filled with water and clamped). The hepatic stellate cells were cultured around aptasensing electrodes inside the microchannels of the device. To stimulate cells for TGF-β1 secretion, mitogenic solution, consisting of 20 ng/mL PDGF spiked in DMEM, was injected into the microchannels. After activation of cells, pressure was released from the control layer raising PDMS cups and exposing the aptasenor to the cytokines released from the activated stellate cells (Scheme 1B). Real-time SWV measurements were performed for up to 20 h to detect the TGF-β1 release from cells by the aptasensor. For the microfluidic electrochemical set up, we used a flow-through Ag/AgCl reference electrode and Pt wire counter electrode connected to the outlet and inlet of the fluid system. A homemade switching system was employed to sample individual electrodes at predefined time intervals. During the electrochemical experiment, the device was kept in a custom-designed environmental box with 5% CO2 and temperature of 37 °C. As shown in Scheme 1, our device contains 8 individually addressable electrodes, one electrode per fluidic channel. However, four microchannels are connected to one inlet and one outlet. We used one set of four channels to measure the TGF-β1 release from PDGF treated stellate cells, whereas the other set of four channels was used to monitor cytokine production in nontreated cells.
Verifying Activation of Stellate Cells by Immunofluorescent Staining and ELISA
Hepatic stellate cells in four of the microfluidic channels were stimulated with 20 ng/mL PDGF for 24 h while the cells in the other four channels were left unstimulated. Cells inside the microfluidic devices were then fixed in 4% paraformaldehyde (Electron Microscopy Sciences) + 0.3% Triton-X100 (Sigma) in PBS for 15 min followed by incubation in blocking solution (1% bovine serum albumin (BSA) in PBS) for 1 h. The channels were washed several times with PBS and exposed to rabbit anti-α-smooth muscle actin (α-SMA, Abcam) antibody for 90 min. The samples were again washed for 5 min and incubated with goat antirabbit IgG conjugated with Alexa-488 (diluted 1:1000) (Invitrogen) for 1 h. After a 5 min washing with PBS, samples were incubated with 4,6-diamidino-2-phenylindole (DAPI) for 15 min. All incubations were performed at room temperature. Fluorescent images were captured by a NIKON Eclipse Ti.
TGF-β1 ELISA was performed using the ELISA kit obtained from R&D Systems. 1.6 × 106 cells/mL stellate cells were seeded in each well of the collagen-coated 6-well plate using the same method used for the microfluidic device. Half of the wells were treated with 20 ng/mL PDGF, and the rest were left with nonstimulated DMEM. Samples were collected from each well after 48 h of PDGF treatment. ELISA was conducted in a 96-well plate using the protocol provided by R&D Systems.
Results and Discussion
The goal of this paper was to develop an electrochemical aptasensor for detection of TGF-β1, an important inflammatory and fibrogenic cytokine. A biosensor specific and sensitive for TGF-β1 was developed, miniaturized, and integrated with microfluidics. This sensing microsystem was then employed to monitor the TGF-β1 release from stellate cells in the process of activation.
SPR and Electrochemical Analysis of Aptamer-TGF-β1 Interactions
In order to arrive at the ideal TGF-β1 aptasensor, we characterized aptamer immobilization and target binding via SPR studies (Supporting Information). These investigations showed 1 μM to be an optimal aptamer concentration for target binding. To determine the binding affinity of TGF-β1 aptamer with recombinant TGF-β1, the aptamer-modified SPR surface was challenged with a series of TGF-β1 concentrations (Figure S1, Supporting Information). Kinetic analysis of the obtained binding data was done via the “Scrubber” software package provided with the SPR instrument to determine the equilibrium dissociation constant (Kd). The value of Kd for aptamer–target binding was found to be 1.07 nM. This data analysis utilizes SPR angle shift, due to the change in refractive index, as a response unit to quantify the binding of macromolecules at the sensor surface.
The surface density (Ntot) of 1 μM TGF-β1 apatmer on the Au surface was determined to be 3.83 × 1012 molecules/cm2 via electrochemistry utilizing eq 1.17
| 1 |
where Iavg(E0) is the average peak current in a voltammogram, n is the number of electrons transferred per redox event (with MB label n = 2), F is the Faraday current, R is the universal gas constant, T is the temperature, E is the peak amplitude, and f is the frequency.
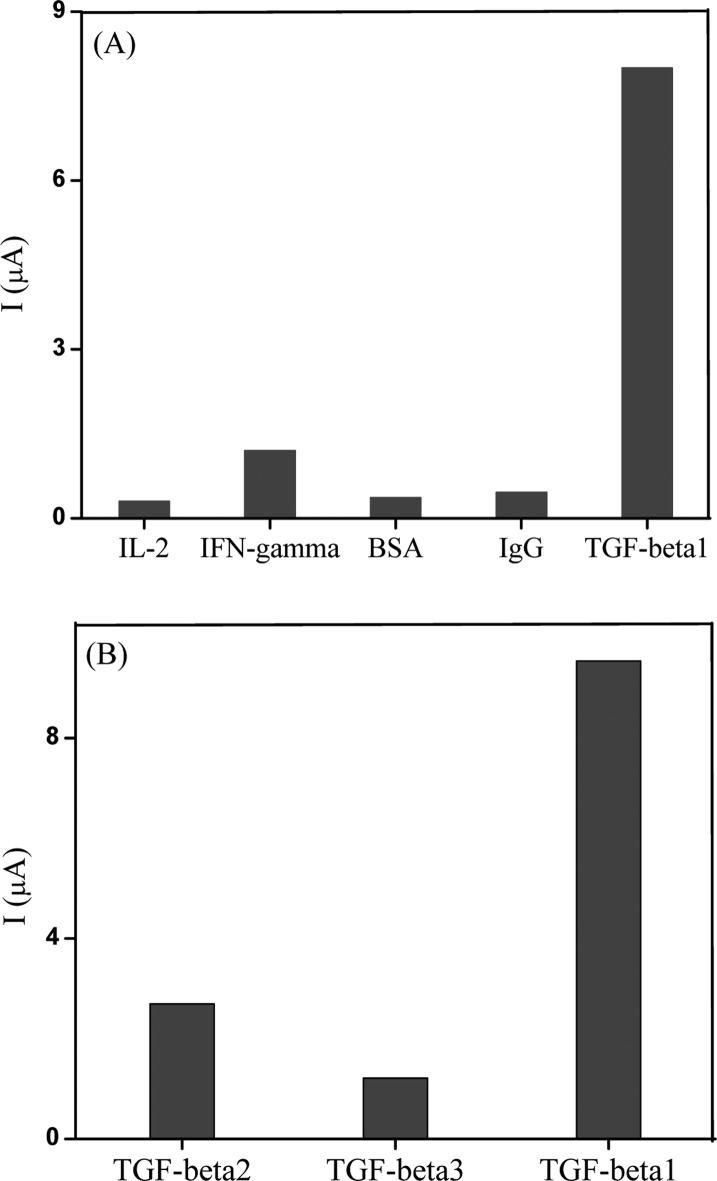
Additional studies to determine specificity of TGF-β1 aptamer were carried out using electrochemistry. Redox-labeled aptamer molecules were assembled on Au electrodes and loaded into a homemade electrochemical cell as described in the previous section of the manuscript. These electrodes were challenged with 50 ng/mL concentrations of IgG, BSA, IL-2, IFN-γ, and TGF-β1. Figure 1A shows that aptasensor response to nonspecific proteins was less than 10% of the signal generated in response to TGF-β1. Comparison of sensor responses to three known isoforms of TGF-β (1, 2, 3) showed ∼15% cross-reactivity with TGF-β3 and 30% cross-reactivity with TGF-β2 (Figure 1B). This observation is not unexpected in light of homology between TGF-β1 and -β3.5
Figure 1.
(A) The results of SWV response of aptasensor toward 50 ng/mL of different nonspecific (IL-2, IFN-γ, BSA, IgG) molecules and 50 ng/mL of the specific target molecule (TGF-β1). Plot showing the change in aptasensor current resulting due to each nonspecific and specific molecule. These results show that the biosensor did not respond to nonspecific proteins but did respond to TGF-β1. (B) Response of the aptasensor toward 50 ng/mL of different isoforms of the TGF-β molecule.
Operation of the Microfluidic Device
In the multilayered microfluidic device, the upper most PDMS layer controls the raising and lowering of the microcups fabricated into the other PDMS layer (Figure 2A). The up and down action of the PDMS layer was regulated by applying positive or negative pressure via the control layer. Water was injected into the control layer to apply positive pressure and was evacuated to create negative pressure.
Figure 2.
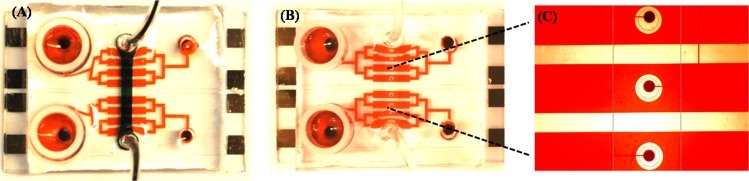
(A) Fluidic channels of the reconfigurable microfluidic device are infused with red dye whereas the control layer is filled with black dye. (B, C) Device is reconfigured, lowering microcups around the electrodes. The channels are filled with red dye to highlight that the area around the electrodes remains free of dye. (C) Higher magnification view of the electrodes being sequestered from the rest of the channel.
In Figure 2, food dye was used to illustrate the working principle of the microfluidic device. In Figure 2A, the PDMS control channel was filled with black dye while the bottom layer was infused with red dye. This image shows the presence of 8 parallel microchannels, each containing a sensing electrode. Figure 2B,C shows the microchannels that were infused with red dye after the cups where lowered (control layer filled with water). Thus, one can see dye-free white regions around the electrodes, surrounded by red liquid.
The ability to reconfigure the microfluidic channel to separate the environment around the electrodes from the rest of the fluidic channel was used to seed cells. Figure 3A,B shows stellate cells 30 min and 12 h after seeding into the microfluidic channel. One can see an annular region around the electrode that was protected by microcups and remained free of cells after seeding. To highlight the importance of protecting electrodes during the seeding of “sticky” anchorage-dependent stellate cells, aptamer-functionalized electrodes were characterized by SWV after seeding cells with or without microcup protection. As shown in Figure 3C, the redox signal of electrodes directly exposed to cell seeding was about 15-fold lower than the electrode protected with microcups. Thus, the reconfigurable microfluidic device allowed us to ensure that aptasensors were not fouled during collagen coating and cell seeding steps and retained the high sensitivity needed for cell secretion monitoring.
Figure 3.
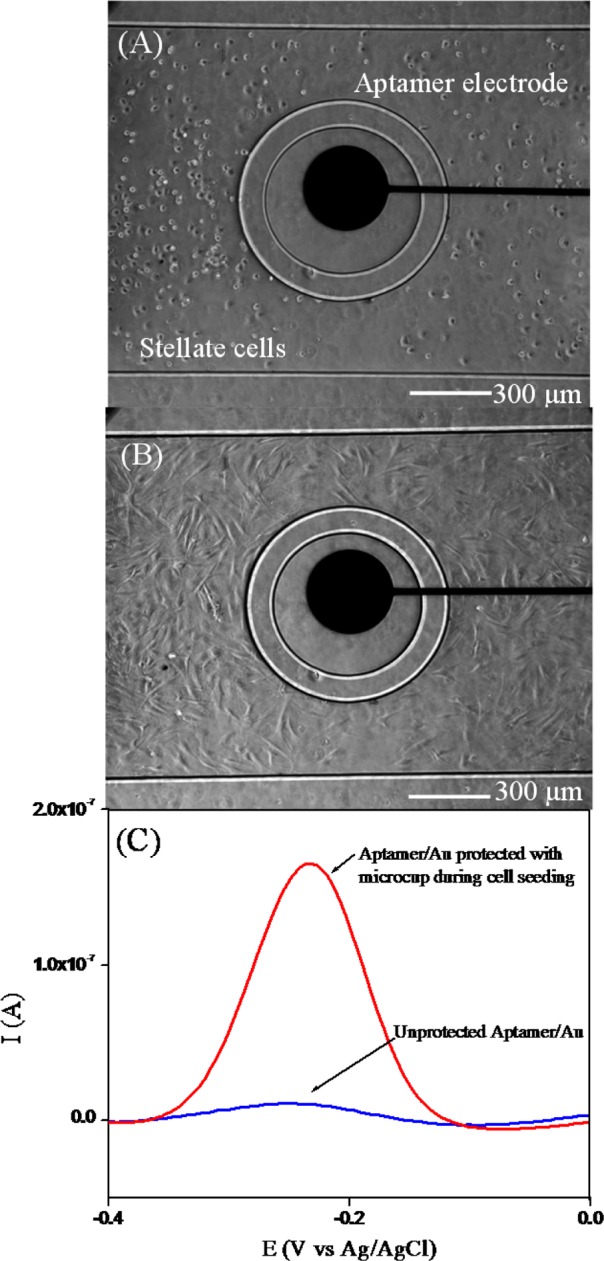
(A) Stellate cells 30 min after seeding around Au electrodes. (B) The same cells 12 h after seeding spread out around the electrode. (C) SWV curves looking at the MB signal obtained from aptasensor when cells were seeded with and without microcup protection. These results show 15-fold higher peak currents for the electrode protected by microcups during cell seeding.
Monitoring of TGF-β1 Release from Stellate Cells
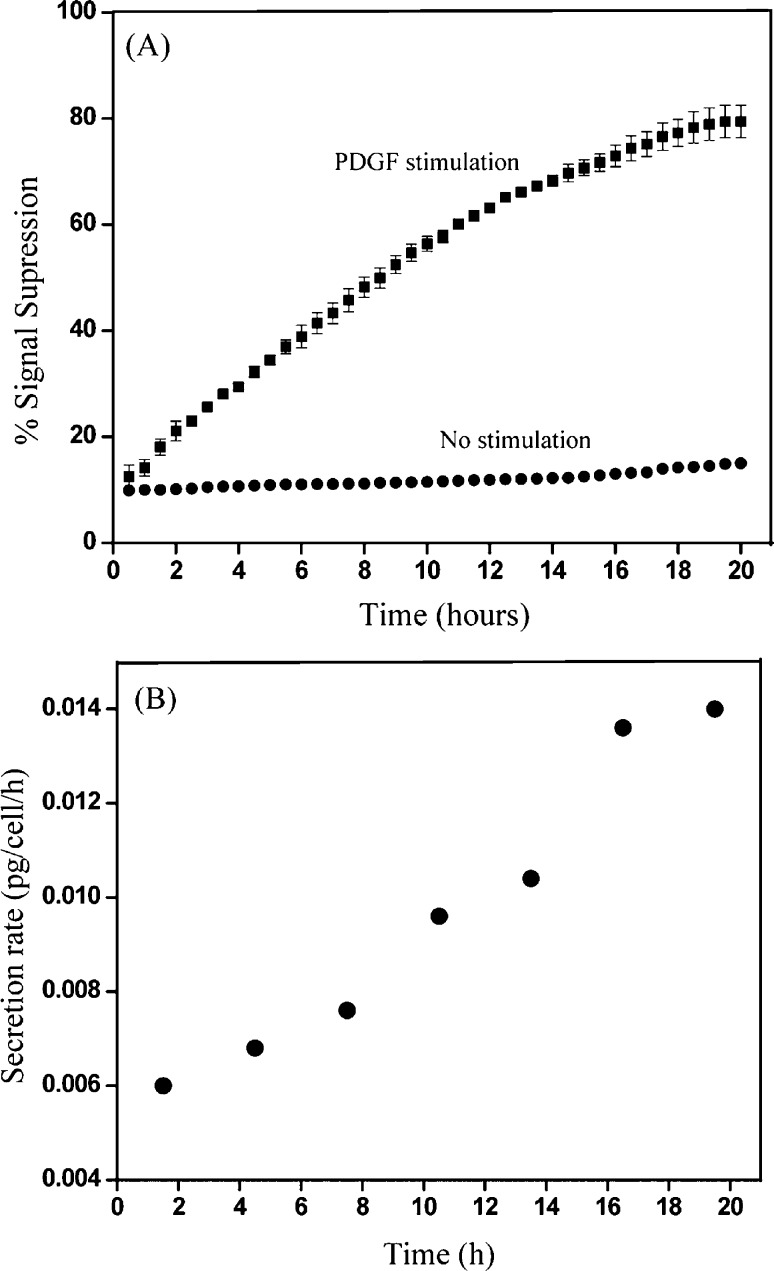
As described in Scheme 1B, it was expected that our biosensor would react to TGF-β1 secreted from stellate cells, providing an electrochemical signal associated with the amount and the rate of cytokine secretion. Prior to carry out cell detection experiments, we wanted to construct calibration curves to determine the linear range of the miniature aptasensors integrated with microfluidics. Figure 4A shows typical response of aptasensor to varying (increasing) concentration of exogenous TGF-β1. The signal decreases with increasing concentration of the analyte and is reported as signal suppression – ((initial current – final current)/initial current). Figure 4B shows a calibration curve of signal suppression vs TGF-β1 concentration. The experimental data shows the linear range for our biosensor from 1 to 250 ng/mL for exogenous TGF-β1.
Figure 4.
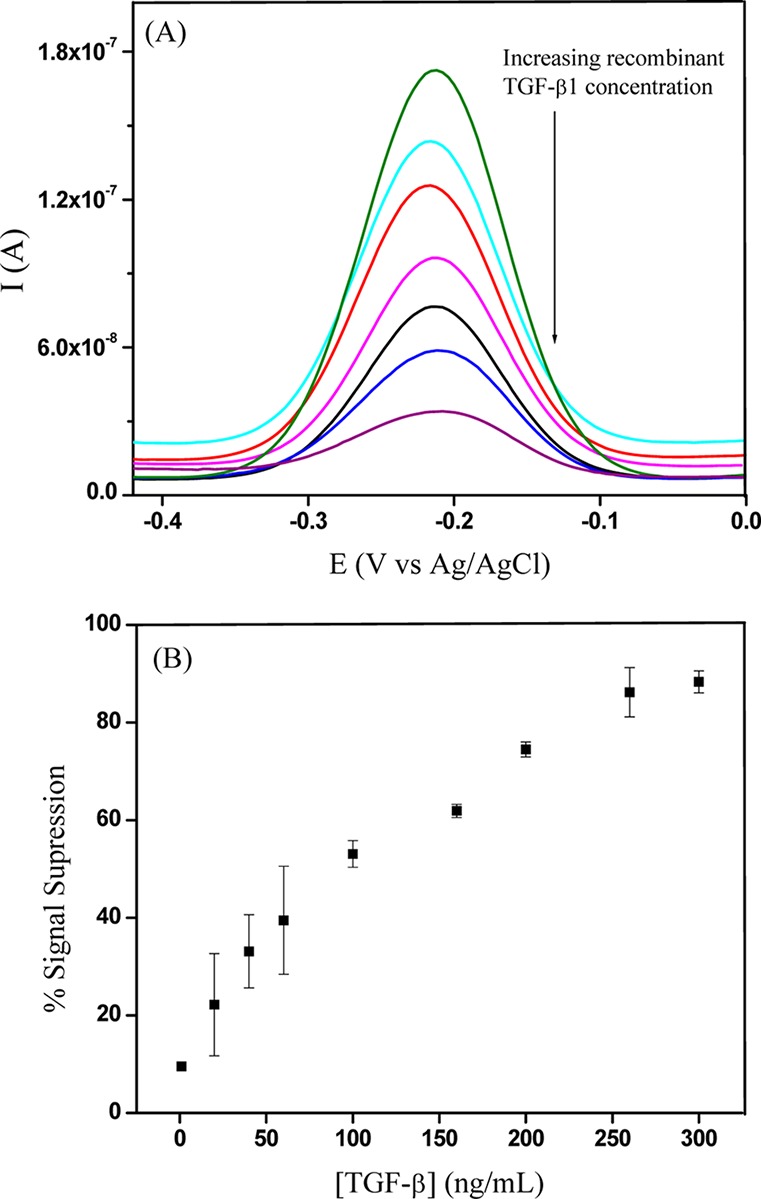
(A) SWV curves obtained for aptamer-modified electrodes challenged with recombinant TGF-β1 ranging in concentration from 1 to 300 ng/mL. (B) Signal suppression plotted against recombinant TGF-β1 concentration. The aptamer shows good response in the cell culture media with signal saturation after 250 ng/mL TGF-β1.
For TGF-β1 release experiments, stellate cells were seeded into microfluidic channels that were reconfigured so as to protect aptasensing electrodes. Upon attaching and spreading, cells were stimulated by infusion of PDGF, growth factor known to activate stellate cells.18−20 The device was reconfigured once again to raise the microcups, allowing for cell secreted factors to diffuse to the nearby sensing electrode. The resultant electrochemical signals were continuously monitored by sequentially addressing aptasensing electrodes. To ensure viability and function, microfluidic devices with stellate cells were kept under physiological temperature and 5% CO2 during the 20 h experiment.
Figure 5A shows TGF-β1 secretion results after PDGF activation. In our experiments, one inlet of the device, connected to four cell seeded microchannels, was infused with stimulant to activate the cells while the other four channels contained quiescent cells. As highlighted by data in Figure 5A, electrodes in proximity to activated stellate cells were recording significantly higher changes in redox current (signal suppression) compared to electrodes near quiescent cells. After 18 h, the electrodes near activated stellate cells reached 80% signal suppression whereas sensors next to quiescent cells showed less than 20% signal suppression. Given that 80% signal suppression corresponds to a saturated aptasensor (see Figure 4B), the electrode sensing activated stellate cells likely became saturated after 18 h of monitoring.
Figure 5.
(A) Continuous monitoring of the TGF-β1 release from stellate cells within the microfluidic device using aptamer modified microfabricated electrodes. The device was kept in the CO2 and temperature control chamber during the SWV experiments. (B) Change of the TGF-β1 secretion rate over time obtained from the reaction-diffusion model analyzed with COMSOL.
To determine cell secretion rates based on the binding curves shown in Figure 5A, we constructed a numerical model for simulating cytokine production, diffusion, convection, and binding using COMSOL multiphysics (COMSOL Inc., Burlington, MA). The geometry and sensor configuration were the actual sensing devices used for the experiments. The main parameters in the simulations are listed in Table 1.
Table 1. List of Parameters Obtained Experimentally and Further Used for Determining Secretion Rates via Simulation.
| diffusion coefficient | 1.3 × 10–6 cm2/s |
| surface binding density | 3.38 × 1012 molecules/cm2 |
| association rate constant | 4.48 × 105 M−1 s−1 |
| dissociation rate constant | 4.82 × 10–4 s−1 |
| flow rate | 0.1 μL/min |
We assumed a constant secretion rate in each 3 h time interval and then determined the secretion rate by the reaction–diffusion–convection model coupled with least-squares approximation (further details provided in the Supporting Information and ref (21)). From the model, the average cytokine production rate from the stimulated cells within the microfluidic channel was determined to be 0.0140 pg/cell/h (Figure 5B) while for quiescent cells, the rate was 0.0009 pg/cell/h. This rate is comparable to the production of TNF-α by activated monocytes reported by our group previously. Our TGF-β production rate over the course of 24 h (3.36 × 10–1 pg/cell) is considerably higher than rates reported for stellate cells activated under standard tissue culture conditions (4.2 × 10–5 to 3.2 × 10–3 pg/cell).1,22,23 This may be explained by the enhanced sensitivity of our approach where local concentrations are being monitored. It is also possible that confinement of stellate cells inside low volume microfluidic channels enhances the rate of TGF-β secretion.
A number of control experiments were carried out to ensure that the signal observed in Figure 5A was indeed due to TGF-β1 secretion. Exposure of stellate cells to culture media without PDGF did not cause an appreciable change in electrochemical signal (Figure 5A, lower curve). Similarly, there was no effect of PDGF containing media on the aptamer modified Au surface without cells (result not shown).
We conducted another control experiment where activated stellate cells residing inside microfluidic channels were bathed with TGF-β1 antibodies during the sensing experiment. The presence of competing antibody molecules abrogated redox signals at the sensing electrodes, demonstrating once again that electrodes were indeed sensing secreted TGF-β1 (Figure S4, Supporting Information).
Characterizing Phenotype and Function of Stellate Cells Using Molecular Biology Approaches
Immunofluorescent staining and ELISA techniques were used to confirm that exposure to PDGF activated stellate cells caused them to release TGF-β1. Figure 6A,B shows expression of α-SMA, one of the benchmarks of stellate cell activation, in the presence and absence of PDGF.24 These results clearly demonstrate increased expression of SMA in cells after a 24 h exposure to PDGF, indicating activation of these cells. Another correlate of stellate cell activation is production of TGF-β1. We collected media from activated and quiescent cells cultured in a well plate for 48 h and performed TGF-β1 ELISA. This experiment (Figure 6C) revealed that stellate cells activated with PDGF produced 6 times more TGF-β1 compared to quiescent cells. While it may not be possible to compare concentrations obtained with ELISA and aptasensors due to differences in analytical methods, the 6-fold enhancement in TGF-β1 activated and resting stellate cells was observed for both approaches (see Figures 5A and 6C for comparison).
Figure 6.
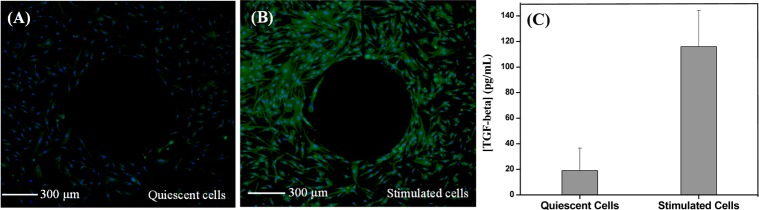
Immunofluorescence images showing α-SMA expression (green) from (A) quiescent stellate cells and (B) PDGF activated stellate cells, seeded around Au microelectrodes inside the device. DAPI (blue) was used for nucleus staining. (C) ELISA analysis of TGF-β1 secretion by stellate cells cultured in a 6-well plate during 2 days.
Conclusions
The paper describes the use of aptamer-modified electrodes for continuous monitoring of the TGF-β1 release from hepatic stellate cells. While our lab has previously demonstrated detection from immune cells,12,25 this study deals with monitoring secretory activity of adhesive stromal cells. Reconfigurable microfluidic devices were used to address electrode fouling problems. Such devices were microfabricated to contain microcups imbedded in the roof of the microfluidic channel. By actuating a control layer, microcups were lowered to protect the electrodes during collagen coating and cell seeding steps. The device was reconfigured once again after seeding and activation of stellate cells, to commence the detection phase of the experiment. These specific and sensitive aptasensors were used to electrochemically monitor TGF-β1 secretion from activated and resting stellate cells and to determine TGF-β1 secretion rates. Furthermore, additional molecular biology assays demonstrated that stellate cells were indeed activated by stimulation with PDGF and that these activated stellate cells produced TGF-β1. In fact, a similar 6-fold ratio of TGF-β1 production between activated and resting cells was obtained using aptasensors inside microfluidic channels and ELISA in 6-well plates. These results point to the fact that stellate cells functioned similarly (vis a vis TGF-β1 secretion) inside microfluidic devices and under standard tissue culture conditions. In combination with aptasensor specificity testing, the TGF-β1 antibody competition experiment showed that detection was indeed specific to secreted TGF-β1. Given the importance of TGF-β1 signaling in inflammation, fibrogenesis, T-cell differentiation, and stem cell development, the ability to locally and continuously monitor production of TGF-β1 has high significance. In the future, the duration of the detection experiment will be extended from 20 h to multiple days. Multiplexed aptamer-based detection of several important trophic factors is also envisioned.
Acknowledgments
The authors thank Dr. Ying Liu (ReLIA Diagnostic Systems Inc), Dr. Dong-Sik Shin, and Prof. Y. Yokobayashi (Department of Biomedical Engineering, UC Davis) for helpful discussions. Authors also thank James Enomoto, Timothy Kwa, and Lydia Kwon for technical help. The financial support for these studies was provided by NSF and NIH. “Research Investments in Science and Engineering (RISE) Program, UC Davis” is acknowledged for providing additional funding.
Supporting Information Available
Text and figures giving details of the surface plasmon resonance (SPR)-based characterization of TGF-β1 aptamer, electrochemical characterization of TGF-β1 aptamer, description of the computational model for secretion rate, and inhibition of TGF-β1 detection via anti-TGF-β1 antibodies. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
† D.P. and Y.G. contributed equally to the work
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- De Bleser P. J.; Xu G.; Rombouts K.; Rogiers V.; Geerts A. J. Biol. Chem. 1999, 2744833881–33887. [DOI] [PubMed] [Google Scholar]
- Bataller R.; Brenner D. A. J. Clin. Invest. 2005, 1152209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsharkawy A. M.; Oakley F.; Mann D. A. Apoptosis 2005, 105927–939. [DOI] [PubMed] [Google Scholar]
- Dooley S.; ten Dijke P. Cell Tissue Res. 2012, 3471245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressner A. M.; Weiskirchen R.; Breitkopf K.; Dooley S. Front. Biosci. 2002, 7, d793–807. [DOI] [PubMed] [Google Scholar]
- Leask A.; Abraham D. J. FASEB J. 2004, 187816–827. [DOI] [PubMed] [Google Scholar]
- Jones C. N.; Tuleuova N.; Lee J. Y.; Ramanculov E.; Reddi A. H.; Zern M. A.; Revzin A. Biomaterials 2010, 31235936–5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonham A. J.; Hsieh K.; Ferguson B. S.; Vallee-Belisle A.; Ricci F.; Soh H. T.; Plaxco K. W. J. Am. Chem. Soc. 2012, 13473346–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Lai R. Y.; Plaxco K. W. Nat. Protoc. 2007, 2112875–2880. [DOI] [PubMed] [Google Scholar]
- Kwa T.; Zhou Q.; Gao Y.; Rahimian A.; Kwon L.; Liu Y.; Revzin A. Lab Chip 2014, 14101695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q.; Kwa T.; Gao Y.; Liu Y.; Rahimian A.; Revzin A. Lab Chip 2014, 142276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Kwa T.; Revzin A. Biomaterials 2012, 33307347–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.; Lee M. S.; Copland J. A.; Luxon B. A.; Gorenstein D. G. Bioorg. Med. Chem. Lett. 2008, 1861835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Tuleouva N.; Ramanculov E.; Revzin A. Anal. Chem. 2010, 82198131–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Stybayeva G.; Macal M.; Ramanculov E.; George M. D.; Dandekar S.; Revzin A. Lab Chip 2008, 8122197–2205. [DOI] [PubMed] [Google Scholar]
- Matharu Z.; Enomoto J.; Revzin A. Anal. Chem. 2013, 852932–939. [DOI] [PubMed] [Google Scholar]
- Ricci F.; Lai R. Y.; Heeger A. J.; Plaxco K. W.; Sumner J. J. Langmuir 2007, 23126827–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira R. K. Arch. Pathol. Lab. Med. 2007, 131111728–1734. [DOI] [PubMed] [Google Scholar]
- Friedman S. L. Physiol. Rev. 2008, 881125–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pabic H.; Bonnier D.; Wewer U. M.; Coutand A.; Musso O.; Baffet G.; Clément B.; Théret N. Hepatology 2003, 3751056–1066. [DOI] [PubMed] [Google Scholar]
- Gao Y.; Zhou Q.; Matharu Z.; Liu Y.; Kwa T.; Revzin A. Biomicrofluidics 2014, 82021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shek F. W.; Benyon R. C.; Walker F. M.; McCrudden P. R.; Pender S. L.; Williams E. J.; Johnson P. A.; Johnson C. D.; Bateman A. C.; Fine D. R.; Iredale J. P. Am. J. Pathol. 2002, 16051787–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Krebs A.; Preimel D.; Popov Y.; Bartenschlager R.; Lohmann V.; Pinzani M.; Schuppan D. Gastroenterology 2005, 1291246–258. [DOI] [PubMed] [Google Scholar]
- Olsen A. L.; Bloomer S. A.; Chan E. P.; Gaca M. D.; Georges P. C.; Sackey B.; Uemura M.; Janmey P. A.; Wells R. G. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 3011G110–G118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yan J.; Howland M. C.; Kwa T.; Revzin A. Anal. Chem. 2011, 83218286–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.