Abstract
Mdm2, an E3 ubiquitin ligase, negatively regulates the tumor suppressor p53. In this study, we utilized a conditional Mdm2 allele, Mdm2FM, and a CreER Tamoxifen inducible recombination system to examine the effects of global Mdm2 loss in adult mice. Two different Tamoxifen injection regimens caused 100% lethality of Mdm2FM/−;CreER mice. Both radio-sensitive and radio-insensitive tissues were impaired. Strikingly, a large number of radio-insensitive tissues, including the kidney, liver, heart, retina and hippocampus, exhibited various pathological defects. Similar Tamoxifen injections in older (16–18 month old) Mdm2FM/−;CreER mice showed abnormalities only in the kidney. In addition, transcriptional activation of p21, Puma and multiple senescence markers in young (2–4 month old) mice following loss of Mdm2 was dampened in older mice. All phenotypes were p53-dependent as Mdm2FM/−;p53−/−;CreER mice subjected to the same Tamoxifen regimens were normal. Our findings implicate numerous possible toxicities in many normal tissues upon use of cancer therapies that aim to inhibit Mdm2 in tumors with wild type p53.
Keywords: Mdm2 inhibition, radio-insensitive tissues, cancer therapy
Introduction
The Mdm2 E3 ubiquitin-ligase mediates ubiquitination of p53 and targets it for degradation [1]. In addition, Mdm2 binds the p53 transactivation domain and physically represses its transcriptional activity [1]. Studies in mice illustrate the importance of this relationship. Mdm2 deficiency in mice results in embryonic lethality due to initiation of apoptosis, which is completely rescued by simultaneous deletion of p53 [2–4]. Using a conditional Mdm2 allele, numerous studies have explored the effects of Mdm2 loss in specific tissues. Deletion of Mdm2 specifically in cardiomyocytes results in absence of heart development at E8.5. [5]. Moreover, Mdm2 deletion in the central nervous system of mouse embryos or smooth muscle cells (SMCs) leads to spontaneous apoptosis of neuronal cells [6–8] and death of SMCs [8], respectively. In addition, Mdm2−/− mice containing two copies of the p53515C allele, which encodes the apoptosis-deficient p53R172P protein, stabilize p53R172P in proliferative cells in multiple tissue compartments. This leads to impairment of progenitor cells in hematopoiesis and cerebellar development [9,10]. Hepatocyte-specific Mdm2 deletion in 6-week old mice led to increased hepatocyte apoptosis and spontaneous liver fibrosis [11]. To date, all of the Mdm2 loss-of-function phenotypes are completely rescued by concomitant removal of p53, indicating that elevated p53 activity determines the detrimental effects of Mdm2 loss in numerous tissues. The role of Mdm2 loss was also examined more globally in mice expressing a p53ERTAM allele. This allele expresses the 4-hydroxytamoxifen-dependent variant of p53 and together with a p53-null allele in an Mdm2-deficient background, shows that increased p53 activity is detrimental to radio-sensitive tissues, which is mainly composed of proliferating cells, whereas radio-insensitive tissues responded indolently [12]. However, the p53ERTAM allele is leaky and expresses some p53 in the absence of Tamoxifen as homogeneous p53ERTAM mice cannot rescue the Mdm2-null phenotype [12]. It therefore remains unclear whether Mdm2 contributes to the regulation of p53 activity independent of the differentiation or proliferation status of the cells.
On the other hand, p53 activity declines with age, as described in a previous study [13]. The efficiency of the p53 response to γ-radiation or other stresses decreased significantly in aged mice, including decreased p53 transcriptional activity and p53-dependent apoptosis. Whether elevated p53 activity due to Mdm2 loss, in and of itself, has similar effects in aged mice is unknown.
The fact that Mdm2 is a critical negative regulator of p53 is of clinical significance for cancer treatment. Many tumors overexpress Mdm2 and maintain wild type p53; thus, an appealing therapeutic strategy is to activate p53 by inhibition of Mdm2, or by interfering with the Mdm2-p53 interaction. Many small molecules, such as Nutlin, RITA or stapled peptides against p53, have been developed to do just that and some are in clinical trials [14]. However, given the cell-lethal phenotypes of Mdm2 loss in mice, potential adverse effects of Mdm2 inhibition in cancer patients should not be overlooked.
To address the above issues, Mdm2 was deleted in adult mice by using the Mdm2FM conditional allele and CreER. Mdm2 is lost only upon exposure to Tamoxifen. Our data clearly showed that loss of Mdm2 is harmful to both radio-sensitive and radio-insensitive tissues in young mice, and has milder effects in aged mice. The deleterious effects of Mdm2 deletion in adult mice are p53-dependent.
Materials and methods
Mice
Mdm2FM and CreER alleles have been published [15,16]. All mouse experiments were performed in compliance with MD Anderson Cancer Center Institutional Animal Care and Use Committee and conform to the guidelines of the United States Animal Welfare Act and the National Institutes of Health. Tamoxifen was injected intrapretoneally into Mdm2FM/−;CreER, Mdm2FM/−, or Mdm2FM/−;p53−/− ;CreER mice at 3mg/40gram body weight three times. Mice were sacrificed 2 days after the last injection.
To assess how efficiently Tamoxifen injection induced recombination of the Mdm2FM allele in different tissues, a pair of primers, p1 (5′ GAAGCTTGATATCGAATTCCG 3′) and p2 (5′ CCGCTCTAGAACTAGTGGAT 3′), was designed to specifically detect the Mdm2Δ5/6 allele and not any other Mdm2 allele (Suppl. Fig. 1A). Mdm2FM/+ mice were mated to CMV-Cre mice to obtain the Mdm2Δ5/6/+ allele that results from the Cre recombination in the germ line. The amount of Mdm2Δ5/6 DNA determined from real-time PCR with primers p1 and p2 from Mdm2Δ5/6/+ tail DNA represents a 100% percent recombination of the Mdm2FM allele, to which the amount of Mdm2Δ5/6 allele DNA in all the tissues from the Mdm2FM/−;CreER mice (with Tamoxifen administered) were compared, generating the recombination efficiency.
Histology, TUNEL staining, and immunohistochemistry
Tissues were fixed in 10% (vol/vol) formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin, or trichrome. Pathology was performed by Carolyn Van Pelt, Mingjian James You and Laura Pageon. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were carried out on paraffin-embedded sections according to the manufacture’s specifications (Calbiochem, Darmstadt, Germany). Antibodies used for immunohistochemistry were: p53, 1:100 (CM5, Novacastra, Norwell, MA, USA); Caspase 3, 1:200 (Cell Signaling Technology, Danvers, MA, USA); and Ki67, 1:100 (Abcam Inc., Cambridge, MA, USA). Visualization was performed using the ABC and DAB kits and counterstaining was performed with nuclear fast red (Vector Laboratories, Burlingame, CA, USA).
Western blot and Real-time PCR analysis
Tissues were homogenized and protein lysates were prepared for western blots. The antibodies used were: α-smooth muscle actin (A-2547), β-actin (AC-15) and γ-tubulin (GTU-88) (Sigma, St. Louis, MO, USA). Total RNA was extracted from the tissues by using TRIzol reagent (Life Technologies, CA, USA). The first-strand cDNA synthesis kit (GE Healthcare, NJ, USA) was used for reverse transcriptase reactions. Real-time PCR was performed according to the manufacturer’s specifications (Life Technologies, CA, USA).
ChIP Assay
Kidneys harvested from young and old Mdm2FM/−;CreER mice were ground into powder and washed with PBS. Chromatin was fixed with formaldehyde, and chromosome immunoprecipitation (ChIP) assays were carried out as described previously [17,18].
Statistical Analysis
Student t-test was performed with Prism 4 software (GraphPad Software, San Diego, CA, USA). Differences were considered significant at a value of p<0.05.
Results
Lethal effects of Mdm2 loss in adult mice
Cre-mediated recombination of the Mdm2FM conditional allele yields the Mdm2Δ5/6 allele, which is functionally null [15]. This Mdm2FM allele thus allows us to examine the effects of Mdm2-deficiency in various adult tissues. To examine the global effects of Mdm2 loss, Mdm2FM/FM mice were mated to Mdm2+/−;CreER mice to generate Mdm2FM/−;CreER and Mdm2FM/− mice. CreER mice express an active Cre recombinase only upon addition of Tamoxifen. These mice were subjected to intraperitoneal injection of Tamoxifen every day for three continuous days (designated as daily administration) at the age of 2–4 months. All Tamoxifen injected Mdm2FM/−;CreER mice became moribund within 1–2 days after injection and were sacrificed. By contrast, all the Mdm2FM/− control mice lacking CreER remained active and healthy throughout Tamoxifen treatments, and thereafter.
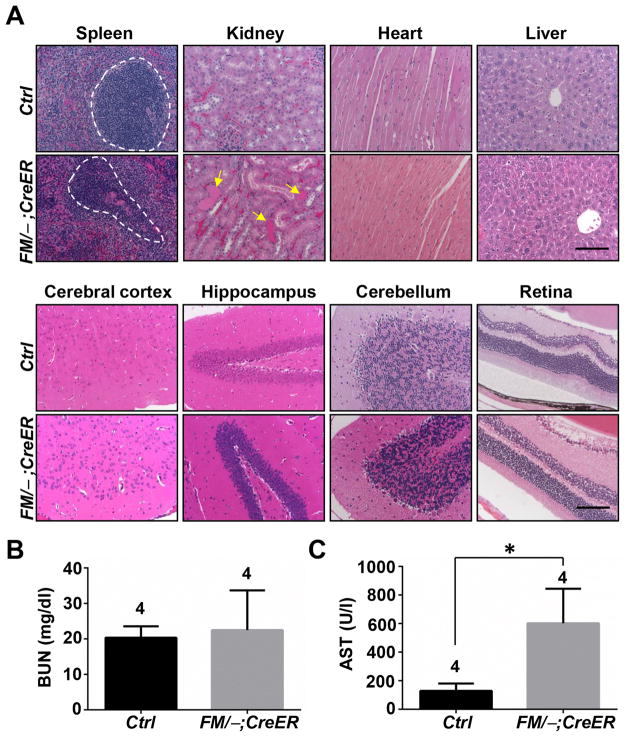
To explore the pathologies of the lethality, tissues from the Mdm2FM/−;CreER mice were harvested and analyzed histologically. Consistent with a previous report [12], multiple radio-sensitive tissues underwent atrophy, as evidenced by extensive absence of white pulp in the spleen (Fig. 1A, Spleen), erythroid depletion in bone marrow, and villus atrophy and apoptotic crypts in the intestine (data not shown). Surprisingly, contrary to previous studies [12], the kidney, a tissue insensitive to radiation, also exhibited defects, as shown by protein casts and tubule dilation (Fig. 1A, Kidney). Nevertheless, other radio-insensitive tissues, including the heart, liver, cerebral cortex, hippocampus, cerebellum and retina remained morphologically normal (Fig. 1A). Furthermore, Masson’s trichrome stain did not detect any fibrosis in the kidney, liver and heart (data not shown).
Figure 1.
The response of Mdm2FM/−;CreER mice to daily Tamoxifen injection. A, H&E staining of paraffin-embedded sections of various tissues. B, Measurement of the blood urine nitrogen concentration (BUN) in blood samples. C, Measurement of the aspartate aminotransferase (AST) concentration in blood samples. Ctrl: the control Mdm2FM/− mice. FM/−;CreER: the Mdm2FM/−;CreER mice. Dashed circles indicate white pulp. Arrows indicate dilated tubules filled with protein cast. Scale bar, 100 μm. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B and C.
To further examine the pathologies, blood was subjected to standard chemistry analysis. The blood urine nitrogen (BUN) concentration, which assays kidney function, remained normal (Fig. 1B), indicating that the morphological defects observed in the kidney were not severe enough to cause functional impairment within this time period. Surprisingly, although morphologically normal, the liver exhibited a functional abnormality, as the level of aspartate aminotransferase (AST) increased by nearly 4 fold compared to control mice (Fig. 1D).
The absence of lesions in most of the radio-insensitive tissues did not seem to be due to a low recombination efficiency of the Mdm2FM allele following Tamoxifen treatment. In fact, in the cerebrum, cerebellum and heart, 40–50% of the Mdm2FM alleles were recombined (Suppl. Fig. 1B), which is comparable to the recombination efficiency of the Mdm2FM allele in the kidney (~75%, Suppl. Fig. 1B). Additionally, the degree of recombination did not correlate with the onset of a phenotype. The spleen had a relatively low recombination efficiency, approximately 20% (Suppl. Fig. 1B), yet displayed absence of white pulp. The liver had a similar recombination frequency and exhibited elevated AST levels (Fig. 1A). [19].
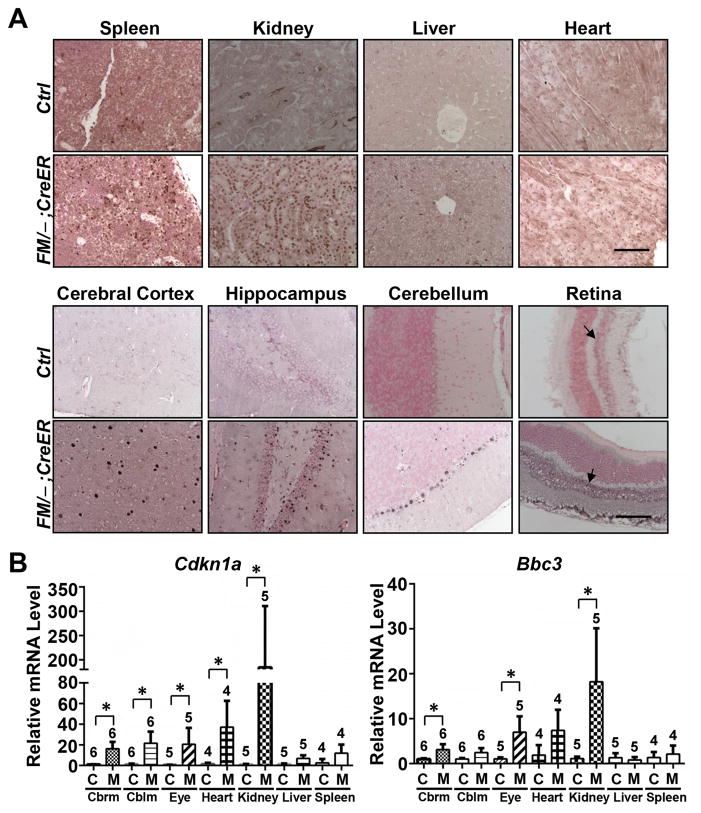
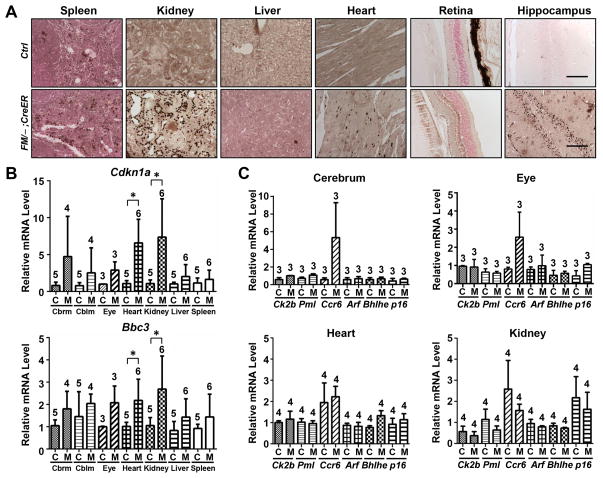
The recombination of the Mdm2FM allele consequently stabilized the p53 protein, not only in the radio-sensitive, but also in radio-insensitive tissues such as kidney, liver, heart, cerebral cortex, hippocampus, cerebellum and retina (Fig. 2A) indicating Mdm2 mediates p53 degradation in all of these tissues. To assess directly p53 activity resulting from loss of Mdm2 in these tissues, we used real-time PCR to assay induction of p53 target genes p21 and Puma. Both genes were significantly induced in tissues including kidney, cerebrum, cerebellum and eye, albeit the induction level varied between tissue types (Fig. 2B). p21 was robustly activated by approximately 190 fold in the kidney, 40 fold in the heart, and 20 fold in cerebellum, cerebrum and eye, but only slightly in the liver and spleen. On the other hand, Puma was elevated approximately 20 fold in the kidney, 7 fold in the eye and heart, and 3 fold in the cerebrum and 2 fold in the cerebellum. Thus, even tissues that failed to show morphological defects upon Mdm2 loss, showed elevated p53 activity.
Figure 2.
p53 activation in the Mdm2FM/−;CreER mice subjected to daily Tamoxifen injection. A, p53 immunohistochemical staining of paraffin-embedded sections of tissues. B, Real-time reverse transcription PCR analysis of p21 and Puma mRNAs in a variety of tissues. Cbrm: cerebrum. Cblm: cerebellum. Ctrl or C: the control Mdm2FM/− mice. FM/−;CreER or M: the Mdm2FM/−;CreER mice. Scale bar, 100 μm. Arrows indicate inner nuclear layer of retina. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B.
Loss of Mdm2 impaired both radiosensitive and radio-insensitive tissues in a p53-dependent manner
Loss of Mdm2 is deleterious to radio-sensitive tissues. Above we demonstrated that kidney, retina and liver, three radio-insensitive tissues, exhibited various abnormalities following Mdm2 deletion (Fig. 1). However, due to the speed at which the mice developed symptoms and had to be sacrificed (1–2 days), it was unclear whether radio-insensitive tissues might also be affected. To potentially decrease lethality, we employed an alternative treatment regimen, in which Tamoxifen was injected intrapretoneally to 2–4 month old Mdm2FM/−;CreER mice every 3 days for a total of three times (designated interrupted regimen). Within 1–2 days after the third dose, this interrupted regimen also induced 100% fatality of the Mdm2FM/−;CreER mice, which exhibited severe loss of balance and less mobility before being sacrificed. By contrast, Mdm2FM/− control animals lacking CreER remained active and healthy throughout Tamoxifen administrations, and thereafter.
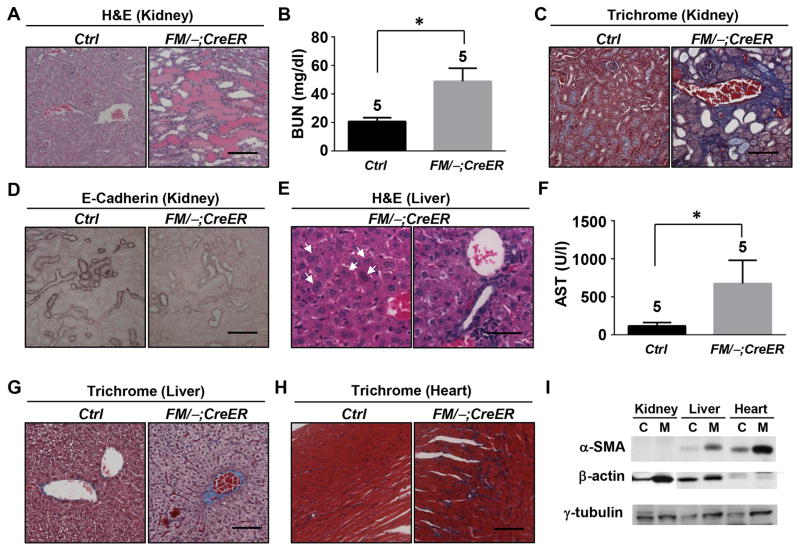
Histological analysis revealed that, consistent with previous reports, all classical radio-sensitive tissues being studied, including spleen, thymus, bone marrow and intestine were damaged (data not shown). Importantly, interrupted Tamoxifen injections affected more radio-insensitive tissues, including kidney, liver, heart, retina and hippocampus, than daily injections. In contrast to its relatively mild phenotype to daily Tamoxifen injections (Fig. 1), the kidney was severely traumatized by interrupted injections, as evidenced by highly extensive tubule dilation and tubular proteinuria (Fig. 3A) and a 2.5 fold increase of BUN levels (Fig. 3B). Interestingly, the kidney also exhibited a fibrosis phenotype (Fig. 3C). Previous studies have indicated the contribution of epithelial-mesenchymal transition (EMT) to fibrosis [20]. Indeed, loss of E-Cadherin, one of the fundamental events in EMT [21], occurred in the kidney of the Mdm2FM/−;CreER mice following interrupted Tamoxifen injections (Fig. 3D). Another impaired radio-insensitive tissue is the liver, which exhibited multinucleate hepatocytes (Fig. 3E, Left) and lymphocytes infiltration into portal tract and sinusoids (Fig. 3E, Right). Accompanying these morphological abnormalities was impaired liver function, as demonstrated by a nearly 6-fold elevation of the AST level compared with the control mice (Fig. 3F). Similar to the kidney, the liver and heart both underwent drastic fibrosis (Fig. 3G and H). However, there was no E-Cadherin loss (data not shown). Instead, increased α-smooth muscle actin expression, which is another biomarker for EMT [21], was detected in both liver and heart, but not in the kidney (Fig. 3I). The β-actin expression, however, was evidently elevated in the kidney, consistent with the notion that there is dynamic actin remodeling during EMT [22]. Together, these data suggest different molecular mechanisms underlie the fibrosis phenotype among the kidney, liver and heart in the Mdm2FM/−;CreER mice after interrupted Tamoxifen treatment.
Figure 3.
Interrupted Tamoxifen injection caused deleterious effects on a variety of radio-insensitive tissues of the Mdm2FM/−;CreER mice. A, H&E staining of paraffin-embedded sections of kidney. B, Measurement of the blood urine nitrogen concentration (BUN) in blood samples. C, Trichrome staining of paraffin-embedded sections of kidney. D, E-Cadherin immunohistochemical staining of paraffin-embedded sections of kidney. E, H&E staining of paraffin-embedded sections of liver. Arrows indicate multinucleated hepatocytes. F, Measurement of the aspartate aminotransferase (AST) concentration in blood samples. G, Trichrome staining of paraffin-embedded sections of liver. H, Trichrome staining of paraffin-embedded sections of heart. I, Western blot analysis of α-SMA and β-actin expressions in protein lysates of kidney, liver and heart. Ctrl or C: the control Mdm2FM/− mice. FM/−;CreER or M: the Mdm2FM/−;CreER mice. α-SMA: α-smooth muscle actin. Scale bar, 100 μm. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B and F.
In the Mdmd2FM/−;CreER mice, the interrupted Tamoxifen injection regimen induced 10–50% recombination of the Mdm2FM alleles in various tissues being studied (Suppl. Fig. 1C), which consequently stabilized p53 expression (Fig. 4A). The stabilized p53 protein in the Mdm2FM/−;CreER mice exhibited a robust activity by increasing the p21 transcription, in both radio-sensitive, such as spleen, and radio-insensitive tissues including kidney, heart, liver, eye, cerebrum and cerebellum (Fig. 4B). Except for the spleen and liver, Puma, another p53 target, was also effectively activated in all other tissues examined.
Figure 4.
Interrupted Tamoxifen injection induced p53 activity in the Mdm2FM/−;CreER mice. A, p53 immunohistochemical staining of paraffin-embedded sections of tissues. B, Real-time reverse transcription PCR analysis of p21 and Puma mRNAs in a variety of tissues. Cbrm: cerebrum. Cblm: cerebellum. Ctrl or C: the control Mdm2FM/− mice. FM/−;CreER or M: the Mdm2FM/−;CreER mice. Scale bar, 100 μm. Arrows indicate inner nuclear layer of retina. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B.
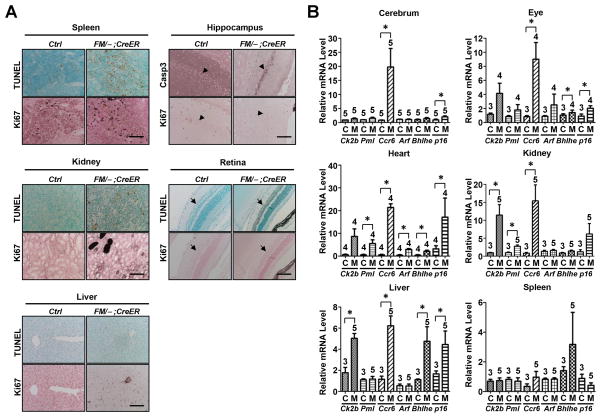
Numerous studies have demonstrated the role of p53 in mediating cellular apoptosis, cell cycle arrest and senescence [23,24]. Therefore, we further explored whether these p53 functions were involved in the phenotypes of Mdm2FM/−;CreER mice following Tamoxifen injections. As expected, the spleen underwent extensive apoptosis, but was accompanied by an aberrant increase of proliferation (Fig. 5A, Spleen), which may explain the relatively low percentage of recombination at the Mdm2FM allele (Suppl. Fig. 1C). Surprisingly, although not widespread, apoptosis was also detected in the kidney and liver of the Mdm2FM/−;CreER mice, and as expected was completely absent in control kidney and liver samples (Fig. 5A, Kidney and Liver). The inner nuclear layer of retina and the hippocampus both underwent evident apoptosis as well (Fig. 5A, Retina and Hippocampus), which may contribute to the loss-of-balance symptoms exhibited by Mdm2FM/−;CreER mice (Suppl. Video). The heart of the Mdm2FM/−;CreER mice did not undergo detectable apoptosis (data not shown). However, activation of the p21 gene, which occurred in the heart and other radio-insensitive tissues (Fig. 4B), suggests that p53 might induce senescence in those tissues. As a validation, in addition to p21, the transcriptional activation profile of multiple senescence markers, including p15, pml, Dcr2, Arf, Dec1 and p16, were examined in tissues from both the Mdm2FM/− and Mdm2FM/−;CreER mice. Notably, two or more of these markers (including p21) were clearly induced in all radio-insensitive tissues examined (Fig. 5B). Interestingly, the activation pattern, in terms of which and how many senescence markers were activated, as well as fold activation, varied tremendously among tissues from the Mdm2FM/−;CreER mice (Fig. 5B), indicating tissue specificity of the molecular mechanisms underlying p53-induced senescence.
Figure 5.
Interrupted Tamoxifen injection induced apoptosis and senescence in the Mdm2FM/−;CreER mice. A, TUNEL staining and ki67 immunohistochemical staining of paraffin-embedded sections of tissues. Caspase 3 staining was alternatively used to detect apoptosis in hippocampus due to extensive non-specific TUNEL staining in the control Mdm2FM/− mouse brain. B, Real-time reverse transcription PCR analysis of the mRNAs of a variety of senescence markers in different tissues. Ctrl or C: the control Mdm2FM/− mice. FM/−;CreER or M: the Mdm2FM/−;CreER mice. Casp3: Caspase 3. Scale bar, 100 μm. Arrowheads indicate hippocampus. Arrows indicate inner nuclear layer of retina. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B.
Although the major function of Mdm2 is to suppress p53 activities, emerging evidence has identified p53-independent roles of Mdm2 [25,26]. To examine whether the phenotypes observed above are p53-dependent, 2–4 month old Mdm2FM/−;p53−/−;CreER mice were subjected to the same daily or interrupted Tamoxifen treatments as described above. Contrary to the lethality of the Mdm2FM/−;CreER mice, mice lacking p53 remained active throughout 7 days of Tamoxifen administration, and thereafter. Consistent with the overall normal physical appearance, there were no pathological defects detected in collected tissues including spleen, kidney, heart, liver, retina and brain (Suppl. Fig. 2). These results unequivocally show that the deleterious effects following Mdm2 deletion in the Mdm2FM/−;CreER mice were p53-dependent.
Mdm2 is essential for mouse viability at all ages
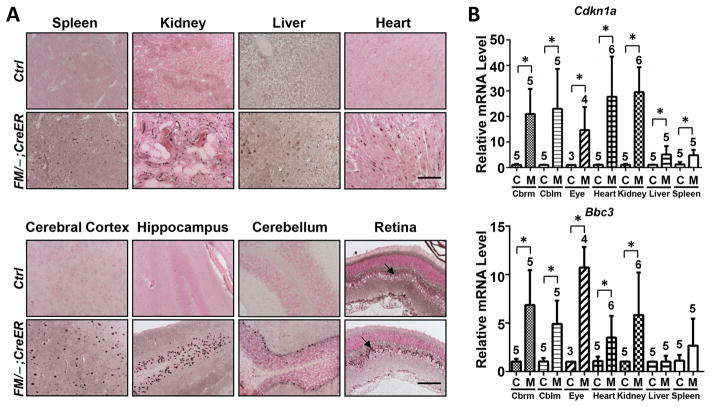
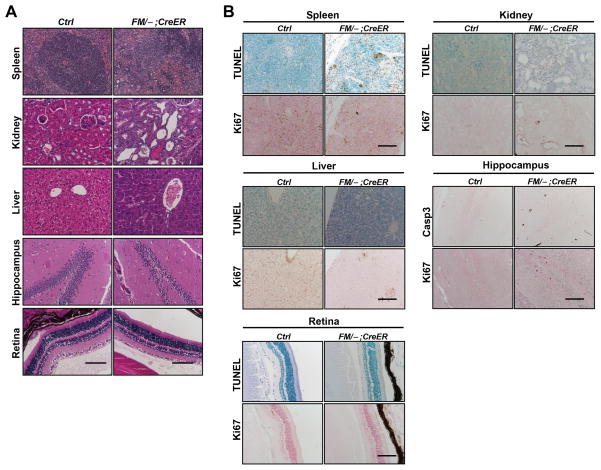
The use of the conditional Mdm2 allele, Mdm2FM, allowed us to determine that Mdm2 was indispensable for mouse viability at young adult stages (2–4 months old). Since human cancer patients that might receive Mdm2 inhibitors as a treatment option vary with respect to age, we examined the necessity of Mdm2 in suppressing p53 activity in aged mice. Therefore, 16–18 month old mice were given the same interrupted Tamoxifen treatment as described above. This treatment induced 100% lethality within 1–2 days after the third dose of Tamoxifen. Extensive atrophy was detected in the spleen (Fig. 6A, Spleen). Tubular dilution and protein cast were also revealed in the kidney (Fig. 6A, Kidney), albeit not as extensive as what was observed in the kidney of young Mdm2FM/−;CreER mice (compare with Fig. 3A). Pathological analysis was unable to detect any defects in the liver (Fig. 6A, Liver). In addition, while the spleen showed apoptosis, none of the radio-insensitive tissues being studied, including the kidney, liver, retina and hippocampus, exhibited apoptosis (Fig. 6B). The milder response to Tamoxifen treatment in the aged Mdm2FM/−;CreER mice was apparently not due to inefficient Mdm2 deletion, since Tamoxifen induced the same percentage of recombined Mdm2FM allele in all tissues of the aged Mdm2FM/−;CreER mice as those of young adult mice (Suppl. Fig. 1C). As a consequence of Mdm2 loss, p53 was stabilized (Fig. 7A), but, surprisingly, did not cause significant activation of p21 or Puma in most tissues examined (Fig. 7B). This is in sharp contrast with the remarkable activation of p21 and Puma in the young Mdm2FM/−;CreER mice (Fig. 4B), and may partially account for the milder tissue phenotypes in these aged Mdm2FM/−;CreER mice (Fig. 6). Furthermore, none of the senescence markers were significantly expressed in the kidney, heart, retina and cerebrum (Fig. 7C), contrary to the expression patterns in young mice (Fig. 5B).
Figure 6.
The response of aged Mdm2FM/−;CreER mice to interrupted Tamoxifen injection. A, H&E staining of paraffin-embedded sections of various tissues. B, TUNEL staining and ki67 immunohistochemical staining of paraffin-embedded sections of tissues. Caspase 3 staining was alternatively used to detect apoptosis in hippocampus due to extensive non-specific TUNEL staining in the control Mdm2FM/− mouse brain. Ctrl: the control Mdm2FM/− mice. FM/−;CreER: the Mdm2FM/−;CreER mice. Casp3: Caspase 3. Scale bar, 100 μm.
Figure 7.
Activation of p53 and senescence markers in the aged Mdm2FM/−;CreER mice subjected to interrupted Tamoxifen injection. A, p53 immunohistochemical staining of paraffin-embedded sections of tissues. B, Real-time reverse transcription PCR analysis of p21 and Puma mRNAs in a variety of tissues. C, Real-time reverse transcription PCR analysis of the mRNAs of a variety of senescence markers in different tissues. Cbrm: cerebrum. Cblm: cerebellum. Ctrl or C: the control Mdm2FM/− mice. FM/−;CreER or M: the Mdm2FM/−;CreER mice. Scale bar, 100 μm. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar in B and C.
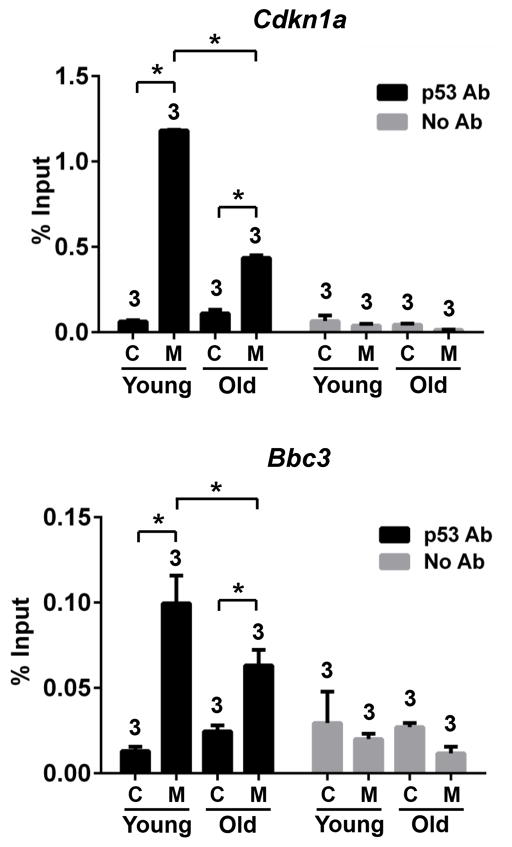
We further explored the mechanism underlying this declining p53 sensitivity by performing Chromatin Immunoprecipitation (ChIP) assays of young and old Mdm2FM/−;CreER kidneys. In response to Mdm2 deletion, p53 binding to p21 and Puma promoters were significantly enhanced in both young and old kidneys (Fig. 8). However, notably, for both target promoters, p53 binding was dampened in old kidneys, suggesting that poorer access of p53 to its target promoters, at least partially, accounts for the decreased sensitivity of p53 signaling in old mice.
Figure 8.
ChIP assays with p53 antibody to examine p53 binding at the p21 and Puma promoters in the kidneys of young or old mice subjected to interrupted Tamoxifen treatment. C: the control Mdm2FM/− mice. M: the Mdm2FM/−;CreER mice. * p<0.05. The number of animals in each sample group is indicated on the top of each respective bar.
In addition to p53, previous studies indicate that Mdm2 also suppresses two other p53 family members, p63 and p73 [27–30]. To examine whether p63 or p73 activates gene transcription differentially in different tissues during aging following Mdm2 deletion, the mRNA levels of two p53-independent transcriptional targets of p63 and p73, BRCA2 and Rad51 [31], in the cerebrum, heart and kidney of young or old Mdm2FM/−;CreER mice were assayed. In all these tissues of young or old mice, loss of Mdm2 did not increase BRCA2 or Rad51 mRNA levels (Suppl. Fig. 3A), indicating that the decreased sensitivity of p53 signaling in old mice is not due to differences in p63 or p73 activation. Mdm4, another p53 negative regulator [1], is not involved in this age-dependent p53 signaling either, as Mdm2 deletion did not alter Mdm4 protein levels in either young or old mice (Suppl. Fig. 3B)
Taken together, our results suggest that, as the mouse ages, p53 signaling becomes sluggish in response to loss of Mdm2. Despite this, since major organs, such as the spleen and kidney, were impaired, older mice with Mdm2 depletion still exhibited lethal phenotypes, indicating that Mdm2 is essential for viability even late in life.
Discussion
Concerns have arisen for potential adverse effects of Mdm2 inhibitor treatment of cancers that overexpress Mdm2 and maintain wild type p53. Although there are studies implying that Mdm2 inhibitors only affect tumor cells and not normal tissues [33–36], these data should be cautiously interpreted, as these studies either used xenograft models or short-term assays. Whether prolonged administration of Mdm2 inhibitors will be detrimental to patients remains unknown. In fact, by using a p53ERTAM allele expressing the 4-hydroxytamoxifen-dependent variant of p53, the Evan laboratory showed that increased p53 activity is detrimental to adult mice [12]. This study provided solid evidence for the deleterious effects of loss of Mdm2 in radio-sensitive tissues. However, as this study was performed in an Mdm2-deficient and p53 heterogeneous background (Mdm2−/−; p53ERTAM/−), it does not mimic the clinical situation whereby cancers overexpress Mdm2 and have two wild type p53 alleles. In contrast, our conditional Mdm2 allele, Mdm2FM, allows us to activate p53 from both alleles after deletion of Mdm2 which, therefore, more faithfully recapitulates the above clinical situation. The intrinsic differences in these two model systems may largely account for the discrepancy in the outcomes. Mdm2FM/−; CreER mice subjected to the interrupted Tamoxifen regimen exhibited defects not only in classical radio-sensitive tissues, but also multiple radio-insensitive tissues, including kidney, heart, liver and tissues with well differentiated neuronal cells such as retina and hippocampus. On the other hand, the treatment with daily injection regimen of Mdm2FM/−; CreER mice in our current study and of Mdm2−/−; p53ERTAM/− mice previously [12], caused less severe phenotypes in both cases compared with the interrupted injection of Mdm2FM/−;CreER mice, suggesting that different treatment regimens may also contribute to the differences in tissue responses. Our study emphasizes the possibility that treating cancer patients with Mdm2 inhibitors may be detrimental and therefore needs to be cautiously monitored. In particular, both radio-sensitive and radio-insensitive tissues should be monitored.
In previous studies, mice with Mdm2 loss specifically in the ureteric bud epithelium or nephron progenitor cells displayed severe renal defects and died after birth or perinatally, respectively, indicating high sensitivity of kidney to p53 dosage during organogenesis [37,38]. In the present study, notably, although with different severity, Mdm2 deletion consistently caused kidney injury, regardless of the Tamoxifen treatment regimen (i.e., daily vs. interrupted injection,) or mouse age (i.e., 2–4 vs. 16–18 months old). These data highlight the necessity of closely monitoring kidney function of patients that receive Mdm2 inhibitors as treatment.
Growing evidence suggests p53-independent roles of Mdm2, but only when Mdm2 is overexpressed or under inflammatory conditions [25,26,39]. In our present study, all the defects observed in the Mdm2FM/−;CreER mice upon Tamoxifen injections were p53-dependent since they were completely absent in the Mdm2FM/−;p53−/−;CreER mice subjected to the same treatment. This clearly suggests that, at basal levels, the key target of Mdm2 is p53.
Mdm2 is required for embryonic development since germ line deletion of Mdm2 is embryonic lethal due to p53-induced apoptosis [2,3]. Although viable, Mdm2−/− mice containing the homozygous p53515C allele, which encodes the apoptosis-deficient p53R172P protein, die within two weeks after birth, suggesting that Mdm2 remains essential for suppressing p53 activity during early postnatal stage [9]. Above, we showed that loss of Mdm2 was indispensable for the viability of both young (2–4 months) and old (16–18 months) adult mice. The necessity of Mdm2 for suppressing the activity of p53 during the whole life span of the mouse has thus been completed. Nevertheless, although the ultimate outcome is the same, loss of Mdm2 caused less severe symptoms in old adult mice compared with those in young mice, which was not due to differences in Cre-mediated recombination efficiency. Fewer numbers of tissues in old mice showed lesions; molecularly, the transcripts of Puma, p21 and all of the senescence markers examined were much less efficiently activated in old mice. This age-association of the deleterious effects following Mdm2 deletion is of clinical significance. Childhood cancers, which are mostly p53 wild-type at the time of diagnosis, have been considered an ideal target for using Mdm2 inhibitors as a therapeutic approach [40]. However, our observation that loss of Mdm2 causes more severe damage to normal tissues in younger mice suggests that extra precaution is necessary when using Mdm2 inhibitors in pediatric cancer patients.
In mice treated with γ-radiation, the age-associated decline in p53 functionality has also been described [13]. In the same study, the function of the Ataxia-telangiectasia mutated (ATM) kinase was found to decline significantly with age as well, which may be responsible for the decline of the p53 response to radiation. It was thus suggested that the age-associated declines in ATM and p53 signaling functions in mice provided a potential mechanism for the age-associated increases in cancer rates. However, since in our study p53 is activated by Cre-mediated Mdm2 deletion instead of radiation, one can speculate that other factors, in addition to the declined ATM activity, are involved in down-regulating p53 functionality in aging mice. Indeed, ChIP assays showed that poorer access of p53 to its binding site on target promoters, possibly due to epigenetic changes in nucleosomes, at least partially accounted for the decreases sensitivity of p53 signaling in old mice (Fig. 8). Additional studies will promote understanding the correlation between tumorigenesis and the aging process.
Supplementary Material
Footnotes
Author contributions
YZ and SBX contributed equally to this study and were responsible for the experimental design, undertaking the experiments, data analysis and interpretation, and manuscript drafting. QL and SH carried out some of the immunohistochemistry staining, real time PCR assay and data analysis. MT carried out some mouse dissections as well as DNA and RNA purifications, and helped with manuscript revision. CVP, MJY and LP performed pathological analysis of HE stained tissue sections. GL conceived the experiments, interpreted data, revised and finalized the manuscript.
The authors declare that there are no conflicts of interest
SUPPLEMENTARY MATERIAL ON THE INTERNET
The following supplementary material may be found in the online version of this article:
Figure S1. Recombination efficiency of the Mdm2FM allele in multiple tissues of the Mdm2FM/− ;CreER mice subjected to Tamoxifen injection.
Figure S2. The response of Mdm2FM/−;p53−/−;CreER mice to interrupted Tamoxifen injection.
Figure S3. The transcriptional activity of p63 and p73, as well as Mdm4 protein levels in young and old mice subjected to interrupted Tamoxifen injection.
References
- 1.Marine JC, Francoz S, Maetens M, et al. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell death and differentiation. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 2.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 3.Jones SN, Roe AE, Donehower LA, et al. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 4.Chavez-Reyes A, Parant JM, Amelse LL, et al. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer research. 2003;63:8664–8669. [PubMed] [Google Scholar]
- 5.Grier JD, Xiong S, Elizondo-Fraire AC, et al. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Molecular and cellular biology. 2006;26:192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francoz S, Froment P, Bogaerts S, et al. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong S, Van Pelt CS, Elizondo-Fraire AC, et al. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3226–3231. doi: 10.1073/pnas.0508500103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boesten LS, Zadelaar SM, De Clercq S, et al. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell death and differentiation. 2006;13:2089–2098. doi: 10.1038/sj.cdd.4401973. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Terzian T, Xiong S, et al. The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. The Journal of pathology. 2007;213:360–368. doi: 10.1002/path.2238. [DOI] [PubMed] [Google Scholar]
- 10.Abbas HA, Maccio DR, Coskun S, et al. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 2010;7:606–617. doi: 10.1016/j.stem.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodama T, Takehara T, Hikita H, et al. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. The Journal of clinical investigation. 2011;121:3343–3356. doi: 10.1172/JCI44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ringshausen I, O’Shea CC, Finch AJ, et al. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Feng Z, Hu W, Teresky AK, et al. Declining p53 function in the aging process: a possible mechanism for the increased tumor incidence in older populations. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Lozano G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grier JD, Yan W, Lozano G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis. 2002;32:145–147. doi: 10.1002/gene.10066. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 17.Pant V, Xiong S, Jackson JG, et al. The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes & development. 2013;27:1857–1867. doi: 10.1101/gad.227249.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson JG, Pereira-Smith OM. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Molecular and cellular biology. 2006;26:2501–2510. doi: 10.1128/MCB.26.7.2501-2510.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada A, Takada Y, Kim U, et al. Bone marrow, spleen, and thymus regeneration patterns in mice after whole-body irradiation. Radiation research. 1971;45:522–535. [PubMed] [Google Scholar]
- 20.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. The Journal of clinical investigation. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer metastasis reviews. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 23.Jin S, Levine AJ. The p53 functional circuit. Journal of cell science. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 24.Jackson JG, Post SM, Lozano G. Regulation of tissue- and stimulus-specific cell fate decisions by p53 in vivo. The Journal of pathology. 2011;223:127–136. doi: 10.1002/path.2783. [DOI] [PubMed] [Google Scholar]
- 25.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell death and differentiation. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 26.Bouska A, Eischen CM. Mdm2 affects genome stability independent of p53. Cancer research. 2009;69:1697–1701. doi: 10.1158/0008-5472.CAN-08-3732. [DOI] [PubMed] [Google Scholar]
- 27.Zeng X, Chen L, Jost CA, et al. MDM2 suppresses p73 function without promoting p73 degradation. Molecular and cellular biology. 1999;19:3257–3266. doi: 10.1128/mcb.19.5.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zdzalik M, Pustelny K, Kedracka-Krok S, et al. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell cycle. 2010;9:4584–4591. doi: 10.4161/cc.9.22.13871. [DOI] [PubMed] [Google Scholar]
- 29.Kadakia M, Slader C, Berberich SJ. Regulation of p63 function by Mdm2 and MdmX. DNA and cell biology. 2001;20:321–330. doi: 10.1089/10445490152122433. [DOI] [PubMed] [Google Scholar]
- 30.Galli F, Rossi M, D’Alessandra Y, et al. MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. Journal of cell science. 2010;123:2423–2433. doi: 10.1242/jcs.061010. [DOI] [PubMed] [Google Scholar]
- 31.Lin YL, Sengupta S, Gurdziel K, et al. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS genetics. 2009;5:e1000680. doi: 10.1371/journal.pgen.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Xiong S, Pant V, Suh YA, et al. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer research. 2010;70:7148–7154. doi: 10.1158/0008-5472.CAN-10-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 34.Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 35.Shangary S, Qin D, McEachern D, et al. Temporal activation of p53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernal F, Wade M, Godes M, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilliard S, Aboudehen K, Yao X, et al. Tight regulation of p53 activity by Mdm2 is required for ureteric bud growth and branching. Developmental biology. 2011;353:354–366. doi: 10.1016/j.ydbio.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hilliard SA, Yao X, El-Dahr SS. Mdm2 is required for maintenance of the nephrogenic niche. Developmental biology. 2014;387:1–14. doi: 10.1016/j.ydbio.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulay SR, Thomasova D, Ryu M, et al. MDM2 (murine double minute-2) links inflammation and tubular cell healing during acute kidney injury in mice. Kidney international. 2012;81:1199–1211. doi: 10.1038/ki.2011.482. [DOI] [PubMed] [Google Scholar]
- 40.Tisato V, Norcio A, Voltan R, et al. MDM2 non-genotoxic inhibitors as innovative therapeutic approaches for the treatment of pediatric malignancies. Curr Med Chem. 2013;20:2226–2236. doi: 10.2174/0929867311320170007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.