Abstract
WWOX was cloned as a putative tumor suppressor gene mapping to chromosomal fragile site FRA16D. Deletions affecting WWOX accompanied by loss of expression are frequent in various epithelial cancers. Translocations and deletions affecting WWOX are also common in multiple myeloma and are associated with worse prognosis. Metanalysis of gene expression datasets demonstrates that low WWOX expression is significantly associated with shorter relapse-free survival in ovarian and breast cancer patients. Although somatic mutations affecting WWOX are not frequent, analysis of TCGA tumor datasets led to identifying 44 novel mutations in various tumor types. The highest frequencies of mutations were found in head and neck cancers, uterine and gastric adenocarcinomas.
Mouse models of gene ablation led us to conclude that Wwox does not behave as a highly penetrant, classical tumor suppressor gene since its deletion is not tumorigenic in most models and its role is more likely to be of relevance in tumor progression rather than in initiation. Analysis of signaling pathways associated with WWOX expression confirmed previous in vivo and in vitro observations linking WWOX function with the TGFβ/SMAD and WNT signaling pathways and with specific metabolic processes. Supporting these conclusions recently we demonstrated that indeed WWOX behaves as a modulator of TGFβ/SMAD signaling by binding and sequestering SMAD3 in the cytoplasmic compartment. As a consequence progressive loss of WWOX expression in advanced breast cancer would contribute to the pro-metastatic effects resulting from TGFβ/SMAD3 hyperactive signaling in breast cancer.
Recently, GWAS and resequencing studies have linked the WWOX locus with familial dyslipidemias and metabolic syndrome related traits. Indeed, gene expression studies in liver conditional KO mice confirmed an association between WWOX expression and lipid metabolism.
Finally, very recently the first human pedigrees with probands carrying homozygous germline loss of function WWOX mutations have been identified. These patients are characterized by severe CNS related pathology that includes epilepsy, ataxia and mental retardation.
In summary, WWOX is a highly conserved and tightly regulated gene throughout evolution and when defective or deregulated the consequences are important and deleterious as demonstrated by its association not only with poor prognosis in cancer but also with other important human pathologies such as metabolic syndrome and CNS related pathologic conditions.
Keywords: WWOX, FRA16D, fragile site, WW domains, cancer, TGFβ, SMAD, WNT, lipid metabolism, metabolic syndrome, epilepsy, ataxia, mental retardation
1. Introduction
By using conventional positional cloning approaches in 2000 we cloned WWOX (WW domain containing oxidoreductase), a gene spanning chromosome region 16q23.3-q24.1, an area that was narrowed down as commonly affected by genomic loses in breast and other cancers (1, 2). WWOX contains two distinct domains; the amino terminus shows high sequence conservation to the WW domain family of proteins displaying two WW domains and a central 283 amino acids domain homologous to the Short-Chain Dehydrogenase/Reductase (SDR) superfamily (Figure 1).
Figure 1.

At the genomic level WWOX spans a region of over 1.1 Mbp in size even though the encoded gene is not large, only 9 exons (NM_016373.2) coding for a 414 AA protein (NP_057457.1). We observed that the very large 5 and 8 introns contain previously identified translocation break points for multiple myeloma, specifically t(14;16)(q32;q23) (3). We concluded that such intronic regions were highly recombinogenic and prone to chromosomal rearrangements and we speculated in our report that likely this was the same locus as that of chromosomal fragile site FRA16D (2). Soon thereafter a second group in search of the gene target for FRA16D cloned FOR that resulted to be the same as WWOX confirming the original speculations (4) and a third group reported the mouse sequence (5). FRA16D, is the second most frequently affected constitutive site of chromosomal fragility in the whole human genome, second only to FRA3B. Common or constitutive chromosomal fragile sites are found in all individuals and are a constant chromosomal feature. These highly recombinogenic chromosomal loci are susceptible to the occurrence of breaks, gaps and rearrangements.
2. WWOX throughout evolution
There is remarkable sequence conservation both at exonic and intronic level at the human and mouse orthologous chromosomal fragile site regions for human FRA16D/WWOX and mouse Fra8E1/Wwox (6). WWOX/Wwox proteins display 94% identity and 96% similarity at the amino acid level (5, 6). Remarkably when comparing with the drosophila homologous protein, identity reaches 49% and 66% similarity. Interestingly, within WWOX introns there is remarkable evolutionary conservation, multiple ultraconserved regions can be found within these intronic regions suggesting that there must exist significant evolutionary selection pressure for preserving not only the coding but also non-coding regions of the WWOX locus. This phenomenon that is also observed in other large fragile site loci such as FRA3B/FHIT, appears to be partially related to sequences encoding essential non-coding RNAs within such large intronic regions (7, 8). An additional reason for preservation of such large non-coding areas at the WWOX locus is the proven existence of various functional enhancer elements within such intragenic regions (9).
Gilad et al. (10) utilized a multispecies cDNA array containing probes for > 1000 genes matching human, chimpanzee, orangutan and rhesus macaque sequences, thus phylogenetically representing approximately 70 million years of evolution, for comparing liver specific gene expression between these species. One of the goals of the study was to identify those genes whose expression has remained constant throughout evolution, i.e. genes that remained evolving under ‘stabilizing selection’. Interestingly, WWOX ranked among the top genes under stabilizing selection, a category mostly enriched by genes that fall under the gene ontology functional classification of ‘regulation of physiological processes’. Furthermore, these authors concluded that precisely focusing on genes such as WWOX, whose expression is tightly conserved among primates, would be helpful for identifying promising candidates for disease-association studies (10).
3. WWOX normal expression and loss of expression in cancer
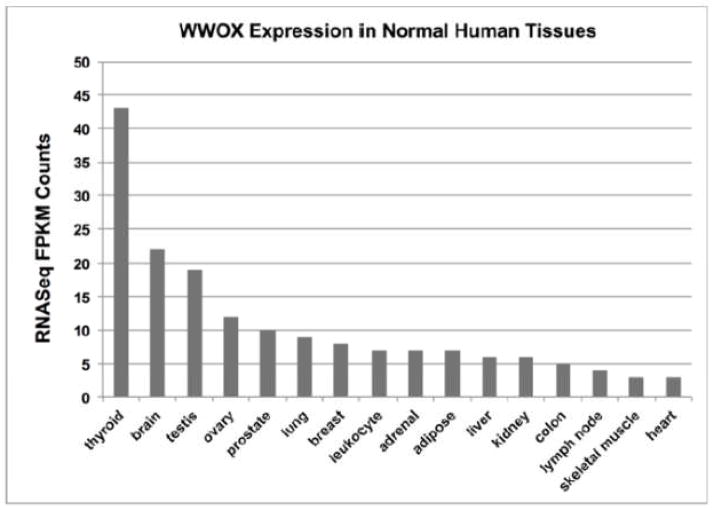
We observed that WWOX protein is ubiquitously expressed in human tissues by means of immunohistochemistry (IHC) studies (11). However it is preferentially highly expressed in secretory epithelia, in reproductive, exocrine and endocrine organs and interestingly also in neuronal bodies throughout the CNS including strong expression in the Purkinje layer of the cerebellum (11). RNASeq analyses from multiple human tissues are quite in agreement with the previous IHC studies and thyroid, brain, testis, ovary and prostate demonstrated to be the tissues with highest expression levels in that order (Figure 2) as per the EMBL-EBI Expression Atlas (http://www.ebi.ac.uk) Illumina Body Map (experiment E-MTAB-513).
Figure 2.
Studies from various laboratories have shown loss of WWOX protein expression in multiple neoplasias such as breast (12, 13), ovarian (14), gastric (15), liver (16), lung (17), thyroid (18), pancreatic (19) and other cancers (20). This topic has been reviewed extensively elsewhere by other authors (21). Importantly, in various cancers loss of WWOX expression is associated with indicators of poor prognosis and outcome, e.g. in breast cancer (12, 13), ovarian cancer (14), non-small cell lung cancer (17) and bladder cancer (22).
It is worth noting that various alternatively spliced mRNA WWOX forms were originally described by us and others as commonly occurring in tumors and cancer lines. The most common of these alternative transcripts displayed deletions of exons 5–8 or 6–8 (4, 23, 24). However, these alternatively spliced forms and others including those reported in GeneBank as isoforms 2 (NM_130791.2) and 3 (NM_130844.2) so far have not been demonstrated to encode viable protein, since they are not detected by Western blot to the best of our knowledge.
The most important mechanism leading to loss of WWOX expression appears to be genomic loss via gross chromosomal deletions and rearrangements as will be discussed in the next section. Additional mechanisms include epigenetic silencing by promoter hypermethylation (25) and degradation. It has been shown that WWOX can be inactivated via polyubiquitination and subsequent proteosomal degradation. ACK1 has been shown to interact with and phosphorylate WWOX at Y287 thus targeting WWOX for rapid degradation (26). WWOX has also been proposed to be a substrate of ubiquitin ligase ITCH (27). Additionally recently it has also been reported WWOX downregulation via targeting by miR-134 in head and neck squamous cell carcinomas (28).
4. New data on WWOX Copy Number Losses and mRNA expression in Cancer
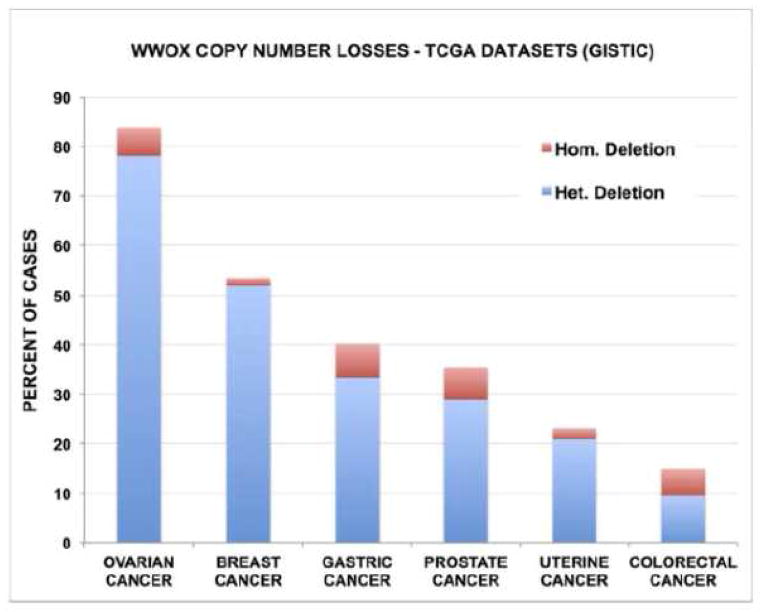
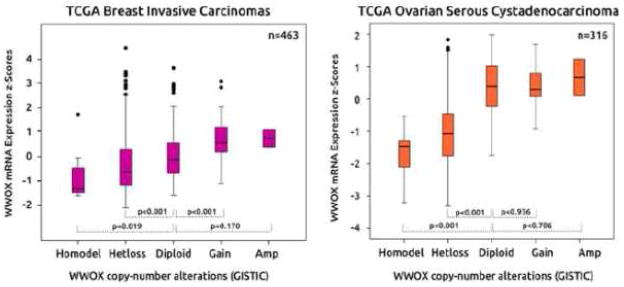
A comprehensive analysis of copy number variation on over 740 cancer lines determined that the WWOX locus (FRA16D) was the third most common site of the whole genome affected by hemi and homozygous losses only after the p16 (CDKN2A) and the FHIT (FRA3B) loci (29). Recently, Beroukhim R et al (30) reported a high resolution study of somatic copy number alterations analyzing over 3000 cancer specimens and identified WWOX among the most commonly affected genes by genomic deletions as per GISTIC (genomic identification of significant targets in cancer) algorithm analyses (31). By using the same algorithm we can now determine genomic losses on each specific tumor dataset reported so far by The Cancer Genome Atlas (TCGA) using the cbioPortal resource (www.cbioportal.org) (32, 33). Thus we identified the tumor types most commonly affected by heterozygous and homozygous WWOX deletions (Freeze February 2014). As can be observed in Figure 3, ovarian serous cystoadenocarcinoma is the tumor type most commonly affected by genomic losses affecting WWOX with 78% of cases showing heterozygous loss and 6% homozygous losses (34) Heterozygous losses were observed in 52% of breast cancers (35), 33% of gastric adenocarcinomas, 29% of prostate adenocarcinomas, 21% of endometrial cancers (36) and 10% colorectal carcinomas (37) (Fig. 3). Gastric adenocarcinomas and prostate carcinomas show a relatively high frequency of homozygous losses affecting 7% of cases. Figure 4 displays the correlation between WWOX copy number status and gene expression levels in ovarian and breast cancer. As can be observed, heterozygous losses affect significantly WWOX expression levels in both tumor types but even more significantly in ovarian cancer.
Figure 3.
Figure 4.
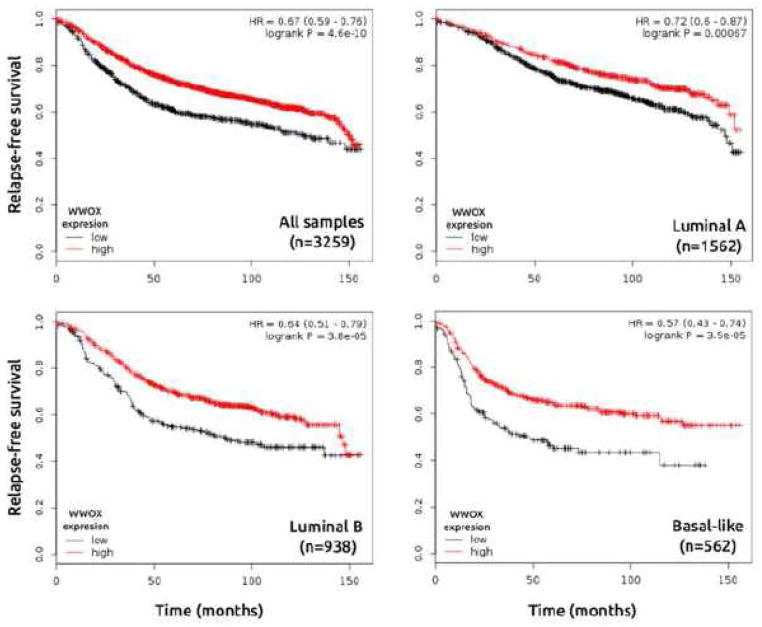
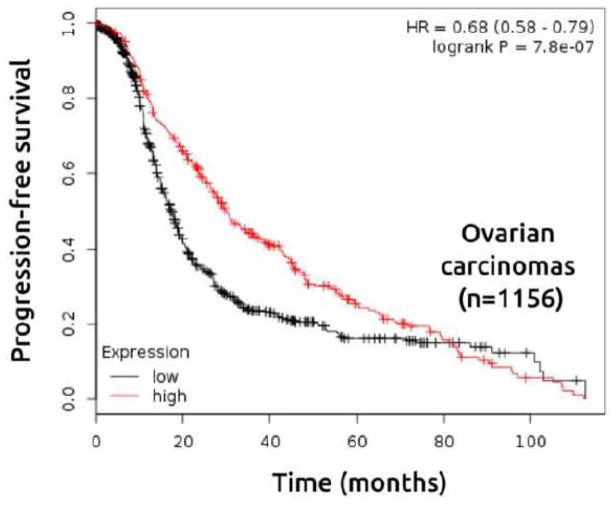
To further explore the prognostic value of WWOX mRNA expression in breast and ovarian carcinomas, we evaluated information from publicly available gene expression data sets (oligo-microarrays) using the Kaplan-Meier Plotter resource (38). First, patients were divided into two groups (high and low WWOX expression) according to the quantile expressions of Affymetrix 219077_s_at and 210695_s_at probes. These groups were then compared using the relapse-free survival and progression-free survival for breast and ovarian cancer datasets respectively. Kaplan–Meier analysis revealed that low expression of WWOX was associated with shorter relapse-free survival in breast cancer patients (n=3259) (Figure 5) and shorter progression-free survival in ovarian cancer patients as well (n=1156) (Figure 6). Interestingly, we identified a significant association between low expression of WWOX and short time relapse-free survival in all breast cancer intrinsic subtypes, but more pronounced in luminal B and basal-like subtypes (Figure 5).
Figure 5.
Figure 6.
5. WWOX Mutations in Cancer
The development of next generation sequencing technologies led to a dramatic accumulation of genomic and expression data in the past three years on multiple tumor types. The data by TCGA gives us now the opportunity to determine the true frequency of somatic mutations affecting WWOX in cancer. Data was obtained using the cbioportal resource (www.cbioportal.org) (32, 33), as per freeze on February 2014.
The frequency of mutations affecting the WWOX coding region across tumor types as expected is not very high. Nevertheless a total of 44 novel WWOX somatic mutations have been identified to date in various tumor types, 5 of these resulted in nonsense mutations (3 in head and neck cancers and 2 in lung cancers), 2 frameshift insertions, 1 frameshift deletion and 36 missense mutations, each specific mutation is depicted in Figure 7 and described in Table 1. Of these, 8 somatic mutations were identified in uterine adenocarcinomas (3.2% of cases, (36), 8 in head and neck cancers (4.1% of cases from Broad (39), and in 2% of TCGA cases), 4 in gastric adenocarcinomas (2.3% of cases), 5 in colorectal cancers (2.2% of cases (37), 6 in melanoma (<2%), 5 in lung adenocarcinomas (<2%), 2 in lung squamous cell carcinomas (<2%), 3 in breast carcinomas (<1%), and one mutation each in a pancreas, a bladder and a renal clear cell carcinoma (Table 1). Of particular interest is that two identical mutations were identified at G260 resulting in frameshift insertions in a uterine adenocarcinoma and in a breast carcinoma, two missense mutations were detected at R254 one in a colorectal and one in a uterine adenocarcinoma (R254C and R254S respectively), two mutations at H147 were also observed resulting in missense mutations in a head and neck carcinoma and a lung adenocarcinoma (H147Q and H147Y respectively).
Figure 7.
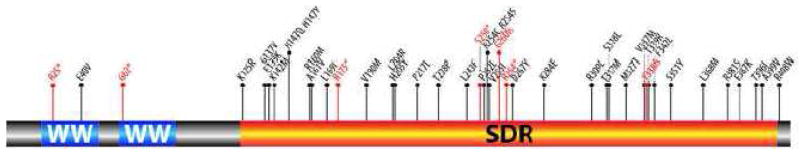
Table 1.
WWOX Mutations in Cancer
| Case ID | Cancer Study | AA Change | Type | Mutation Assessor | |
|---|---|---|---|---|---|
| TCGA-AX-A05Z | Uterine (TCGA ) (36) | D267Y | Missense | Low | |
| TCGA-AX-A2H5 | Uterine (TCGA ) (36) | G260fs | FS ins | ||
| TCGA-AP-A051 | Uterine (TCGA ) (36) | L169I | Missense | Low | |
| TCGA-AP-A0LM | Uterine (TCGA ) (36) | L368M | Missense | Medium | |
| TCGA-BS-A0UL | Uterine (TCGA ) (36) | R254S | Missense | Medium | |
| TCGA-BS-A0UF | Uterine (TCGA ) (36) | R381S | Missense | Medium | |
| TCGA-AX-A0J1 | Uterine (TCGA ) (36) | R408W | Missense | Low | |
| TCGA-N7-A4Y0 | Uterine CS (TCGA) | L204R | Missense | High | |
| HN_62671 | Head & neck (Broad) (39) | K142M | Missense | Medium | |
| HN_63080 | Head & neck (Broad) (39) | R175* | Nonsense | ||
| HN_62814 | Head & neck (Broad) (39) | V190M | Missense | High | |
| TCGA-CV-7178 | Head & neck (TCGA ) (in revision) | H147Q | Missense | Neutral | |
| TCGA-CN-4731 | Head & neck (TCGA ) (in revision) | R25* | Nonsense | ||
| TCGA-CQ-5325 | Head & neck (TCGA ) (in revision) | S351Y | Missense | High | |
| TCGA-CN-4730 | Head & neck (TCGA ) (in revision) | Y338fs | FS del | ||
| TCGA-D6-6516 | Head & neck (TCGA ) (in revision) | R264* | Nonsense | ||
| MEL-Ma-Mel-48 | Melanoma (Broad) (104) | S318L | Missense | Medium | |
| ME011 | Melanoma (Broad) (104) | T396I | Missense | Neutral | |
| TCGA-ER-A197 | Melanoma (TCGA provisional | K284E | Missense | Neutral | |
| TCGA-FW-A3R5 | Melanoma (TCGA provisional | L243F | Missense | Medium | |
| TCGA-FW-A3R5 | Melanoma (TCGA provisional | P217L | Missense | Medium | |
| TCGA-EE-A2MN | Melanoma (TCGA provisional | P252L | Missense | Medium | |
| TCGA-BR-4256 | Stomach (TCGA provisional) | F342L | Missense | Low | |
| TCGA-BR-4368 | Stomach (TCGA provisional) | M327T | Missense | Neutral | |
| TCGA-CG-5721 | Stomach (TCGA provisional) | T317M | Missense | Medium | |
| TCGA-BR-6452 | Stomach (TCGA provisional) | V337M | Missense | Low | |
| TCGA-AA-3854 | Colorectal (TCGA) (37) | A399V | Missense | Low | |
| TCGA-AG-A02N | Colorectal (TCGA) (37) | K125R | Missense | Low | |
| TCGA-AA-3972 | Colorectal (TCGA) (37) | R254C | Missense | High | |
| TCGA-AA-A01Z | Colorectal (TCGA) (37) | T339R | Missense | Neutral | |
| TCGA-AA-A00N | Colorectal (TCGA) (37) | E139K | Missense | Medium | |
| LUAD-74TBW | Lung adeno (Broad) (105) | E40V | Missense | Low | |
| LUAD-B00915 | Lung adeno (Broad) (105) | R160M | Missense | Medium | |
| TCGA-55-7576 | Lung adeno (TCGA) (in press) | H147Y | Missense | Neutral | |
| TCGA-05-4396 | Lung adeno (TCGA) (in press) | G62* | Nonsense | ||
| TCGA-05-4389 | Lung adeno (TCGA) (in press) | S250* | Nonsense | ||
| TCGA-60-2720 | Lung squ (TCGA) (106) | E387K | Missense | Neutral | |
| TCGA-66-2787 | Lung squ (TCGA) (106) | G137V | Missense | High | |
| BR-M-105 | Breast (Broad) (107) | G260fs | FS ins | ||
| TCGA-A2-A0CX | Breast (TCGA) (35) | H205Y | Missense | Medium | |
| TCGA-A8-A0A6 | Breast (TCGA) (35) | T228P | Missense | Medium | |
| ICGC_0037_TD | Pancreas (ICGC) (108) | V255I | Missense | Neutral | |
| TCGA-AK-3428 | Renal clear cell (TCGA) (109) | R309L | Missense | High | |
| TCGA-GV-A3QF | Bladder (TCGA) (110) | A161V | Missense | Low | |
The detected nonsense and frameshift mutations all have obvious severe impact in protein functionality and of the 36 missense mutations 18 (50%) are predicted to affect protein function based on the mutation assessor algorithm (40), noted in bold as high and medium functional impact in Table 1. In summary, of all somatic mutations detected so far affecting WWOX in human tumors 59% (26 of 44) are very likely to affect protein function, suggesting that is unlikely these mutations represent simple background mutational events.
6. WWOX in Multiple Myeloma
The frequent involvement of WWOX genomic and expression abnormalities in multiple myeloma (MM) deserves a separate section. In 1998, prior to the cloning of WWOX, Chesi et al (3), reported the molecular characterization of a novel translocation in MM, t(14;16)(q32;q23). The 16q23 region was observed translocated to an IgH locus in 20–25% of MM lines and fresh MM samples. It was concluded then that the consequence of the t(14;16)(q32;q23) is mostly directed to increase MAF expression (an oncogene located 3′ of WWOX’s locus) (3). However, we know now that an additional consequence of such translocation is the destruction of a functional WWOX allele. Analysis of the genomic area spanned by WWOX revealed that the t(14;16)(q32;q23) translocation breakpoints identified as KMS11, MM.1, JJN3 and ANBL6 in ch16q23 all map within intron 8 of the WWOX gene (2, 6, 41). Translocation t(14;16)(q32;q23) is an indicator of poor prognosis and usually associates with monosomy 13 (42). Further pointing to the highly recombinogenic nature of the WWOX locus, very recently it was reported the generation of chimeric transcripts involving WWOX and the PVT1 gene (a gene next to the MYC locus) in the RPMI8226 cell line harboring der(16)t(16;22)ins(16;8)(q23;q24), in which PVT1 exon 1 was fused to WWOX exon 9 (43). Additionally deletion of the WWOX locus is observed in a significant percentage of MM patients (20–35%), as determined by array CGH studies (44) and SNP arrays (45–47). Furthermore, it was determined that del(16q) was an independent adverse prognostic indicator (45). It was further determined that LOH at 16q23.1 was significantly associated with loss of WWOX expression in MM cases (46). Dickens and coworkers characterized regions of homozygous deletion in MM and WWOX was one of 29 genes identified as affected by homozygous deletions in significant numbers of MM cases (48).
In summary, loss of WWOX expression in MM is frequent and occurs as the consequence of multiple mechanisms including gross chromosomal rearrangements such as deletion and translocations. Additionally, t(14;16)(q32;q23) and 16q losses are both associated with poor prognosis in this disease.
7.1 Mouse models of WWOX ablation
Since expression of WWOX is lost in many types of cancer, mouse models of Wwox ablation have been generated to mimic this loss and characterize the subsequent effects on development and tumorigenesis. To this end, a mouse Wwox hypomorphic model (49) and two full knockout lines (50, 51) have been generated by different laboratories. The Wwox hypomorphic model was generated by means of gene trap insertion technology targeting the Wwox gene (Wwoxgt/gt). Wwox expression was undetectable in embryos (10.5 dpc) and most adult tissues. However, this model was deemed a functional hypomorph because low levels of protein expression could be detected in some tissues such as testes (49). Phenotypic analysis revealed that Wwoxgt/gt mice were viable but had a significantly shorter lifespan than their wild type (WT) counterparts (p=0.02). During the first 18 months of life 23% of Wwoxgt/gt died compared to WT and it was not possible to determine the causes of death. However, at the end of the survival experiment Wwoxgt/gt female hypomorphs displayed a significant increase in malignant neoplasias compared to WT littermates (p=0.015), with the majority of tumors consisting of B-cell lymphomas. Reduced fertility was also observed due to testicular atrophy in hypomorphic males (49).
Two full Wwox knockout mouse models have been generated using either a targeting construct to delete large regions of the Wwox gene (50) or using mice harboring loxP sites flanking exon 1 of the gene (Wwoxfl/fl) crossed to mice expressing Cre-recombinase under control of the adenovirus EIIA promoter (EIIA-Cre)(51). Both full knockout models displayed complete ablation of Wwox protein in all tissues examined with heterozygotes expressing ~50% of WT levels. It was also found that Wwox KO mice die postnatally as early as 72 hours after birth with none living longer than 3–4 weeks. The lifespan of the Wwox heterozygotes was indistinguishable from WT mice. Further phenotypic characterization revealed that Wwox ablation resulted in significant growth retardation (dwarfism) that was noticeable at birth. Serum chemistries of Wwox KO mice revealed hypoglycemia, hypocalcemia as well as signs of metabolic acidosis and kidney failure (50, 51). Interestingly, bone histomorphometry showed that Wwox KO mice have severe defects in bone formation in part due to increased osteoclastic activity (50, 51). Much like the hypomorphic Wwoxgt/gt mouse, Wwox KO mice were found to have gonadal abnormalities such as testicular hypoplasia in males and reduced ovarian size in females. These effects were suggested to be the result of impaired steroidogenesis (52).
One of the principal aims of ablating Wwox in mouse tissues was to determine whether or not this gene behaves as a classical tumor suppressor. Analysis of spontaneous tumor formation in the two short-lived full Wwox KO models differed. Aqeilan et al reported spontaneous formation of focal lesions along the diaphysis of Wwox KO mouse limbs that appeared neoplastic and were reported as suggestive of either periosteal or chondroid osteosarcoma (50), but this was observed in only 4 pups out of 13 analyzed. In contrast, analysis of EIIA-Cre; Wwox KO mice found no evidence of spontaneous neoplasia in any tissue examined. This was also the case for EIIA-Cre+;Wwox+/flox heterozygous mice indicating that loss of one Wwox allele (i.e. haploinsufficiency) appears not to be deleterious or carcinogenic in the longer-lived heterozygous mice (51). Increase tumorigenicity was reported when Wwox heterozygous mice are exposed to chemical carcinogens (50, 53) or when backcrossed into a C3H mammary tumor susceptible genetic background (54).
7.2 Conditional Targeted Wwox Deletion
Due to the early postnatal death of full Wwox KO mice the phenotypic analysis of Wwox ablation in adult tissues was impossible. To this end conditional, tissue-specific ablation of Wwox was achieved by utilizing Wwoxfl/fl mice and crossing them with either mice expressing Cre recombinase under control of the bovine keratin 5 promoter (BK5-Cre) or under control of the MMTV promoter (MMTV-Cre) for ablation in the mammary gland and other epithelial tissues (55).
Microscopic and morphometric analyses on the effects of Wwox ablation in both conditional deletion models revealed a significant defect in mammary branching morphogenesis in virgin females. However, this defect did not impede the mammary epithelium from reaching functional maturity during pregnancy as these mice were eventually able to lactate normally. By transcriptome analyses we determined that expression of the non-canonical Wnt pathway ligand Wnt5a, a protein known to inhibit ductal extension and lateral branching (56), was significantly upregulated in Wwox KO mammary epithelium. In addition we observed a significant upregulation and activation of the IL6-Jak-STAT3 pathway and levels of phospho-STAT3 were greatly increased in Wwox KO mammary epithelium from virgin mice (55). This increase in Jak-STAT3 signaling could be one of the mechanisms by which Wnt5a is transcriptionally upregulated (57) and subsequently inhibits ductal morphogenesis. Interestingly however, as will be discussed in following sections, Wnt5a is also a direct transcriptional target of the TGFβ/SMAD signaling pathway (56).
BK5-Cre;Wwox KO mice were viable, fertile and females were able to lactate. Unexpectedly, these mice died prematurely (68–117 days of age) independent of gender for yet undetermined reasons. In contrast, Wwox ablation via MMTV-Cre had no effect on survival or natural aging. After 1.5 years of follow up of the MMTV-Cre;Wwox KO mice we determined that loss of Wwox expression is not carcinogenic in any tissue. Similarly, we did not detect any evidence of malignant development in the shorter-lived BK5-Cre;Wwox KO mice. Furthermore, no evidence of increase proliferation or development of premalignant lesions was observed. Consistent with the findings in the EIIA-Cre model, we found that loss of a single Wwox allele (i.e. haploinsufficiency) did not have any observable phenotypic effect in the mammary gland (55).
We further analyzed conditional Wwox ablation in additional tissues such as in bone (osteoblasts) using Osterix-Cre (Osx-Cre) mice and in liver using Albumin-Cre (Alb-Cre) mice. Similarly to the described mammary gland models we found no evidence of spontaneous tumor formation in the targeted tissues (unpublished observations and (58). The observations with the various models of Wwox ablation are summarized in Table 2.
Table 2.
Rodent models of WWOX ablation
| Model | Target Tissue | Observed phenotypes | Tumors | Reference |
|---|---|---|---|---|
| Wwoxgt/gt hypomorph mouse | All tissues |
|
B-cell lymphomas in older female mice | 49 |
| Wwox −/− mouse | All tissues |
|
Lesions compatible with periosteal osteosarcomas in 4 of 13 KO mice | 50,52 |
| EIIA-Cre;Wwoxfl/fl mouse | All tissues |
|
No | 51 |
| Ide/Ide rat, spontaneous Wwox mutation | All tissues |
|
No | 103 |
| BK5-Cre;Wwoxfl/fl mouse | Skin, mammary gland and other Keratin5 expressing epithelia |
|
No | 55 |
| MMTV-Cre; Wwoxfl/fl mouse | Mammary & salivary glands |
|
No | 55 |
| Alb-Cre; Wwoxfl/fl mouse | Liver |
|
No | 58 |
| Osx-Cre; Wwoxfl/fl mouse | Bone (osteoblasts) |
|
No | Aldaz Lab, unpublished |
8.1 WWOX in Cellular Pathways
As earlier mentioned, the WWOX gene encodes a 46 kD cytoplasmic protein containing two functional protein domains, two amino-terminal WW-domains and a carboxy-terminal short chain dehydrogenase domain (SDR) (Fig. 1). The SDR is an enzymatic domain that is predicted to carry out NADP(H)-dependent dehydrogenase reactions with yet to be identified substrates.
We determined that the WWOX protein resides predominantly in the perinuclear region overlapping with the Golgi region (23). To more precisely define the sequences responsible for perinuclear targeting, we used site directed mutagenesis on GFP-WWOX fusion proteins, to generate large exonic deletions of the SDR domain while keeping the WW domains intact and vice versa deleting the WW domains and preserving the SDR domain. We concluded that proper sub-cellular localization is affected by disruption of the SDR domain (59). Furthermore, we generated GFP-WWOX proteins containing single point mutations (S281A, Y293F, and K297A) destroying WWOX’s catalytic activity. Remarkably, we observed that the amino acids predicted to be required for WWOX enzymatic activity were also necessary for perinuclear localization (Aldaz laboratory unpublished observations).
In contrast to our observations Chang et al. reported that WWOX localizes to mitochondria and to the nucleus and these investigators speculated that WWOX translocates between these compartments depending on specific stimuli (5, 60). So far we were unable to detect WWOX protein in the nuclear compartment or in mitochondria (23, 59) the reasons for this discrepancy remain unclear. A possibility exists however, that WWOX may translocate from different cellular compartments upon very specific signals and at a very rapid rate making difficult its detection in the nucleus or mitochondria.
WW domains are involved in protein-protein interactions with modular proline-rich recognition motifs. The highly compact WW domains (35–45AA in length) are characterized by the presence of a pair of conserved tryptophans (W). These two signature W residues are spaced approximately 20–22 AA apart and play a fundamental role in the structure and function of the domain. WW domains have a very diverse proline-containing sequence preference. Based on such binding preference four groups of WW domains have been described. Two are major and more common, Groups I and II and two less common, Groups III and IV (61). Group I binds the minimal core consensus Pro-Pro-x-Tyr (PPxY). Examples of WW domain containing proteins from Group I include YAP1, Dystrophin and NEDD4 family members. We and others demonstrated that WWOX WW1 is a Group I WW domain that specifically binds peptides containing the consensus PPxY motif (62, 63). The WWOX WW2 is not a classical WW domain due to the replacement of the second signature tryptophan with a tyrosine at position 85. In a recent report that analyzed the interaction between WWOX WW domains and ERBB4-CTF, it was shown that the WW2 domain does not bind to consensus PPxY motifs but does augment the ability of WW1 to do so (64).
Several proteins containing the PPxY consensus motif have been identified as WWOX interacting partners such as p73, AP2γ, JUN, EZRIN, ERBB4 (CTF), MET (CTF) and RUNX2. The possible consequences and cancer related implications of these various interactions have been reviewed elsewhere (65). Several of these proteins are transcription factors and due to the primarily perinuclear localization of WWOX these binding events usually result in the cytoplasmic sequestration and subsequent inhibition of nuclear activity. For instance WWOX has been shown to suppress the transcriptional activity of two p53, homologues, p73 and ΔNp63α. Inhibition of p73 was reported to result in an increase in apoptosis achieved by sequestering p73 in the cytoplasm (66). The WWOX interaction with ΔNp63α has been suggested to result in stabilizing ΔNp63α by antagonizing the function of the E3 ubiquitin ligase ITCH while also inhibiting ΔNp63α nuclear translocation and transactivation. It was also reported that inhibition of ΔNp63α function by WWOX reverses the resistance of cancer cells to cisplatin (67). More recently it was suggested a functional role of WWOX as an inhibitor of the Wnt/beta catenin pathway also due to interaction and cytoplasmic sequestration of Dishevelled family member proteins (68).
Although the physiological relevance of several of the described in vivo interactions requires further research, it is clear that a role for WWOX is emerging as a regulator of transcription by limiting transcription factor access to the nucleus through cytoplasmic sequestration. It is also very likely that these interactions and effects of WWOX on downstream target gene expression may be dependent on cell and tissue context.
8.2 Signaling Pathways and Biofunctions Associated with WWOX
Some authors originally proposed that WWOX behaves as a pro-apoptotic molecule (5). Furthermore, WWOX overexpression has been reported to induce apoptosis in lung (69), prostate (70) and breast (71) cancer lines among others; in most of these cases the delivery of WWOX was via adenoviral vectors. On the other hand we and others, never observed or reported apoptosis induction in cancer lines from breast, liver, lung or ovarian origin either transiently or stably transfected for WWOX expression (23, 72, 73). In agreement with Gourley et al (72), we hypothesize that WWOX per se is not pro-apoptotic when overexpressed in cancer cells devoid of WWOX expression and the reported pro-apoptotic effect is highly dependent on using delivery vectors that induced significant cellular stress such as adenoviral vectors. Thus the physiological biofunctions and cellular pathways associated with WWOX remain to be better defined.
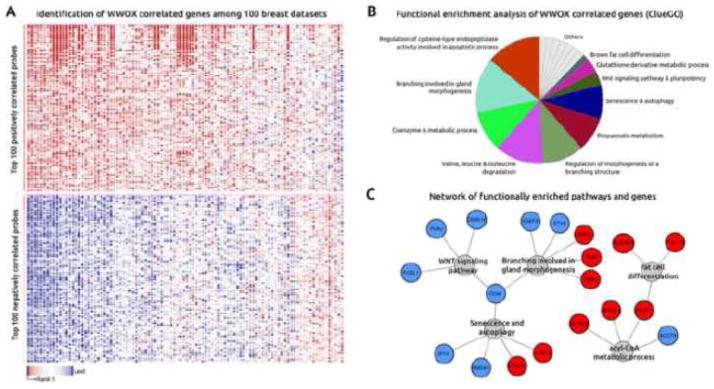
To identify signaling pathways and biofunctions associated with WWOX expression, we employed the “guilt by association” principle, which states that gene co-expression might indicate shared regulatory mechanisms and roles in related biological processes (74, 75). First, we identified the top WWOX co-expressed genes among 100 breast datasets (approximately 4800 samples, including normal, tumor and breast cancer lines) by using the Multiexperiment Matrix (http://biit.cs.ut.ee/mem/) bioinformatics resource. Briefly, we identified the 200 top positively and negatively correlated genes based on the expression profiles of the most reliable WWOX probe sets according to the JetSet (http://www.cbs.dtu.dk/biotools/jetset/) scoring approach (219077_s_at and 210695_s_at probes) (Figure 8A). Second, we used ClueGO (76) and CluePedia Cytoscape’s plug-in (77) for functional analyses, network generation and visualization of the WWOX co-expressed genes. In addition, we employed the Enrichr online resource (http://amp.pharm.mssm.edu/Enrichr) for ChIP enrichment analysis (ChEA) (78). Among the top enriched biofunctions, we found regulation of mammary gland morphogenesis and branching, coenzyme A metabolic process and regulation of cysteine-type endopeptidase activity among other metabolic processes. WNT signaling pathway, senescence/autophagy and fat cell differentiation were also identified as enriched biofunctions associated with the list of WWOX co-expressed genes (Figure 8B,C). These results are consistent with previous studies that showed the correlation between WWOX and ER-alpha expression in human breast carcinomas (12, 13) and the effects of WWOX ablation impairing mouse mammary branching morphogenesis (55). Importantly and as discussed in previous sections of this review, recent studies indeed suggested that WWOX is a novel modulator of the canonical and non-canonical WNT signaling pathways (55, 68). In addition, ChIP enrichment analysis allowed us to identify a set of transcription factors that are the most likely to have regulated WWOX associated gene expression changes. We detected a statistically significant enrichment of SOX2 and SMAD transcription factors that are likely responsible for many of the gene sets co-expressed with WWOX. Interestingly and linking the previous in vivo (55) and these in silico observations, mammary gland branching morphogenesis is intimately link as well with the TGFβ/SMAD signaling pathway (79, 80). It is worth noting, as earlier mentioned, that indeed Wnt5a is a transcriptional target of the TGFβ/SMAD signaling pathway (56).
Figure 8.
8.3 WWOX As a Regulator of TGFβ Signaling
We recently demonstrated that ablation of WWOX in normal breast cells resulted in significant overexpression of a number of TGFβ/SMAD3 target genes known to be essential for cancer progression such as ANGPTL4, PTHLH, FST and SERPINE1 (81). We ultimately showed that WWOX acts as an inhibitor of SMAD3 transcriptional activity by sequestering it in the cytoplasm upon TGFβ stimulation and inhibiting its occupation of target gene promoters. This inhibition relies upon a WW1 domain-dependent interaction with the SMAD3 181PPGY184 motif.
TGFβ signaling in breast cells has been an intense and ongoing area of research due to the effects of this signaling pathway being paradoxical. In normal mammary epithelium TGFβ signaling acts in a tumor suppressive fashion inhibiting cell growth (79) by activating the expression of cyclin dependent kinase inhibitors such as CDKN2B (p15) and CDKN1A (p21) and repressing genes involved in cell proliferation such as MYC (82–84). However, the effect and transcriptional output of TGFβ signaling changes dramatically when breast cancers progress to advanced stages, becoming pro-oncogenic and activating subsets of genes involved in cell proliferation, epithelial-to-mesenchymal transition and metastasis (85–87). The key switches explaining the so-called ‘TGFβ paradox’ as tumors progress are still not well understood (88).
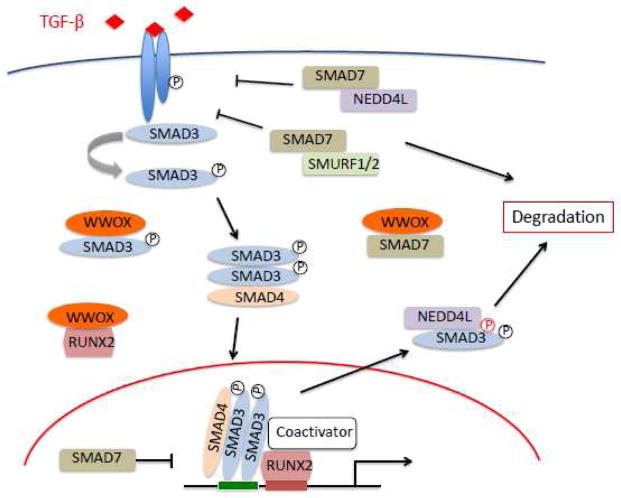
We hypothesize that WWOX is a key component contributing to the regulation and homeostasis of the TGFβ pathway likely in two critical ways: Firstly by binding and preventing the nuclear translocation of receptor-regulated SMADS (R-SMADs) that all contain PPxY motifs (PY box) such as SMAD3 (regardless of their phosphorylation status) and of key co-activators such as RUNX2, thus preventing the transcriptional activation of pro-metastatic genes. Secondly by competing with WW domain containing NEDD4 family ubiquitin ligases such as NEDD4L in charge of degradation of R-SMADs but more importantly also of inhibitory SMAD7 that contains a PY box as well (89). In yet unreported studies we indeed observed a direct interaction between WWOX and SMAD7. In other words when WWOX is available inhibitory SMAD7 would be protected from degradation, thus serving as a potential reservoir of this important inhibitory protein (Figure 9).
Figure 9.
In summary we propose that the progressive loss of WWOX expression in advanced breast cancer would contribute to deregulating the TGFβ pathway and may explain, at least in part, the pro-metastatic effects resulting from TGFβ/SMAD3 hyperactive signaling in breast cancer. In support of this hypothesis meta-analysis of three independent breast cancer datasets representing a total of 819 patient samples showed a significant negative correlation between WWOX and ANGPTL4. Furthermore, the WWOXlo/ANGPTL4hi subgroup was enriched in basal-like and triple-negative breast cancer (81).
9. Emerging Role of WWOX in Other Human Pathologies
9.1 WWOX in Lipid Disorders and Metabolic Syndrome Related Conditions
Genome wide association studies (GWAS) from various laboratories led to the identification of a region-wide significant association between low levels of high-density lipoprotein cholesterol (HDL-C) and the WWOX gene in dyslipidemic families and low HDL-C cases and controls, as well as in population-based cross sectional and prospective cohorts. Low HDL-C is a well- known major risk factor for coronary artery disease. One particular SNP, rs2548861 in intron 8 of WWOX, demonstrated a very significant association (p= 6.9×10−7) with low HDL-C levels (90). Other studies observed not only and association with HDL-C but with triglycerides levels as well (91, 92). Very recently resequencing studies by Iatan et al. (58) identified 8 genomic variants significantly associated and segregating with the low-HDL trait in two French Canadian dyslipidemic families. Furthermore in the same study by gene expression profiling we determined that liver specific Wwox conditional KO mice displayed significantly altered lipid metabolic pathways and increased plasma triglyceride levels in female mice, suggesting a significant role for WWOX in modulating HDL and lipid metabolism (58). The association of the Wwox locus with lipid metabolism is also supported by mouse quantitative trait loci (QTL) maps (93).
Interestingly, multiple studies have implicated the WWOX locus with other metabolic syndrome related conditions such as type 2 diabetes (94, 95), hypertension susceptibility (96), coronary artery calcification (97), obesity (98), and left ventricular thickness (99). Furthermore, by exploring the Phenotype Genotype Integrator resource (PheGenI, http://www.ncbi.nlm.nih.gov/gap/phegeni) that merges NHGRI GWAS data with various databases including dbGAP, it is possible to observe an emerging theme of metabolic syndrome related traits significantly associated with SNPs mapping to the WWOX locus including: body mass index, body weight, C-reactive protein levels, insulin levels, cardiovascular disease, cholesterol, blood pressure and various traits asocciated with cardiovascular parameters among others. Most of these data derives from the NHLBI’s Framingham SNP health Association Resource and the Family Heart Study datasets among other sources (Supplementary Table 1).
9.2. Germline Loss-of-function WWOX mutations lead to Epilepsy, Ataxia and Mental Retardation
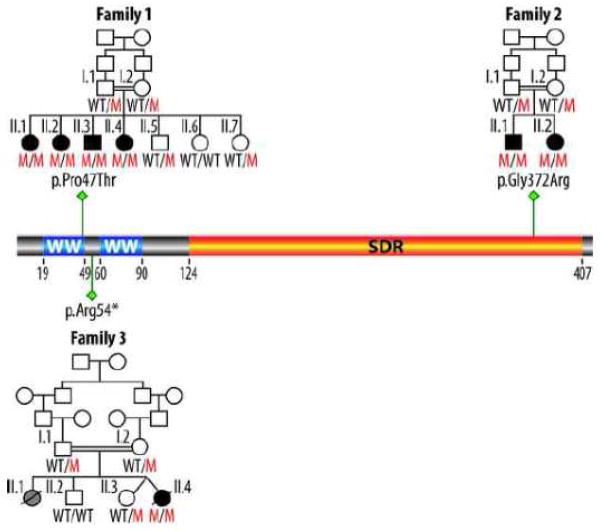
In 2007 Gribaa et al. reported a new form of childhood onset autosomal recessive cerebellar ataxia and epilepsy in a large consanguineous family from Saudi Arabia with four siblings affected and mapped its locus to chromosome 16q21-q23 (100), SCAR12, MIM# 614322). Very recently by exome sequencing it was determined that WWOX is homozygously mutated in all four children affected by the described phenotype (101). The mutation Pro47Thr affects an extremely conserved proline residue that is a critical component of the first WW domain consensus sequence of WWOX. We demonstrated in the same report that this mutation renders WWOX unable of binding PPxY target motifs in interacting partner proteins (101). A second consanguineous family was identified carrying a highly conserved homozygous mutation Gly372Arg of the WWOX protein and the phenotype of the two affected siblings in this family also showed early-onset generalized tonic-clonic epilepsy, metal retardation and ataxia. In this case the mutation affects a glycine found at the C-terminus in the oxidoreductase domain of WWOX and its functional consequences are less clear (101). Interestingly, an independent report very recently confirmed these observations by describing a new case from another consanguineous family affected by a very similar although more severe phenotype characterized by severe growth retardation, microcephaly, epileptic seizures, retinopathy and early death at 16 months of age. In this case the homozygous WWOX germline alteration is a nonsense mutation at arginine 54 (pArg54*) leading to a virtual WWOX human full knockout case (102). The homozygous mutations leading to the described human syndromes are summarized in Figure 10.
Figure 10.
Previously Suzuki et al. (103) reported a phenotype reminiscent to that now observed in humans by analyzing the Ide/Ide rats that are characterized by dwarfism, postnatal lethality, male hypogonadism and epilepsy (Table 2). Interestingly, these authors concluded that the Ide mutation is a 13 bp deletion in exon 9 of Wwox leading to a frameshift of the last 44 codons of Wwox and replacement with a novel open reading frame of 54 codons. These authors could not detect Wwox protein by means of western blot suggesting that this mutation leads to full knockout of the protein.
In the above described report of Mallaret et al. (101) we also demonstrated that Wwox full KO mice display an extremely similar phenotype to that observed in humans and rats, since Wwox KO mice in their short life-span of 3–4 weeks develop spontaneous and audiogenic epileptic seizures as well (Fig. 11).
Figure 11.
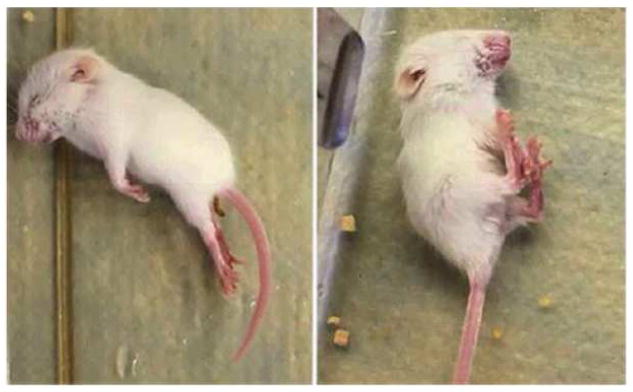
In summary, although homozygous mutations affecting WWOX in humans are likely to be rare the described findings underscore the likelihood that WWOX plays a critical role in normal CNS development and physiology.
Concluding Remarks
In this review we revisited the status of WWOX in cancer and described its emerging role in other important human pathologies. Much still remains to be learned on the physiological role of WWOX, although numerous interacting protein partners have been identified we understand very little on the enzymatic role of WWOX, the true substrate/s of this oxidoreductase remain to be identified.
The mechanistic association of WWOX loss of function with tumor progression warrants further investigation as does the potential role of this protein as modulator of lipid metabolism and its association with metabolic syndrome related conditions.
Finally, the recent identification of the first humans carrying homozygous germline WWOX mutations characterized as suffering early onset epilepsy, ataxia and mental retardation, undoubtedly indicates a key function of this protein in the CNS. The mechanisms on how loss of WWOX function causes such severe neurological syndromes remain to be defined.
Supplementary Material
Highlights.
WWOX deletions and loss of expression associate with poor prognosis in cancer
Mouse KO models indicate that WWOX does not behave as a classical tumor suppressor
WWOX behaves as a modulator of the TGFβ/SMAD and WNT signaling pathways
WWOX modulates lipid metabolism and is associated with metabolic syndrome traits
Human germline WWOX mutations cause ataxia with epilepsy and mental retardation
Acknowledgments
Financial Support: This study was supported by the National Institutes of Health/National Cancer Institute [grant number R01 CA102444-8] to [CMA]; Research Training in Carcinogenesis and Mutagenesis [grant number T32CA009480] to [BWF] and Cancer Center Support Grant [grant number CA016672].
Footnotes
Conflict of Interest Statement: The authors disclose no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen T, Sahin A, Aldaz CM. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res. 1996;56(24):5605–9. [PubMed] [Google Scholar]
- 2.Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–24.1, a region frequently affected in breast cancer. Cancer Res. 2000;60(8):2140–5. [PubMed] [Google Scholar]
- 3.Chesi M, Bergsagel PL, Shonukan OO, Martelli ML, Brents LA, Chen T, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91(12):4457–63. [PubMed] [Google Scholar]
- 4.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, et al. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet. 2000;9(11):1651–63. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- 5.Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, et al. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J Biol Chem. 2001;276(5):3361–70. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- 6.Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer. 2002;34(2):154–67. doi: 10.1002/gcc.10047. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer cell. 2007;12(3):215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 9.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444(7118):499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 10.Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature. 2006;440(7081):242–5. doi: 10.1038/nature04559. [DOI] [PubMed] [Google Scholar]
- 11.Nunez M, Ludes-Meyers J, Aldaz C. WWOX protein expression in normal human tissues. J Mol Hist. 2006;37:115–25. doi: 10.1007/s10735-006-9046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guler G, Uner A, Guler N, Han SY, Iliopoulos D, Hauck WW, et al. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer. 2004;100(8):1605–14. doi: 10.1002/cncr.20137. [DOI] [PubMed] [Google Scholar]
- 13.Nunez MI, Ludes-Meyers J, Abba MC, Kil H, Abbey NW, Page RE, et al. Frequent loss of WWOX expression in breast cancer: correlation with estrogen receptor status. Breast Cancer Res Treat. 2005;89(2):99–105. doi: 10.1007/s10549-004-1474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nunez MI, Rosen DG, Ludes-Meyers JH, Abba MC, Kil H, Page R, et al. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC cancer. 2005;5:64. doi: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, et al. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10(9):3053–8. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 16.Park SW, Ludes-Meyers J, Zimonjic DB, Durkin ME, Popescu NC, Aldaz CM. Frequent downregulation and loss of WWOX gene expression in human hepatocellular carcinoma. Br J Cancer. 2004;91(4):753–9. doi: 10.1038/sj.bjc.6602023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donati V, Fontanini G, Dell’Omodarme M, Prati MC, Nuti S, Lucchi M, et al. WWOX expression in different histologic types and subtypes of non-small cell lung cancer. Clin Cancer Res. 2007;13(3):884–91. doi: 10.1158/1078-0432.CCR-06-2016. [DOI] [PubMed] [Google Scholar]
- 18.Dias EP, Pimenta FJ, Sarquis MS, Dias Filho MA, Aldaz CM, Fujii JB, et al. Association between decreased WWOX protein expression and thyroid cancer development. Thyroid. 2007;17(11):1055–9. doi: 10.1089/thy.2007.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloomston M, Kneile J, Butterfield M, Dillhoff M, Muscarella P, Ellison EC, et al. Coordinate loss of fragile gene expression in pancreatobiliary cancers: correlations among markers and clinical features. Ann Surg Oncol. 2009;16(8):2331–8. doi: 10.1245/s10434-009-0507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paige AJ, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, et al. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. Proc Natl Acad Sci U S A. 2001;98(20):11417–22. doi: 10.1073/pnas.191175898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: Clinical significance and future implications. Exp Biol Med. 2014;239(3):253–63. doi: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- 22.Ramos D, Abba M, Lopez-Guerrero JA, Rubio J, Solsona E, Almenar S, et al. Low levels of WWOX protein immunoexpression correlate with tumour grade and a less favourable outcome in patients with urinary bladder tumours. Histopathology. 2008;52(7):831–9. doi: 10.1111/j.1365-2559.2008.03033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, et al. WWOX, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61(22):8068–73. [PubMed] [Google Scholar]
- 24.Driouch K, Prydz H, Monese R, Johansen H, Lidereau R, Frengen E. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at high levels in human breast tumors. Oncogene. 2002;21(12):1832–40. doi: 10.1038/sj.onc.1205273. [DOI] [PubMed] [Google Scholar]
- 25.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, et al. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24(9):1625–33. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 26.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65(22):10514–23. doi: 10.1158/0008-5472.CAN-05-1127. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, et al. Characterizing WW Domain Interactions of Tumor Suppressor WWOX Reveals its Association with Multiprotein Networks. J Biol Chem. 2014 Mar 28;289(13):8865–80. doi: 10.1074/jbc.M113.506790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CJ, Shen WG, Peng SY, Cheng HW, Kao SY, Lin SC, et al. miR-134 induces oncogenicity and metastasis in head and neck carcinoma through targeting WWOX gene. Int J Cancer. 2014;134(4):811–21. doi: 10.1002/ijc.28358. [DOI] [PubMed] [Google Scholar]
- 29.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104(50):20007–12. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1, 1–19. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 39.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergsagel PL, Kuehl WM. Chromosome translocations in multiple myeloma. Oncogene. 2001;20(40):5611–22. doi: 10.1038/sj.onc.1204641. [DOI] [PubMed] [Google Scholar]
- 42.Sawyer JR, Lukacs JL, Munshi N, Desikan KR, Singhal S, Mehta J, et al. Identification of new nonrandom translocations in multiple myeloma with multicolor spectral karyotyping. Blood. 1998;92(11):4269–78. [PubMed] [Google Scholar]
- 43.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72(19):4954–62. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 44.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer cell. 2006;9(4):313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110(9):3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 46.Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48(7):603–14. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- 47.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116(15):e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 48.Dickens NJ, Walker BA, Leone PE, Johnson DC, Brito JL, Zeisig A, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16(6):1856–64. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludes-Meyers JH, Kil H, Nunez MI, Conti CJ, Parker-Thornburg J, Bedford MT, et al. WWOX hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer. 2007;46(12):1129–36. doi: 10.1002/gcc.20497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aqeilan R, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, et al. Targeted deletion of WWOX reveals a tumor suppressor function. Proc Natl Acad Sci U S A. 2007;104(10):3949–54. doi: 10.1073/pnas.0609783104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludes-Meyers JH, Kil H, Parker-Thornburg J, Kusewitt DF, Bedford MT, Aldaz CM. Generation and characterization of mice carrying a conditional allele of the Wwox tumor suppressor gene. PLoS ONE. 2009;4(11):e7775. doi: 10.1371/journal.pone.0007775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, Gaudio E, et al. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology. 2009;150(3):1530–5. doi: 10.1210/en.2008-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aqeilan RI, Hagan JP, Aqeilan HA, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res. 2007;67(12):5606–10. doi: 10.1158/0008-5472.CAN-07-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdeen SK, Salah Z, Maly B, Smith Y, Tufail R, Abu-Odeh M, et al. Wwox inactivation enhances mammary tumorigenesis. Oncogene. 2011;30(36):3900–6. doi: 10.1038/onc.2011.115. [DOI] [PubMed] [Google Scholar]
- 55.Ferguson BW, Gao X, Kil H, Lee J, Benavides F, Abba MC, et al. Conditional Wwox deletion in mouse mammary gland by means of two Cre recombinase approaches. PLoS ONE. 2012;7(5):e36618. doi: 10.1371/journal.pone.0036618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roarty K, Serra R. Wnt5a is required for proper mammary gland development and TGF-beta-mediated inhibition of ductal growth. Development. 2007;134(21):3929–39. doi: 10.1242/dev.008250. [DOI] [PubMed] [Google Scholar]
- 57.Katoh M. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer (Review) Int J Mol Med. 2007;19(2):273–8. [PubMed] [Google Scholar]
- 58.Iatan I, Choi HY, Ruel I, Reddy MP, Hyunsuk K, Lee J, et al. The WWOX Gene Modulates HDL and Lipid Metabolism. Circ Cardiovasc Genet. 2014 May 28; doi: 10.1161/CIRCGENETICS.113.000248. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet Genome Res. 2003;100(1–4):101–10. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang NS, Doherty J, Ensign A, Schultz L, Hsu LJ, Hong Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem. 2005;280(52):43100–8. doi: 10.1074/jbc.M505590200. [DOI] [PubMed] [Google Scholar]
- 61.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103(7):1001–4. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 62.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene. 2004;23(29):5049–55. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the AP-2gamma transcription factor. Cancer Res. 2004;64(22):8256–61. doi: 10.1158/0008-5472.CAN-04-2055. [DOI] [PubMed] [Google Scholar]
- 64.Schuchardt BJ, Bhat V, Mikles DC, McDonald CB, Sudol M, Farooq A. Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase. Biochemistry. 2013;52(51):9223–36. doi: 10.1021/bi400987k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salah Z, Aqeilan R, Huebner K. WWOX gene and gene product: tumor suppression through specific protein interactions. Future Oncol. 2010;6(2):249–59. doi: 10.2217/fon.09.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci U S A. 2004;101(13):4401–6. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salah Z, Bar-mag T, Kohn Y, Pichiorri F, Palumbo T, Melino G, et al. Tumor suppressor WWOX binds to DeltaNp63alpha and sensitizes cancer cells to chemotherapy. Cell Death Dis. 2013;4:e480. doi: 10.1038/cddis.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bouteille N, Driouch K, Hage PE, Sin S, Formstecher E, Camonis J, et al. Inhibition of the Wnt/beta-catenin pathway by the WWOX tumor suppressor protein. Oncogene. 2009;28(28):2569–80. doi: 10.1038/onc.2009.120. [DOI] [PubMed] [Google Scholar]
- 69.Fabbri M, Iliopoulos D, Trapasso F, Aqeilan R, Cimmino A, Zanesi N, et al. WWOX gene restoration prevents lung cancer growth in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102(43):15611–6. doi: 10.1073/pnas.0505485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, et al. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66(13):6477–81. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 71.Iliopoulos D, Fabbri M, Druck T, Qin HR, Han SY, Huebner K. Inhibition of breast cancer cell growth in vitro and in vivo: effect of restoration of Wwox expression. Clin Cancer Res. 2007;13(1):268–74. doi: 10.1158/1078-0432.CCR-06-2038. [DOI] [PubMed] [Google Scholar]
- 72.Gourley C, Paige AJ, Taylor KJ, Ward C, Kuske B, Zhang J, et al. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin alpha3. Cancer Res. 2009;69(11):4835–42. doi: 10.1158/0008-5472.CAN-08-2974. [DOI] [PubMed] [Google Scholar]
- 73.Becker S, Markova B, Wiewrodt R, Hoffarth S, Hahnel PS, Pleiner S, et al. Functional and clinical characterization of the putative tumor suppressor WWOX in non-small cell lung cancer. J Thorac Oncol. 2011;6(12):1976–83. doi: 10.1097/JTO.0b013e31822e59dd. [DOI] [PubMed] [Google Scholar]
- 74.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–55. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 75.Oti M, van Reeuwijk J, Huynen MA, Brunner HG. Conserved co-expression for candidate disease gene prioritization. BMC Bioinformatics. 2008;9:208. doi: 10.1186/1471-2105-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–3. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–44. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silberstein GB, Daniel CW. Reversible inhibition of mammary gland growth by transforming growth factor-beta. Science. 1987;237(4812):291–3. doi: 10.1126/science.3474783. [DOI] [PubMed] [Google Scholar]
- 80.Silberstein GB, Flanders KC, Roberts AB, Daniel CW. Regulation of mammary morphogenesis: evidence for extracellular matrix-mediated inhibition of ductal budding by transforming growth factor-beta 1. Dev Biol. 1992;152(2):354–62. doi: 10.1016/0012-1606(92)90142-4. [DOI] [PubMed] [Google Scholar]
- 81.Ferguson BW, Gao X, Zelazowski MJ, Lee J, Jeter CR, Abba MC, et al. The cancer gene WWOX behaves as an inhibitor of SMAD3 transcriptional activity via direct binding. BMC cancer. 2013;13:593. doi: 10.1186/1471-2407-13-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110(1):19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 83.Feng XH, Liang YY, Liang M, Zhai W, Lin X. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGF-beta-mediated induction of the CDK inhibitor p15(Ink4B) Mol Cell. 2002;9(1):133–43. doi: 10.1016/s1097-2765(01)00430-0. [DOI] [PubMed] [Google Scholar]
- 84.Pardali K, Kurisaki A, Moren A, ten Dijke P, Kardassis D, Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21(Waf1/Cip1) regulation by transforming growth factor-beta. J Biol Chem. 2000;275(38):29244–56. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- 85.Barcellos-Hoff MH, Akhurst RJ. Transforming growth factor-beta in breast cancer: too much, too late. Breast cancer research : BCR. 2009;11(1):202. doi: 10.1186/bcr2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moses H, Barcellos-Hoff MH. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb Perspect Biol. 2011;3(1):a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Padua D, Massague J. Roles of TGFbeta in metastasis. Cell Res. 2009;19(1):89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 88.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aragon E, Goerner N, Xi Q, Gomes T, Gao S, Massague J, et al. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-beta Pathways. Structure. 2012;20(10):1726–36. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JC, Weissglas-Volkov D, Kyttala M, Dastani Z, Cantor RM, Sobel EM, et al. WW-domain-containing oxidoreductase is associated with low plasma HDL-C levels. Am J Hum Genet. 2008;83(2):180–92. doi: 10.1016/j.ajhg.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40(2):161–9. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saez ME, Gonzalez-Perez A, Martinez-Larrad MT, Gayan J, Real LM, Serrano-Rios M, et al. WWOX gene is associated with HDL cholesterol and triglyceride levels. BMC medical genetics. 2010;11:148. doi: 10.1186/1471-2350-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leduc MS, Lyons M, Darvishi K, Walsh K, Sheehan S, Amend S, et al. The mouse QTL map helps interpret human genome-wide association studies for HDL cholesterol. J Lipid Res. 2011;52(6):1139–49. doi: 10.1194/jlr.M009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang YC, Chiu YF, Liu PH, Shih KC, Lin MW, Sheu WH, et al. Replication of genome-wide association signals of type 2 diabetes in Han Chinese in a prospective cohort. Clin Endocrinol. 2012;76(3):365–72. doi: 10.1111/j.1365-2265.2011.04175.x. [DOI] [PubMed] [Google Scholar]
- 95.Sakai K, Imamura M, Tanaka Y, Iwata M, Hirose H, Kaku K, et al. Replication study for the association of 9 East Asian GWAS-derived loci with susceptibility to type 2 diabetes in a Japanese population. PLoS ONE. 2013;8(9):e76317. doi: 10.1371/journal.pone.0076317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang HC, Liang YJ, Chen JW, Chiang KM, Chung CM, Ho HY, et al. Identification of IGF1, SLC4A4, WWOX, and SFMBT1 as hypertension susceptibility genes in Han Chinese with a genome-wide gene-based association study. PLoS ONE. 2012;7(3):e32907. doi: 10.1371/journal.pone.0032907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Polfus LM, Smith JA, Shimmin LC, Bielak LF, Morrison AC, Kardia SL, et al. Genome-wide association study of gene by smoking interactions in coronary artery calcification. PLoS ONE. 2013;8(10):e74642. doi: 10.1371/journal.pone.0074642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang K, Li WD, Zhang CK, Wang Z, Glessner JT, Grant SF, et al. A genome-wide association study on obesity and obesity-related traits. PLoS ONE. 2011;6(4):e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vasan RS, Glazer NL, Felix JF, Lieb W, Wild PS, Felix SB, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. JAMA. 2009;302(2):168–78. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gribaa M, Salih M, Anheim M, Lagier-Tourenne C, H’Mida D, Drouot N, et al. A new form of childhood onset, autosomal recessive spinocerebellar ataxia and epilepsy is localized at 16q21-q23. Brain. 2007;130(Pt 7):1921–8. doi: 10.1093/brain/awm078. [DOI] [PubMed] [Google Scholar]
- 101.Mallaret M, Synofzik M, Lee J, Sagum CA, Mahajnah M, Sharkia R, et al. The tumour suppressor gene WWOX is mutated in autosomal recessive cerebellar ataxia with epilepsy and mental retardation. Brain. 2014;137(Pt 2):411–9. doi: 10.1093/brain/awt338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abdel-Salam G, Thoenes M, Afifi HH, Korber F, Swan D, Bolz HJ. The supposed tumor suppressor gene WWOX is mutated in an early lethal microcephaly syndrome with epilepsy, growth retardation and retinal degeneration. Orphanet J Rare Dis. 2014;9(1):12. doi: 10.1186/1750-1172-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suzuki H, Katayama K, Takenaka M, Amakasu K, Saito K, Suzuki K. A spontaneous mutation of the Wwox gene and audiogenic seizures in rats with lethal dwarfism and epilepsy. Genes Brain Behav. 2009;8(7):650–60. doi: 10.1111/j.1601-183X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 104.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.