Clinical implementation of genomic medicine is gaining momentum. Since 2003, there has been a 25% yearly increase in the number of genes linked to specific diseases or treatments. Genetic testing is quickly and steadily being incorporated into clinical practice guidelines across a wide range of health indications, including cancer, cardiology, infectious diseases, mental health, and primary care. Within the Veterans Health Administration (VHA), from 2011 through 2013, more than 80 000 veterans underwent at least 1 of 110 different genetic tests. Tests that have high levels of utilization within the VHA include Factor V Leiden gene to identify patients at risk for venous thrombosis, HLA-B*5701 to identify patients at risk for a hypersensitivity reaction to abacavir (a treatment for HIV), IL28B to evaluate potential responsiveness to peginterferon-α-2b treatment for HCV, and several tests to inform risk, prognosis, or treatment for cancer, including BCR-ABL1, FLT3, JAK2, BRAF, BRCA, c-KIT, EGFR, EML4-ALK, KRAS, MLH1, MSH2, MSH6, among others.
BARRIERS TO GENOMIC MEDICINE FOR MINORITY PATIENTS
There are multiple barriers to genomic medicine for minorities. Clinical trial research that informs biomarker and drug development continues to lack participation by minorities. This presents the biggest problem for patients from African ancestries because, unlike Europe and Asia where genomic research is robust and useful data are provided on those populations, the genomic research infrastructure in the African continent is underdeveloped. Most large scale genome wide association studies have been conducted on populations of European ancestry.1 For commercially available genomic innovations, diffusion emanates from academic medical centers.2 Even though most of these centers are located in metropolitan counties with high percentages of minorities, the majority of minorities seek care in community hospitals. Research that informs clinical practice guidelines is based on disease processes and treatment responses of mostly populations of European descent.3 Finally, there continue to be patient and site of care variations in delivery of guideline-recommended care.
RACE AND ANCESTRY IN REPORTING GENETIC VARIATIONS
Understanding individual differences in risk of disease, progression of illness, and response to treatment is the cornerstone of genomic medicine. Expanded use of genetic testing has increased reporting of variations in gene alteration or gene expression by race and ancestry. Several large-scale genome-wide association studies have identified genetic variations that provide insight on biologic mechanisms behind disease progression and responses to treatment.1,4,5 These studies have found that ancestry influences presence of these variations. There are limitations and opportunities when using race or ancestry in genomic clinical care and research, particularly in the context of analyzing health disparities. Race and knowledge of patient ancestry tell us very little about an individual patient’s specific genetic makeup. Yet, for purposes of public health genomics research, race is a crucial surveillance tool to measure access, utilization, and potential health differences among groups of patients.
The majority of racial differences in health in the United States are explained by socioeconomic status and factors surrounding access to medical care. However, to fully capture the role of race in health, it is important to consider the contribution of genetic factors.6 Recent studies have found that self-reported race is a reliable indicator for Western African ancestry.7 Most US-born African Americans are descendants of Western Africans. Yet, there are increasing numbers of African immigrants, who differ significantly in income bracket, education level, and ancestry. African immigrants may not experience the poorer health outcomes associated with lower socioeconomic status that African Americans experience. The significant complexity and potential for confounding relationships between variables require researchers to be cautious and thorough when conducting and reporting research that links specific genetic variations to race or ancestry. It also requires accurate and consistent data about patient race or ancestry. Experience teaches that when multiple methods are used for entering patient race into electronic health record (EHR), or when patients are repeatedly asked to report this information, without an explanation about how it will be used, different self-reported race and ethnicity designations are entered into the EHR for the same patient. Use of race and ancestry information in relation to genetic variations can benefit patients and thus increases the demand for accurate and consistent data on patient race.
RACE, GENOTYPE, AND RESPONSE TO TREATMENT
In several studies, researchers describe variation in gene alteration or gene expression by race or ancestry. In some cases, studies that report racial variations are adequately populated with patients from diverse ancestries and researchers use rigorous methodologies to control for potential confounding and bias. In other cases, reported variations are based on small sample sizes and results are prematurely reported and not challenged by colleagues.
For example, the first study to identify a genetic basis for persistence of HCV in some African American patients identified an association between the IL28B genotype and responsiveness to peginterferon-α-2b.4 This study included 1671 patients, 299 (18.5%) of whom were African American. The study found that patients with two copies of the C allele (CC vs CT or TT) were more than twice as likely to respond to peginterferon-α-2b treatment. Moreover, this study also had a healthy, multiethnic control group to compare the prevalence of the CC genotype in the US population to the prevalence reported by the International HapMap project. Results of the association between IL28B genotype and responsiveness to peginterferon-α-2b were quickly confirmed and extended by numerous other researchers.
By contrast, studies of racial variations in the incidence of EGFR mutations in lung cancer were initially derived from 2 small, single-institution studies, which analyzed a total of 94 patients who self-identified as African American.8,9 Each study reported that 1 African American patient tested positive for the EGFR mutation. Those results contributed to the mistaken perception that African Americans don’t get EGFR mutations and resulted in oncologists not ordering the test for African American patients. It was five years before those findings were challenged by appropriately powered studies.10 Several factors contributed to premature reporting and inaccurate conclusions from racial associations in EGFR mutations in lung cancer. The main cause was inadequate number of tissue samples from African Americans in lung cancer genomic research.
FEDERAL EFFORTS TO ACCELERATE TRANSLATION
There have been several federally funded initiatives designed to accelerate translation of genomic advances to all patients. Much of this funding has focused on cancer because it is the most mature application of genomic medicine. Efforts such as The Cancer Genome Atlas (TCGA) are designed to improve the efficiency and reduce the time between discovery of genetic targets and development of clinically validated applications. However, TCGA relies on voluntary contributions of patient tissue, mostly from large academic institutions. The Director of TCGA reported that TCGA has had challenges obtaining adequate representation of tissue from minorities. In lung cancer, only eight percent of the tissue is from African Americans. When the majority of research is conducted in patients of European ancestry, genomic medicine risks becoming a factor that compounds, rather than reduces, existing racial health disparities.
VA’s ROLE AND RESPONSIBILITY TO FACILITATE EQUITY
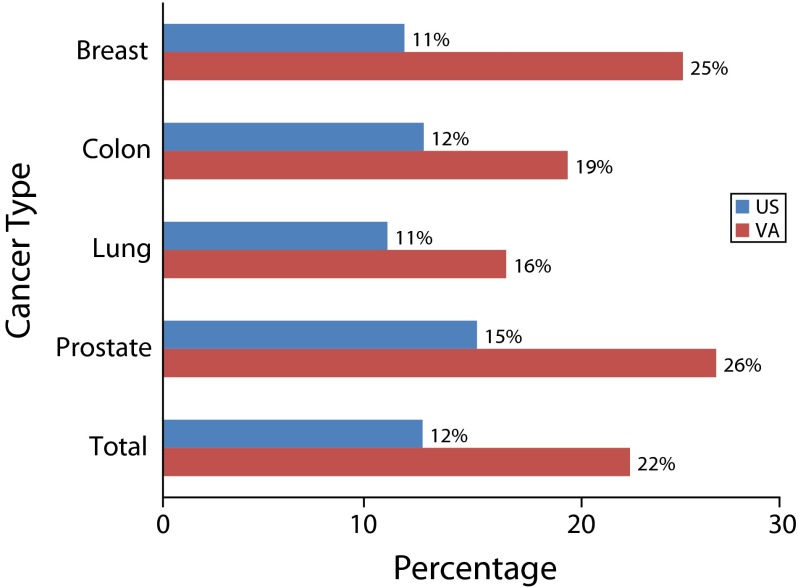
The volume of genetic test utilization highlights the crucial role of VHA clinicians and researchers in genomic medicine. The National Center for Veterans Analysis and Statistics reported that in 2012, 6.33 million veterans were treated in the VHA, of whom 753 270 (11.9%) were African American, 386 130 (6.1%) were Hispanic, and 189 900 (3%) were other minorities, including American Indian, American Native, Asian, Native Hawaiian, Pacific Islander, and mixed races. The VHA has a unique responsibility and opportunity to facilitate equity in access to genomic medicine. As illustrated in Figure 1, in diseases such as breast, colon, lung, and prostate cancers, for which genetics play a significant role in diagnosis, disease progression, and treatment, the percentage of African American patients in the VHA population is more than double that of the national population. In prostate cancer, veterans represent 7.45% of the US prostate cancer population.
FIGURE 1—
Percentage of patients that are African American by cancer type: United States, 2010.
Note. VA = US Department of Veterans Affairs.
Source. National Cancer Institute and the Centers for Disease Control and Prevention, http://statecancerprofiles.cancer.gov; Veterans Health Administration data: VHA Cancer Care Registry, 2010 (VACCR).
The VHA is responding to the challenge of achieving equity in genomic medicine in several ways, including:
Developing a centralized model for delivery of genomic medicine. The Genomic Medicine Service employs a team of 8 genetic counselors who are available to consult with all VHA clinicians about genetic tests requested by patients or providers. These genetic counselors are also available to provide telegenetics consulting services for patients at 60 Veterans Affairs Medical Centers.
Clinical leadership and the Office of Information Technology are developing a database and mobile application called VA GDx, which contains clinical genetic test orders and results from the majority of laboratories providing genetic testing services.
The Million Veteran Program, launched by the Office of Research and Development, has enrolled and collected blood DNA samples from more than 200 000 veterans, 21% of these veterans are minorities, 26 280 are African American and 15 000 are other races.
Health services researchers are collaborating with commercial laboratories such as Genomic Health and Myriad to study the impact of clinically validated gene expression tests on treatment decisions for veterans. These laboratories have clinically validated, regulatory-approved assays. However minorities have been underrepresented in their validation studies. VHA studies will provide important information on the validity and clinical utility of these gene expression algorithms in all veterans.
Clearly, the VHA is ahead of many hospital systems in developing clinical care processes that will facilitate equity in genomic medicine. However, given the veteran population and the VHA’s ability to link large volumes of genetic data to clinical outcomes, it can, and should, do more. Developing a genomic medicine module within the EHR would greatly improve the ability to measure delivery of genomic medicine. It would facilitate the development of pragmatic clinical trials, enabling researchers to study genomic medicine in all patients via the EHR. As illustrated by the example of the EGFR assay in lung cancer, lack of minority participation in research makes it crucial to develop clinical informatics systems capable of studying real world implementation of genomic medicine. With the increased use of high throughput sequencing, this becomes especially important because there will be many genetic variants identified that have unknown clinical significance. By linking clinical genetic testing to the EHR, the VHA can make important contributions to understanding gene variants, particularly those that may appear more frequently in African American populations. There are several federal efforts aimed at cataloging and making available data from large-scale sequencing of human tissue. VHA participation in these initiatives would greatly improve representation of minorities in genomic research and it would generate data that allow us to deliver personalized genomic care for all veterans. Lastly, in diseases where veterans represent a significant percentage of the total population, the VHA needs to be an active participant in any federal initiative to analyze equity.
Acknowledgments
J. Lynch is supported by the Veterans Affairs postdoctoral fellowship program and an interagency agreement from National Cancer Institute. S. A. Ibrahim is supported in part by a K24 Mid-Career Development Award from the National Institute of Arthritis and Musculoskeletal and Skin Disorders (K24AR055259).
Note. The views expressed in this editorial are those of the authors and do not represent those of the Department of Veterans Affairs, the National Institute of Arthritis and Musculoskeletal and Skin Disorders, or the National Institutes of Health.
References
- 1.Haiman CA, Chen GK, Blot WJ et al. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43(6):570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch JA, Khoury MJ, Borzecki A et al. Utilization of epidermal growth factor receptor (EGFR) testing in the United States: a case study of T3 translational research. Genet Med. 2013;15(8):630–638. doi: 10.1038/gim.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sundi D, Ross AE, Humphreys EB et al. African American men with very low–risk prostate cancer exhibit adverse oncologic outcomes after radical prostatectomy: should active surveillance still be an option for them? J Clin Oncol. 2013;31(24):2991–2997. doi: 10.1200/JCO.2012.47.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ge D, Fellay J, Thompson AJ et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 5.Purrington KS, Slager S, Eccles D et al. Genome-wide association study identifies 25 known breast cancer susceptibility loci as risk factors for triple-negative breast cancer. Carcinogenesis. 2014;35(5):1012–1019. doi: 10.1093/carcin/bgt404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams DR, Mohammed SA, Leavell J, Collins C. Race, socioeconomic status, and health: complexities, ongoing challenges, and research opportunities. Ann N Y Acad Sci. 2010;1186:69–101. doi: 10.1111/j.1749-6632.2009.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaeger R, Avila-Bront A, Abdul K et al. Comparing genetic ancestry and self-described race in African Americans born in the united states and in Africa. Cancer Epidemiol Biomarkers Prev. 2008;17(6):1329–1338. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SH, Mechanic LE, Yang P et al. Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res. 2005;11(6):2106–2110. doi: 10.1158/1078-0432.CCR-04-1853. [DOI] [PubMed] [Google Scholar]
- 9.Leidner RS, Fu P, Clifford B et al. Genetic abnormalities of the EGFR pathway in African American patients with non-small-cell lung cancer. J Clin Oncol. 2009;27(33):5620–5626. doi: 10.1200/JCO.2009.23.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinersman JM, Johnson ML, Riely GJ et al. Frequency of EGFR and KRAS mutations in lung adenocarcinomas in African Americans. J Thorac Oncol. 2011;6(1):28–31. doi: 10.1097/JTO.0b013e3181fb4fe2. [DOI] [PMC free article] [PubMed] [Google Scholar]