Abstract
Recessive dystrophic epidermolysis bullosa (RDEB) is an inherited blistering skin disorder caused by mutations in the COL7A1 gene-encoding type VII collagen (Col7), the major component of anchoring fibrils at the dermal-epidermal junction. Individuals with RDEB develop painful blisters and mucosal erosions, and currently there are no effective forms of therapy. Nevertheless, some advances in patient therapy are being made, and cell-based therapies with mesenchymal and hematopoietic cells have shown promise in early clinical trials. To establish a foundation for personalized, gene-corrected, patient-specific cell transfer, we generated induced pluripotent stem cells from three subjects with RDEB (RDEB iPS cells). We found that Col7 was not required for stem cell renewal and that RDEB iPS cells could be differentiated to both hematopoietic and non-hematopoietic lineages. The specific epigenetic profile associated with de-differentiation of RDEB fibroblasts and keratinocytes into RDEB iPS cells was similar to that observed in wild-type iPS cells. Importantly, human wild-type and RDEB iPS cells differentiated in vivo into structures resembling skin. Gene-corrected RDEB iPS cells expressed Col7. These data identify the potential of RDEB iPS cells to generate autologous hematopoietic grafts and skin cells with the inherent capacity to treat the skin and mucosal erosions that typify this genodermatosis.
Keywords: Recessive dystrophic epidermolysis bullosa, induced pluripotent stem cells, type VII collagen
INTRODUCTION
The severe blistering disease recessive dystrophic epidermolysis bullosa (RDEB) is caused by loss-of-function mutations in the type VII collagen gene, COL7A1 (Dang and Murrell, 2008; Shimizu et al., 1996; Varki et al., 2007). Type VII collagen (Col7) deficiency is responsible for diminished or absent formation of homotrimers of Col7, necessary to form anchoring fibrils that normally insert into the lamina densa of the skin basement membrane and provide adhesion to interstitial dermal collagen. Without anchoring fibrils, skin integrity is lost and blistering occurs below the lamina densa. Individuals with RDEB exhibit extensive painful trauma-induced skin and mucosal blisters and erosions. Due to extreme skin fragility, recurrent injury, and aberrant tissue repair, subjects with severe generalized RDEB are prone to develop esophageal strictures, mutilating scarring, pseudosyndactyly, joint contracture,s and highly malignant skin squamous cell carcinomas (Fine et al., 2008). Until recently, treatment options have been limited to palliative measures such as analgesia, bandaging, nutritional support, esophageal dilatation, and treatment of infections (Abercrombie et al., 2008; Mellerio et al., 2007). At present, however, several promising interventions are being developed, including systemic interventions using gene therapy, protein therapy, and cellular transplantation (Chen et al., 2002; Chino et al., 2008; Conget et al., 2010; De Luca et al., 2009; Ferrari et al., 2006; Fivenson et al., 2003; Mecklenbeck et al., 2002; Ortiz-Urda et al., 2003; Ortiz-Urda et al., 2002; Tolar et al., 2009; Wong et al., 2008; Woodley et al., 2004a; Woodley et al., 2004b; Woodley et al., 2007; Yan and Murrell, 2010). Cellular transplantation studies using mesenchymal stem cells or hematopoietic stem and progenitor cells have been shown to provide a source of normal donor Col7 and to ameliorate many of the disease manifestations (Chino et al., 2008; Conget et al., 2010; Tolar et al., 2009). However, these therapies have focused on using allogeneic cells, which maybe targeted for elimination by the host immune system. In addition, for allogeneic hematopoietic cell transplantation, potentially toxic chemoradiotherapy and immune-suppressive agents need to be used to prepare the host to receive the allogeneic donor graft.
Patient-specific stem cells, such as the recently identified induced pluripotent stem (iPS) cells (Park et al., 2008b; Takahashi and Yamanaka, 2006a; Yu et al., 2007), present an opportunity to use the progeny of autologous rather than allogeneic cells to correct RDEB. An advantage of gene-corrected autologous RDEB iPS cells is the possibility of simultaneously deriving Col7-producing ectodermal tissues (such as skin) and mesodermal tissue (such as hematopoietic stem and progenitor cells). Because the systemic use of bone marrow and cord blood transplantation combined with local injections of skin fibroblasts (FBs) may be synergistic in the wound treatment of RDEB patients, autologous gene-corrected iPS cells would represent a novel approach to RDEB treatment by providing a virtually limitless supply of both cell types, while avoiding the need for potent immune-suppressive therapy to prevent an allogeneic immune response. Induction of iPS cells from RDEB subjects may also provide a means of better understanding the sequence of downstream events initiated by Col7 deficiency during the development of epithelial stem and progenitor cells, as well as their progeny, as assayed both in vitro and in vivo. Such knowledge may lead to new treatment approaches that could benefit individuals with RDEB, other related inherited skin diseases, and perhaps additional extracellular matrix disorders.
Here, we show that iPS cells can be obtained from both skin FBs and keratinocytes (KCs) of individuals with RDEB (RDEB iPS cells). We report that the RDEB iPS cells can be differentiated to both hematopoietic and non-hematopoietic cells. These data underscore the potential of RDEB iPS cells in modeling RDEB in the development and future use of gene-corrected RDEB iPS cells to generate autologous hematopoietic grafts for systemic therapy, as well as generating skin cells with the potential to treat the individual skin and mucosal lesions that underscore RDEB.
RESULTS
RDEB iPS cells can be generated from both fibroblasts and keratinocytes
Col7 can be secreted by both FBs and KCs. Therefore, as both cell phenotypes are likely to be relevant to RDEB pathogenesis, we isolated FBs and KCs from the skin of three individuals with RDEB (Table 1). The cultured primary FBs and KCs had the characteristic spindle-shaped and cobblestone appearances, respectively and, as expected, KCs expressed Col7, type XVII collagen, keratin 1 (K1), keratin 5 (K5), and keratin 15 (K15). (Figure S1).
Table 1.
Patient and iPS cell characteristics.
| Patient | Type VII collagen mutations |
Age (years) |
Cell type | Number of iPS colonies per 100,000 cells |
|---|---|---|---|---|
| P1 | c.6176A>G (p.Glu2059Gly) IVS5+1G>A |
6.2 | FB | 3 |
| P2 | c.4919delG (p.Gly1640fsX70) c.8254_8255delAG (p.Arg2751fsX20) |
6.9 | KC | 7 |
| P3 | c.6781C>T (p.Arg2261X) IVS110-1 G>C |
14.5 | FB | 2 |
| P3 | c.6781C>T (p.Arg2261X) IVS110-1 G>C |
14.5 | KC | 2 |
| WT | N/A | 9 | KC | 2 |
P, patient; KC, keratinocytes; FB, fibroblasts; N/A, not applicable.
To derive RDEB iPS cells, RDEB FBs (from patient 1 [P1] and patient 3 [P3]) and RDEB KCs (from patient 2 [P2] and P3) (Table 1) were transduced with retroviral vectors carrying four known reprogramming transcription factors (OCT4, SOX2, KLF4, and c-MYC) that are typically associated with pluripotency. Transduced cells were cultured on a supportive stroma of irradiated murine embryonal fibroblasts (MEFs). After 6–7 weeks, flat colonies of RDEB iPS cells with clear-cut, round edges emerged from the two-dimensional MEF culture (Figure 1 A–O). Even though both FBs and KCs have been used successfully to generate iPS cells, KCs have been preferred by some because they appeared to reprogram faster than FBs (Maherali et al., 2008a). All iPS colonies in this study, however, appeared 45±4 days after viral transduction, and with roughly similar efficiency in FB and KC cultures (Table 1).
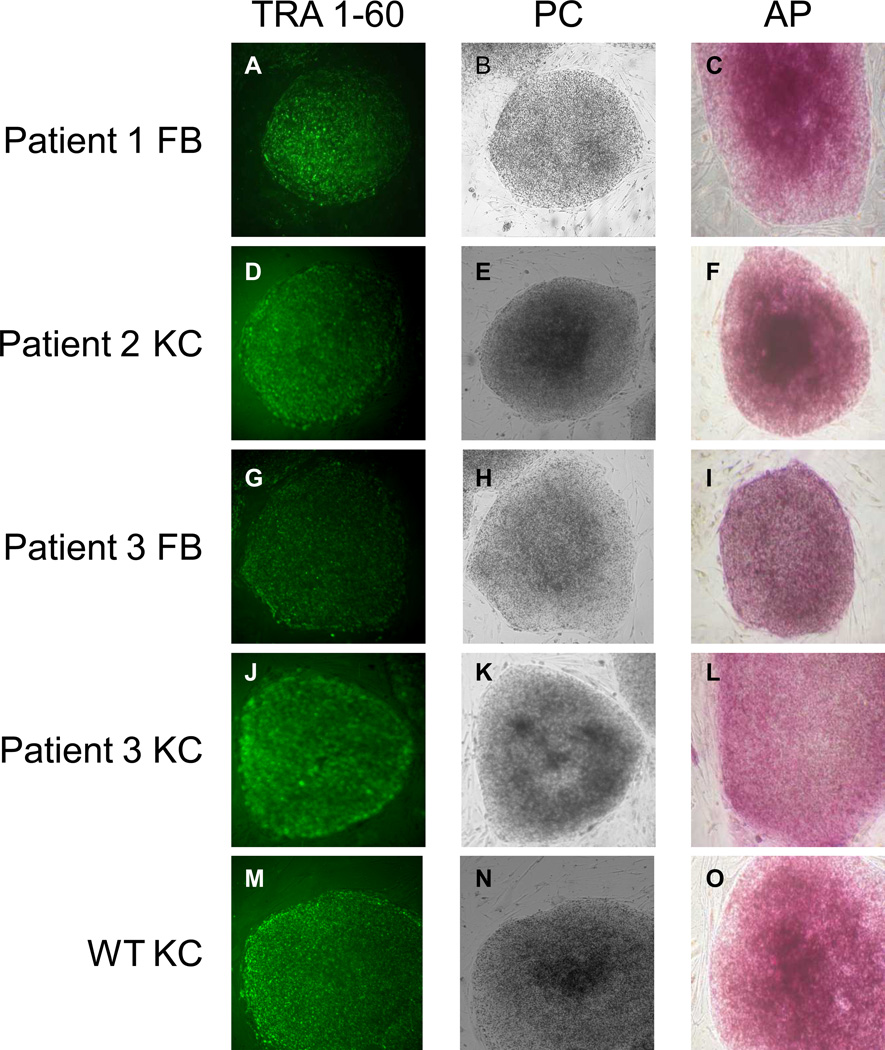
Figure 1. Induction of RDEB skin cells into RDEB iPS cells.
A, D, G, J, M: Live human iPS cultures stained with TRA-1-60 antibody four weeks after transduction. B, E, H, K, N: Phase contrast image of the same iPS colonies. C, F, I, L, O: Alkaline phophatase stain. P: Quantitative RT-PCR analysis of OCT4, SOX2, NANOG, KLF4, c-MYC, LIN28, and DNMT3b expression levels in RDEB KC iPS cells from P2 and P3 relative to expression levels in parental RDEB KCs from P2 and P3. Q: Quantitative RT-PCR analysis of the same genes in WT KC- iPS cells relative to expression levels in parental WT KCs. All values were normalized against endogenous GAPDH expression. FB, fibroblasts; KC, keratinocytes; WT, wild type; PC, phase contrast; AP, alkaline phosphatase; iPS, induced pluripotent stem cells.
When compared to parental RDEB KCs, the RDEB iPS cells showed persistent mRNA expression of SOX2, used for reprogramming, as well as genes associated with pluripotency in iPS cells (NANOG, LIN28, and DNMT3b). Moreover, there was transient mRNA expression of OCT4, c-MYC, and KLF4, used to accomplish reprogramming. The mRNA profile seen in RDEB iPS cells is as would be expected to occur in the wild-type (WT) iPS (WT iPS) cells (Figure 1 P and 1 Q). RDEB iPS cells also expressed protein markers characteristic of reprogrammed iPS and embryonic stem (ES) cells: TRA-1-81, stage-specific embryonic antigens (SSEA)-3 and -4, OCT4, and NANOG (Figure 2 and Figure S2).
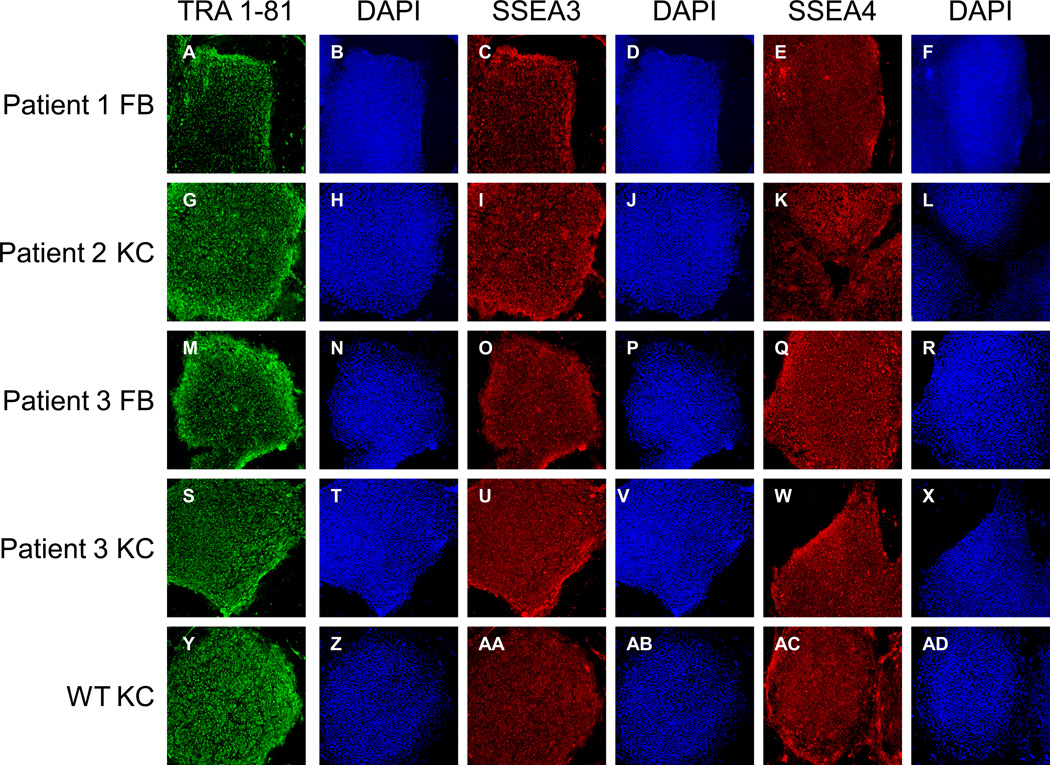
Figure 2. Protein expression profile of RDEB iPS cells.
To confirm their ability to express ES cell proteins, human iPS cells derived from skin of all RDEB patients and WT control were immunostained with TRA-1-81 (A, G, M, S, Y), SSEA-3 (C, I, O, U, AA), and SSEA-4 (E, K, Q, W, AC). Corresponding images stained with 4,6-diamidino-2-phenylindole show nuclei of individual cells in the colonies (B, H, N, T, Z, D, J, P, V, AB, F, L, R, X, AD). Isotype controls are shown in Supplemental Figure 8. Magnification 10×. FB, fibroblasts; KC, keratinocytes; WT, wild type; DAPI, 4,6-diamidino-2-phenylindole.
These reprogramming factors are thought to activate a network of transcriptional factors, which in turn induce epigenetic changes (Aasen et al., 2008; Freberg et al., 2007; Maherali et al., 2008a; Okita et al., 2008; Park et al., 2008a). Therefore, we used bisulfite sequencing to confirm the methylation status of OCT4 and NANOG promoters in RDEB iPS cells. A methylated pattern is indicative of gene silencing, whereas an unmethylated pattern indicates the potential for robust gene expression. Individual colonies of cells from RDEB iPS cells were analyzed by bisulfite sequencing that showed a pattern characteristic of iPS cells in which OCT-4 and NANOG promoter sequences are unmethylated. In contrast, mature progeny such as FBs and KCs had the expected pattern of both methylated and unmethylated sequences (Park et al., 2008b; Raya et al., 2009) (Figure 3 and Figure S3).
Figure 3. Epigenetic profile of RDEB iPS cells.
(A) Bisulfite sequencing of the OCT4 and NANOG promoters in WT FB, RDEB FB, and FB-derived RDEB iPS cells. (B) Bisulfite sequencing of the OCT4 and NANOG promoters in WT KC, RDEB KC and KC-derived RDEB iPS cells. Open circles denote unmethylated CpGs, and filled circles represent methylated CpGs. CpG position relative to the downstream transcriptional start site is shown above each column. Sequencing reactions of specific amplicons are represented by each row of circles. WT, wild type; FB, fibroblasts; KC, keratinocytes; iPS cells, induced pluripotent cells; RDEB, recessive dystrophic epidermolysis bullosa.
Both RDEB FB- and RDEB KC-derived RDEB iPS cells from the three individuals, as well as the one WT control, had normal female karyotypes as determined by high-resolution chromosomal G-banding (Figure S4 and data not shown). Because the purity of the iPS cultures is critical for our downstream experimentation, we formally demonstrated a lack of contamination of stromal cell (MEF) feeder cells used to generate RDEB iPS cells. Competitive PCR of variable number tandem repeats indicated that all RDEB iPS cell lines were fully host-derived (data not shown). Collectively, the transcription profile and cellular phenotype of RDEB iPS cells from all subjects were consistent with morphological and phenotypical gain of pluripotency.
RDEB iPS cells widely differentiate in vivo
To provide further evidence of the identity of iPS cells on a functional level, KC-derived RDEB iPS cells were injected intramuscularly into immune-deficient mice. Within 6–8 weeks, well-differentiated cystic teratomas were observed, confirming the phenotype-defining ability of iPS cells to differentiate in vivo into a wide array of cell lineages encompassing cells of endodermal, mesodermal, and ectodermal origin (Figure 4).
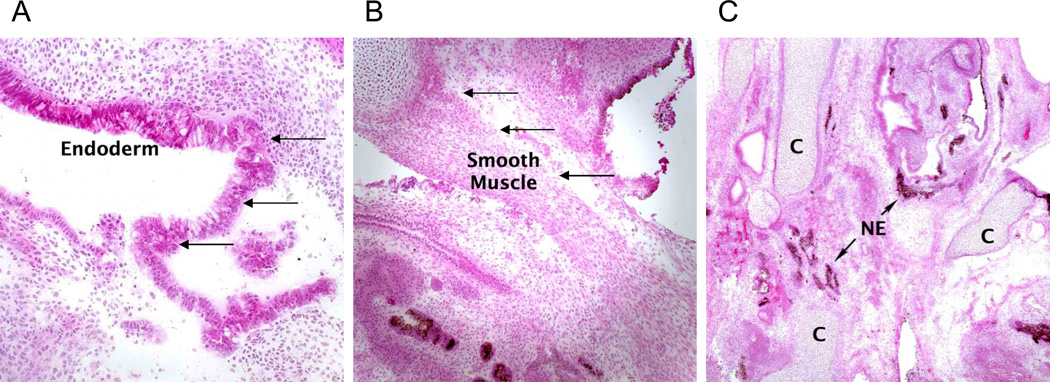
Figure 4. Non-hematopoietic differentiation of RDEB iPS cells.
Histological examination of mature teratoma from an immunodeficient mice injected with KC RDEB iPS cells revealed (A) columnar epithelium of endodermal origin (arrows), (B) smooth muscle of mesodermal origin (arrows), and (C) melanocytes of ectodermal origin (arrows). NE, neuroectoderm; C, cartilage. Similar mature teratomas with contribution of ectodermal-, mesodermal-, and endodermal-derived cells formed after injection of KC WT iPS cells (data not shown). Magnification 20×. Hematoxylin-eosin stain.
RDEB iPS cells differentiate hematopoietic cells
As we have shown that cells contained in bone marrow and cord blood can home to RDEB skin and produce Col7, one of the mesodermal lineages, hematopoietic, is directly relevant to our ultimate goal of generating gene-corrected cells for autologous hematopoietic cell transplantation. The hematopoietic potential of the RDEB iPS cells was tested by using embryoid bodies (Figure S5 A). Embryoid bodies mimic early stages of embryonic development and serve to provide three-dimensional architecture, which promotes cell differentiation. Embryoid bodies derived from RDEB iPS cells were enzymatically dissociated into single cells. After seeding in low-attachment dishes, cells were induced to differentiate in medium containing human hematopoietic growth factors: stem cell factor, Flt3-ligand, interleukin-3, interleukin-6, granulocyte-colony stimulating factor, and bone morphogenetic protein 4. To define the commitment of these cells to hematopoietic lineage, we assessed their capacity to function as colony-forming units (CFUs). Colony-forming capacity is a standard measure to assess both the quality and quantity of blood-forming stem cells and progenitor cells. The colony-forming capacity of the hematopoietic progeny of RDEB iPS cells was comparable to the colony-forming capacity of the hematopoietic progeny of WT iPS cells, and both erythroid and myeloid colonies formed (Figure S5 B, C, and D). In addition to the quantitative measures, such as the ability to form colonies in the semisolid medium, cells with phenotypic characteristics of myeloid and erythroid progenitors were observed in equal distribution on cytospins generated from WT iPS cells and RDEB iPS cell colonies grown in methylcellulose-enriched medium (data not shown). In aggregate, these data confirm that RDEB iPS cells can differentiate into cells with hematopoietic potential.
RDEB iPS cells differentiate into skin-like structures
With our primary focus on skin pathology relevant to RDEB subjects, we wished to identify teratoma-derived structures resembling skin. To delineate this ectodermal-to-skin transition, we used staining with K5 antibody to reveal epidermis-like layers of KCs. Remarkably, in the WT iPS cell-derived teratomas, Col7 was expressed as a continuous band mimicking Col7 expression at the basement membrane in normal skin (Figure 5 A–D and Figure S6 A). In contrast, and consistent with the Col7 deficiency in RDEB individuals, no Col7 was detected in the skin-like structures derived from RDEB iPS cells (Figure 5 E–H and Figure S6 B). In order to demonstrate that RDEB iPS cells can be gene-corrected with exogenous Col7 DNA, RDEB iPS cells were transfected with expression plasmid harboring WT human Col7 gene. Two days after transfection, Col7 protein was expressed in the gene-corrected RDEB iPS cultures similarly to the pattern of Col7 expression observed in the WT iPS cells (Figure S7). In summary, these data suggest that WT and RDEB iPS cells can provide a new system to study skin pathology in a patient-specific fashion.
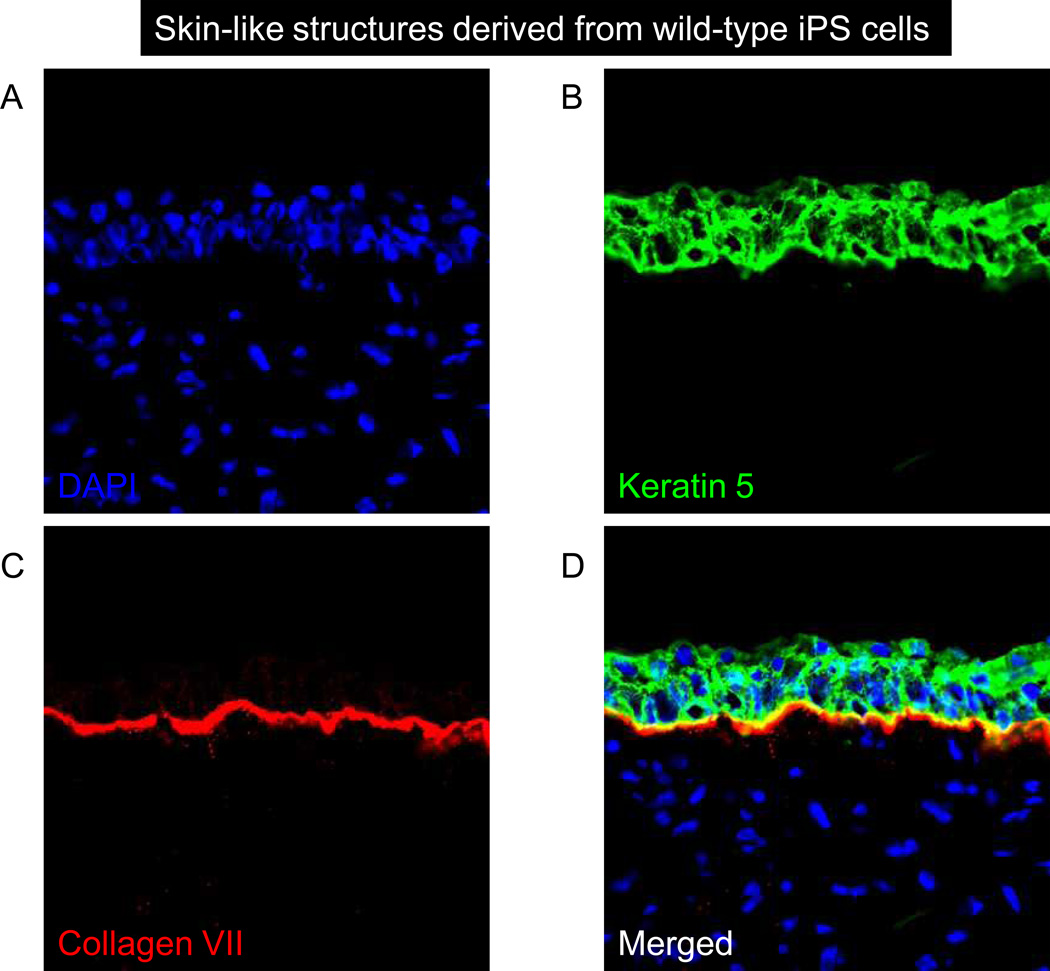
Figure 5. Skin-like structures derived from WT and RDEB iPS cells.
(A–D) K5 (green) and Col7 (red) are co-expressed in WT KC iPS cell-derived teratomas. (E–H) Similar structures form in the RDEB KC iPS cell-derived teratomas, yet Col7 is undetectable. Magnification 20×. DAPI, 4,6-diamidino-2-phenylindole.
DISCUSSION
To our knowledge these are the first data to report that autologous iPS cells can be obtained from RDEB subjects. We found that iPS cells can be generated with a similar efficacy of reprogramming using either skin FBs or KCs of individuals with RDEB. These RDEB iPS cells can be induced to differentiate into hematopoietic and non-hematopoietic cells. Our observation that the RDEB iPS cells have the capacity to form skin-like structures with no Col7 deposition underscores the potential of RDEB iPS cells in modeling RDEB in development, in uncovering clinically relevant compensatory changes triggered by the absence of Col7, and in the search for effective therapeutic interventions for RDEB patients. Relevant to this, we found that the NANOG promoter in RDEB FBs and KCs is hypomethylated. This suggests that the increased proliferative signaling engaged in aberrant skin repair in individuals with RDEB could, in theory, engage cell cycle regulation common to rapid growth, self-renewal, and pluripotency states. Lastly, we found that the RDEB iPS cells express Col7 after transfection of WT Col7. In the future, gene-corrected RDEB iPS cells could be used not only to generate an autologous hematopoietic graft but also to generate non-hematopoietic skin cells with the potential to treat individual skin and mucosal wounds. Independently or together, these two strategies have the potential to decrease the complications or lack of sustained improvement observed with currently used therapies and to result in improved survival and quality of life for people with RDEB.
To the best of our knowledge, this is the first described human iPS cell model of an extracellular matrix disorder. As the RDEB iPS cells exhibit disease-relevant phenotype, they provide a novel model system to study the pathogenesis of RDEB and related disorders. When compared to studies of differentiated human cells and animal models of RDEB, iPS cell studies have the added benefit of offering the opportunity for a more extensive analysis of the cellular effects of Col7 deficiency independent of the secondary effects on systemic inflammatory responses that would occur in vivo due to the erosions and blister formation characteristic of RDEB. Because nearly all cell types can be derived from the iPS cells, analysis of human RDEB iPS cells may have advantages over analysis of murine animal models that are confounded by the obvious differences in development, life span, and species-specific consequences of cellular pathology in skin and mucosal membranes affected by Col7 deficiency. Relevant to this and in contrast to other congenital disorders, such as those affecting DNA repair genes in Fanconi anemia (Raya et al., 2009; Tulpule et al., 2010), Col7 does not appear to be necessary for stem cell self-renewal and generation of iPS cells.
The complexity of RDEB, as evidenced by the plethora of biochemical events downstream of the Col7 pathology and by the varied phenotypes of human disease, is perhaps matched by the versatility and promise of reprogramming cell technology (Ebert et al., 2009; Maherali et al., 2008b; Park et al., 2008c; Takahashi and Yamanaka, 2006b; Vierbuchen et al., 2010). An unresolved question, however, is whether the potential risks of iPS cell-derived therapy, such as those related to (1) genotoxicity of reprogramming using ES cell-specific transcription factors and the methods by which gene augmentation of mutated genes is accomplished (e.g., viral vector delivery) (Kohn et al., 2003), (2) fidelity of acquisition and maintenance of pluripotent phenotype and lineage-specific cell fates, and (3) cell purity of the on-demand differentiated iPS cell progeny in the absence of undifferentiated and potentially tumorigenic iPS cells, can be minimized or avoided altogether.
Nevertheless, even at present RDEB iPS cells offer a new model of RDEB disease and may help to understand the pathogenic cascades underlying the inherent complexity of RDEB. These insights, and the use of the cellular progeny of gene-corrected autologous RDEB iPS cells, present an opportunity for future clinical translation in a manner that may preclude the immunologic complications of allogeneic transplantation.
METHODS
Patients
After obtaining consent as approved by the Institutional Review Board of the Human Subjects Committee at the University of Minnesota, skin or marrow cells were collected from healthy volunteers and from individuals with RDEB.
Induced pluripotent stem (iPS) cells
Detailed methods describing isolation of FBs and KCs are included as Supplemental Materials. For induction of iPS cells, four reprogramming factors, OCT4, SOX2, KLF4, and c-MYC were used to produce retroviral supernatants. Each of the viral supernatants was produced by transfecting 293T/17 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with three plasmids: one cargo plasmid (containing the reprogramming gene), a plasmid expressing the VSV-G envelope gene, and a helper plasmid with the retroviral Gag/Pol gene. The plasmids were obtained from Addgene (Cambridge, MA). Forty-eight to 72 hours after transfection, viral supernatants were harvested, centrifuged at 400 × g for 15 minutes, and filtered through a 0.45 µm filter.
About 50,000 KCs or 100,000 FBs per well of a 6-well plate were plated and infected with a 1:1:1:1 mix of retroviral supernatants of pMIG containing OCT4, SOX2, KLF4, and c-MYC in the presence of 5 µg/mL of protamine sulfate. Transduction consisted of a 45-minute spinfection at 1800 rpm at room temperature; supernatants were left in contact with the cells for 24 hours at 37°C and 5% CO2. The next day, cells were spinfected for a second time. Cells were placed in plates coated using a Coating Matrix Kit (Cascade Biologics, Portland, OR) and maintained in KC medium (EpiLife Medium, Cascade Biologics: 0.06 mM calcium chloride, plus human KC growth supplement) at 37°C, 5% CO2. Five days later, cells were trypsinized and seeded onto feeder layers of irradiated CF1 MEFs. After 24 hours, the medium was changed to human ES cell medium, consisting of DMEM/F12 (Invitrogen) supplemented with 10% KnockOut Serum Replacement (Invitrogen), 2 mM GlutaMAX (Invitrogen), 50 µM 2-mercaptoethanol (Invitrogen), 1× non-essential amino acids (Invitrogen), 50 U/mL penicillin, 50 mg/mL streptomycin, and 10 ng/mL basic FB growth factor (R&D Systems, Minneapolis, MN). Cultures were maintained at 37°C, 5% CO2, with daily medium changes. Starting one week after plating onto MEFs, medium was supplemented with 1 µM PD0325901 and 1 µM CT 99021 (both from Stemgent, San Diego, CA) for one week. Colonies were picked based on morphology 30–60 days after the initial infection.
We identified iPS cell colonies using live staining with TRA-1-60 antibody (1:400, Millipore, Billerica, MA) and secondary antibody Alexa 488–conjugated anti–mouse IgM (1:400, Invitrogen) diluted in hES medium, and added into the culture plate. TRA-1-60+ colonies were identified under a fluorescence microscope (Leica DMI6000B). To confirm that the iPS cells were not contaminated with donor cells, competitive PCR analysis of variable tandem repeat regions was performed as described (Scharf et al., 1995).
To confirm their cellular phenotype, the iPS cells were fixed with 4% paraformaldehyde for 20 minutes. If nuclear permeation was needed, cells were treated with 0.2% TritonX (Sigma-Aldrich, St. Louis, MO) in phosphate-buffered saline (PBS) for 30 minutes, blocked in 3% bovine serum albumin (BSA) in PBS for 2 hours, and incubated with primary antibody overnight at 4°C. Antibodies targeting the following antigens were used: TRA1-60 (MAB4360, 1:400), TRA1-81 (MAB4381, 1:400), SSEA4 (MAB4304, 1:200), and SSEA3 (MAB-4303, 1:200), all from Chemicon (Billerica, MA); OCT3/4 (AB27985, 1:200) from Abcam (Cambridge, MA); and NANOG (EB068601:100) from Everest (Upper Heyford, Oxfordshire, UK). Cells were incubated with secondary Alexa Fluor Series antibodies (all 1:500, Invitrogen) for two hours at room temperature and then with 4,6-diamidino-2-phenylindole (DAPI, 1 µg/mL, Invitrogen) for 10 minutes. Images were examined using a Leica DMI6000B microscope equipped with Q-Imaging Retiga 2000R Camera and Q-Capture software. Direct alkaline phosphatase activity was analyzed per the manufacturer’s recommendations (Millipore).
RT-PCR
RNA was isolated using a GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) and treated with TURBO DNA-free (Ambion, Austin, TX). First-strand cDNA was synthesized using a Superscript III First Strand Synthesis SuperMix for qRT-PCR (Invitrogen). RT-PCR was performed using TaqMan Gene Expression Assays and TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Carslbad, CA) per manufacturer’s protocol.
TaqMan Gene Expression Assays: Col7a1 (Hs01574733_g1), Col17 (Hs00166711_m1), Ker1 (Hs00196158_m1), Ker15 (Hs00267035_m1), POU5F1 Hs 00999634_gH; SOX2 Hs 00602736_s1; NANOG Hs 02387400_g1; KLF4 Hs 00358836_m1; MYC Hs 00153408_m1; LIN28 Hs 00702808_s1; DNMT3 Hs 01003405_m1; with GAPDH (Hs99999905_m1) used as an endogenous control. Expression levels were measured in duplicate. For genes with expression below the CT fluorescence threshold, the CT was set to 40 to calculate the relative expression. Analysis was done utilizing an ABI PRISM 7500 sequence detection system (Applied Biosystems).
Bisulfite genomic sequencing
Genomic DNA was isolated using a PureLink Genomic DNA Mini Kit (Invitrogen). Bisulfite treatment was done using an EpiTect Bisulfite kit (Qiagen, Valencia, CA). Converted DNA was PCR-amplified using OCT4-specific primer sets and NANOG-specific primer sets (Freberg et al., 2007). PCR products were gel purified using a PureLink Quick Gel Extraction and PCR Purification Combo Kit (Invitrogen), and cloned into bacteria using the TOPO TA Cloning Kit for Sequencing (Invitrogen).
Differentiation of iPS cells
Detailed methods describing isolation of hematopoietic differentiation of iPS cells are included as Supplemental Materials. For teratoma formation, NOG young adult mice were injected with 1 million cells resuspended in a mixture of DMEM/F12, Matrigel (BD Biosciences, San Jose, CA), and collagen (ratio 2:1:1, 40 µL per mouse) into the right quadriceps muscle. Tumors were harvested in 4–8 weeks and cryopreserved at −80°C in Optimal Cutting Temperature medium (Sakura Finetek USA, Torrance, CA). Six µm-thick sections were cut and mounted on Superfrost Plus glass slides (Thermo Fisher Scientific, Waltham, MA). Tissues were then fixed in acetone for five minutes at room temperature. After rehydrating the samples in PBS for five minutes, blocking solution of 10% normal donkey serum was applied for one hour at room temperature. The tissues were then incubated with mouse anti-human Col7 antibody (1:250, BD Biosciences), and rabbit anti-human K5 antibody (1:600, Covance, Emeryville, CA,) and then incubated with secondary antibodies, anti-mouse Cy3 (1:500, Jackson ImmunoResearch, West Grove, PA,) and goat anti-rabbit Alexa Fluor 488 (1:800, Invitrogen). Tissues were rinsed three times with PBS and coverslipped with hardset mounting medium with DAPI (Vector Laboratories, Burlingame, CA). Optical parameters used for light microscopy are included as Supplemental Materials.
Correction of the RDEB iPS cells
The COL7A1-COL7A1 expression cassette, in which the full-length COL7A1 cDNA expression is driven by a short COL7A1 promoter (Titeux et al., 2010), was cloned into the pEGFP-1 plasmid backbone (Clontech, Mountain View, CA) as folows: 1) pEGFP-1 was digested by HindIII and NotI to excise the EGFP open reading frame, 2) the pCMS-COL7A1-COL7A1 retroviral backbone (Titeux et al., 2010) was digested by HindIII and NotI to release the COL7A1-COL7A1 expression cassette, 3) the pEGFP-1 (Δ-EGFP) and the 10 kb COL7A1-COL7A1 fragments were ligated to generate the pCOL7A1-COL7A1-pA vector (13.4 kb). RDEB iPS cells were nucleofected with this Col7 cDNA using the Human Stem Cell Nucleofector Kit 1 (program A-023, Amaxa/Lonza, Walkersville, MD) with 1.2 mg of DNA per 1 million cells. Nucleofected cells were plated in AggreWell plates (StemCell Technologies, Vancouver, BC, Canada). After 48 hours, embryoid bodies formed and were transferred to four-chamber glass slides coated with Matrigel. To assess Col7 expression 72 hours after nucleofection, cells were fixed with a 1:1 ratio of ice-cold methanol/acetone for seven minutes, washed with 1× PBS, blocked with 10% normal donkey serum for one hour, and stained with primary Col7 antibody (1:250, BD Biosciences) for two hours at room temperature. Slides were washed and secondary antibody donkey anti-mouse cy3 (1:500, Jackson Immunoresearch, West Grove, PA) was applied for one hour. DAPI was added, and slides were imaged by confocal microscopy using Olympus BX61 FluoView 500 confocal microscope and FluoView software.
Data analysis
Differences between measurements were evaluated using Student’s t test, with p values less than 0.05 considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank Dr Gerry O’Sullivan for examination of teratomas, and Marianna Wong, Jess Lorenz, Trevor Keyler, and Pavlina Chuntova for preparing laboratory data. We are grateful to Dr John McGrath for stimulating discussions on developing new therapies for RDEB.
GRANT SUPPORT
Supported in part by grants from DEBRA International, Epidermolysis Bullosa Research Fund, University of Minnesota Academic Health Center, and by the Liao Family Epidermolysis Bullosa Fund, Sarah Rose Mooreland Epidermolysis Bullosa Fund, and Children’s Cancer Research Fund, Minneapolis, Minnesota.
Abbreviations used
- AP
alkaline phosphatase
- BSA
bovine serum albumin
- Col7
type VII collagen
- CFU
colony-forming unit
- CFU-E
CFU-erythroid
- CFU-GM
CFU-granulocyte/macrophage
- Col17
type XVII collagen
- DAPI
4,6-diamidino-2-phenylindole
- ES
embryonic stem
- FB
fibroblast
- FBS
fetal bovine serum
- iPS cell
induced pluripotent stem cell
- KC
keratinocyte
- K1
keratin 1
- K5
keratin 5
- K15
keratin 15
- MEF
murine embryonal fibroblast
- PBS
phosphate-buffered saline
- PC
phase contrast
- PCR
polymerase chain reaction
- RDEB
recessive dystrophic epidermolysis bullosa
- RNA
ribonucleic acid
- RT
reverse transcriptase
- WT
wild type
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Abercrombie EM, Mather CA, Hon J, Graham-King P, Pillay E. Recessive dystrophic epidermolysis bullosa. Part 2: care of the adult patient. Br J Nurs. 2008;17 doi: 10.12968/bjon.2008.17.sup3.28911. S6, S8, S10 passim. [DOI] [PubMed] [Google Scholar]
- Chen M, Kasahara N, Keene DR, Chan L, Hoeffler WK, Finlay D, et al. Restoration of type VII collagen expression and function in dystrophic epidermolysis bullosa. Nat Genet. 2002;32:670–675. doi: 10.1038/ng1041. [DOI] [PubMed] [Google Scholar]
- Chino T, Tamai K, Yamazaki T, Otsuru S, Kikuchi Y, Nimura K, et al. Bone marrow cell transfer into fetal circulation can ameliorate genetic skin diseases by providing fibroblasts to the skin and inducing immune tolerance. Am J Pathol. 2008;173:803–814. doi: 10.2353/ajpath.2008.070977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conget P, Rodriguez F, Kramer S, Allers C, Simon V, Palisson F, et al. Replenishment of type VII collagen and re-epithelialization of chronically ulcerated skin after intradermal administration of allogeneic mesenchymal stromal cells in two patients with recessive dystrophic epidermolysis bullosa. Cytotherapy. 2010;12:429–431. doi: 10.3109/14653241003587637. [DOI] [PubMed] [Google Scholar]
- Dang N, Murrell DF. Mutation analysis and characterization of COL7A1 mutations in dystrophic epidermolysis bullosa. Exp Dermatol. 2008;17:553–568. doi: 10.1111/j.1600-0625.2008.00723.x. [DOI] [PubMed] [Google Scholar]
- De Luca M, Pellegrini G, Mavilio F. Gene therapy of inherited skin adhesion disorders: a critical overview. Br J Dermatol. 2009;161:19–24. doi: 10.1111/j.1365-2133.2009.09243.x. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose JFF, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–281. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Pellegrini G, Matsui T, Mavilio F, De Luca M. Gene therapy in combination with tissue engineering to treat epidermolysis bullosa. Expert Opin Biol Ther. 2006;6:367–378. doi: 10.1517/14712598.6.4.367. [DOI] [PubMed] [Google Scholar]
- Fine JD, Eady RA, Bauer EA, Bauer JW, Bruckner-Tuderman L, Heagerty A, et al. The classification of inherited epidermolysis bullosa (EB): Report of the Third International Consensus Meeting on Diagnosis and Classification of EB. J Am Acad Dermatol. 2008;58:931–950. doi: 10.1016/j.jaad.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Fivenson DP, Scherschun L, Choucair M, Kukuruga D, Young J, Shwayder T. Graftskin therapy in epidermolysis bullosa. J Am Acad Dermatol. 2003;48:886–892. doi: 10.1067/mjd.2003.502. [DOI] [PubMed] [Google Scholar]
- Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–1553. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn DB, Sadelain M, Glorioso JC. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat Rev Cancer. 2003;3:477–488. doi: 10.1038/nrc1122. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008a;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008b;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecklenbeck S, Compton SH, Mejia JE, Cervini R, Hovnanian A, Bruckner-Tuderman L, et al. A microinjected COL7A1-PAC vector restores synthesis of intact procollagen VII in a dystrophic epidermolysis bullosa keratinocyte cell line. Hum Gene Ther. 2002;13:1655–1662. doi: 10.1089/10430340260201743. [DOI] [PubMed] [Google Scholar]
- Mellerio JE, Weiner M, Denyer JE, Pillay EI, Lucky AW, Bruckner A, et al. Medical management of epidermolysis bullosa: Proceedings of the IInd International Symposium on Epidermolysis Bullosa, Santiago, Chile, 2005. Int J Dermatol. 2007;46:795–800. doi: 10.1111/j.1365-4632.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urda S, Lin Q, Green CL, Keene DR, Marinkovich MP, Khavari PA. Injection of genetically engineered fibroblasts corrects regenerated human epidermolysis bullosa skin tissue. J Clin Invest. 2003;111:251–255. doi: 10.1172/JCI17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Urda S, Thyagarajan B, Keene DR, Lin Q, Fang M, Calos MP, et al. Stable nonviral genetic correction of inherited human skin disease. Nat Med. 2002;8:1166–1170. doi: 10.1038/nm766. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008a;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008b;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008c;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf SJ, Smith AG, Hansen JA, McFarland C, Erlich HA. Quantitative determination of bone marrow transplant engraftment using fluorescent polymerase chain reaction primers for human identity markers. Blood. 1995;85:1954–1963. [PubMed] [Google Scholar]
- Shimizu H, McGrath JA, Christiano AM, Nishikawa T, Uitto J. Molecular basis of recessive dystrophic epidermolysis bullosa: genotype/phenotype correlation in a case of moderate clinical severity. J Invest Dermatol. 1996;106:119–124. doi: 10.1111/1523-1747.ep12329600. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006a;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006b;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Titeux M, Pendaries V, Zanta-Boussif MA, Decha A, Pironon N, Tonasso L, et al. SIN Retroviral Vectors Expressing COL7A1 Under Human Promoters for Ex Vivo Gene Therapy of Recessive Dystrophic Epidermolysis Bullosa. Mol Ther. 2010;18:1509–1518. doi: 10.1038/mt.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Ishida-Yamamoto A, Riddle M, McElmurry RT, Osborn M, Xia L, et al. Amelioration of epidermolysis bullosa by transfer of wild-type bone marrow cells. Blood. 2009;113:1167–1174. doi: 10.1182/blood-2008-06-161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule A, Lensch MW, Miller JD, Austin K, D'Andrea A, Schlaeger TM, et al. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki R, Sadowski S, Uitto J, Pfendner E. Epidermolysis bullosa. II. Type VII collagen mutations and phenotype-genotype correlations in the dystrophic subtypes. J Med Genet. 2007;44:181–192. doi: 10.1136/jmg.2006.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1042. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128:2179–2189. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- Woodley DT, Keene DR, Atha T, Huang Y, Lipman K, Li W, et al. Injection of recombinant human type VII collagen restores collagen function in dystrophic epidermolysis bullosa. Nat Med. 2004a;10:693–695. doi: 10.1038/nm1063. [DOI] [PubMed] [Google Scholar]
- Woodley DT, Keene DR, Atha T, Huang Y, Ram R, Kasahara N, et al. Intradermal injection of lentiviral vectors corrects regenerated human dystrophic epidermolysis bullosa skin tissue in vivo. Mol Ther. 2004b;10:318–326. doi: 10.1016/j.ymthe.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Woodley DT, Remington J, Huang Y, Hou Y, Li W, Keene DR, et al. Intravenously injected human fibroblasts home to skin wounds, deliver type VII collagen, and promote wound healing. Mol Ther. 2007;15:628–635. doi: 10.1038/sj.mt.6300041. [DOI] [PubMed] [Google Scholar]
- Yan WF, Murrell DF. Fibroblast-based cell therapy strategy for recessive dystrophic epidermolysis bullosa. Dermatol Clin. 2010;28:367–370. xii. doi: 10.1016/j.det.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.