Abstract
Hydroxynitrile lyases (HNLs) are powerful carbon–carbon bond forming enzymes. The reverse of their natural reaction – the stereoselective addition of hydrogen cyanide (HCN) to carbonyls – yields chiral cyanohydrins, versatile building blocks for the pharmaceutical and chemical industry. Recently, bacterial HNLs have been discovered, which represent a completely new type: HNLs with a cupin fold. Due to various benefits of cupins (e.g. high yield recombinant expression in Escherichia coli), the class of cupin HNLs provides a new source for interesting, powerful hydroxynitrile lyases in the ongoing search for HNLs with improved activity, enantioselectivity, stability and substrate scope. In this study, database mining revealed a novel cupin HNL from Acidobacterium capsulatum ATCC 51196 (AcHNL), which was able to catalyse the (R)-selective synthesis of mandelonitrile with significantly better conversion (97%) and enantioselectivity (96.7%) than other cupin HNLs.
Keywords: Hydroxynitrile lyase, Manganese-dependent HNL, Biocatalysis, Cyanohydrin synthesis, Mandelonitrile
Hydroxynitrile lyases (HNLs) catalyse the reversible stereoselective addition of hydrogen cyanide (HCN) to aldehydes and ketones. Both, (R)- and (S)-selective HNLs are well-established industrial biocatalysts for the synthesis of chiral cyanohydrins, which are important building blocks for pharmaceuticals and agrochemicals [1–3]. Hydroxynitrile lyases represent a diverse group of mainly plant enzymes with members in several protein folds [4]: FAD-dependent oxidoreductases, α/β-hydrolase family, serine carboxypeptidases, Zn2 +-dependent alcohol dehydrogenases, and very recently, bacterial cupins. They descend from distinct ancestors and differ in their enantioselectivity, co-factor dependence and substrate scope [5,6]. For industrial application, in particular, activity and stability at acidic pH and solvent stability are desired to reduce the unselective non-enzymatic background reaction. Further demands made on HNLs are high expression levels in an easy to handle expression host and a broad substrate scope. Although a lot of effort has already been made to improve and tailor HNLs for industrial requirements by protein, substrate and reaction engineering [5,7–9], there is still a clear need for further enhancement of HNLs or discovery of new HNLs exhibiting improved properties. A new door has recently opened with the discovery of bacterial hydroxynitrile lyases with a cupin fold, e. g. GtHNL from Granulicella tundricola, by our group [10,11]. Cupins are small stable beta barrel proteins, which are ubiquitous in nature and very diverse in function [12]. In most cupins metal ions are bound to the active site [13]. Cupin HNLs are characterised by high expression levels in Escherichia coli and an already remarkable stability under various conditions, however, so far relatively low activity compared to other HNLs [6,10,11]. The goal of the present study was to expand the class of cupin HNLs by identifying new members with higher activity and enantioselectivity and broader substrate scope via database search. Out of ten chosen targets, one enzyme, AcHNL from Acidobacterium capsulatum ATCC 51196 exhibited improved activity and was characterised in more detail.
The basic approach comprised a Basic Local Alignment Search Tool (BLAST, [14]) search with the sequence of GtHNL (Uniprot: E8WYN5, [10]) focused on protein sequences originating from acidophilic and/or thermophilic organisms. The search was restricted to protein sequences with identical metal-binding amino acids as GtHNL (His53, His55, Gln59 and His94). Moreover, only sequences containing His96 and His106 located in the active site (numbering according to GtHNL) were considered, because previous site-directed mutagenesis studies showed that they were essential for activity [10]. Out of hundreds of sequences with high similarity to GtHNL, a set of ten mainly hypothetical proteins was selected (Table S1) originating from acidophilic and/or thermophilic bacteria or bacteria with high resistance to environmental hazards. The respective enzymes share between 38% and 84% sequence identity with GtHNL (Fig. 1).
Fig. 1.

Multi sequence alignment of CH_1: Methylocella silvestris, CH_2: Acidobacterium capsulatum ATCC 51196, CH_3: Acidiphilium cryptum JF-5, CH_4: Paenibacillus sp. JDR-2, CH_5: Deinococcus deserti, CH_6: Burkholderia phymatum STM815, CH_7: Sinorhizobium medicae WSM419, CH_8: Bradyrhizobium sp. WSM471, CH_9: Alicyclobacillus acidocaldarius subsp. acidocaldarius Tc-4-1, and CH_10: Caldicellulosiruptor saccharolyticus DSM 8903 with GtHNL. The alignment was generated with Clustal W [15] and visualized with the programme ESPript 3.0 [16].
The genes were codon optimised for E. coli and ordered as synthetic DNA, recloned into the pET26b(+) expression vector and transformed into E. coli BL21(DE3) Gold (see Supplemental information). Preliminary activity screenings were performed on colony level towards (R/S)-mandelonitrile. As GtHNL was found to be dependent on manganese [10], the activities of the selected enzymes expressed with and without various bivalent ions in the expression media were compared. As expected, all enzymes were able to cleave the substrate in the presence of MnCl2 (Fig. 2), however to a different extent. The addition of other metal ions (Cu2 +, Co2 +, Zn2 +, Ni2 +, Fe2 +) to the media did not show any impact on the activity (data not shown). In comparison to GtHNL especially the enzyme from A. capsulatum ATCC 51196 (CH_2) developed stronger blue spots on the HCN-sensitive detection paper after the same period of time.
Fig. 2.
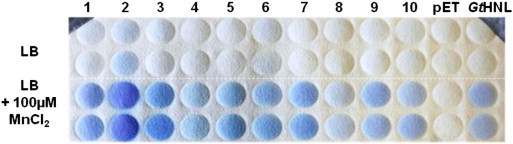
Influence of MnCl2 addition on the cyanogenesis activity. The assay was performed with an adapted version of the colony HNL-assay developed by Krammer et al. [17] (see supplemental information) using 12 mM (R/S)-mandelonitrile in 100 mM citrate phosphate buffer, pH 3.5. Upper two rows: activity of cupin HNLs expressed without MnCl2; lower two rows: activity of cupin HNLs expressed in the presence of 100 μM MnCl2. Negative control: pET26b-vector; positive control: GtHNL. The numbering refers to CH_1–10.
In accordance with the screening results, all target enzymes except CH_8 were expressed in high yield as soluble proteins in E. coli (~ 50% of total soluble protein) (Figure S1).
As the chemical background reaction is significantly reduced at low pH, an “ideal” HNL should exhibit high stability and activity at low pH. Hence, the cyanogenesis activity of E. coli lysate containing the expressed enzymes was tested at pH 3.5, 4.0, 4.5 and 5.0 in 100 mM sodium oxalate buffer or citrate phosphate buffer or at pH 5.6 in 100 mM MES oxalate buffer. All HNLs (except expectedly CH_8) were active at pH 5.0 or higher to variable degrees and also at pH 4.5 the enzymes exhibited low activity; especially CH_2 showed a promising signal at pH 4.5. No activity was detected at pH 3.5 (data not shown).
Subsequently, the stability of CH_2 at different pH values was examined by incubation of cleared CH_2 lysate in the respective buffers. In accordance with the results from the assay, the SDS-PA gel revealed that at pH 4.5 most of CH_2 was still found in the soluble fraction (Fig. 3); the amount, however, decreased significantly at lower pH values.
Fig. 3.
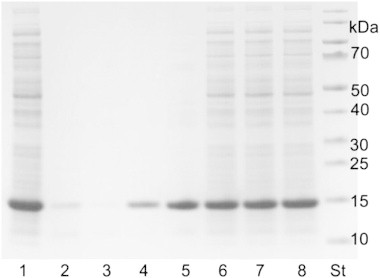
SDS-PA gel of cleared lysates (~ 10 μg) of CH_2 (pH 7.0, lane 1 as reference) and soluble fractions of CH_2 after incubation of the cleared lysates in 100 mM citrate phosphate buffer for 30 min and subsequent centrifugation, pH 3.0 (lane 2), 3.5 (lane 3), 4.0 (lane 4), 4.5 (lane 5), 5.0 (lane 6), 5.5 (lane 7) and 100 mM MES oxalate buffer pH 5.6 (lane 8). (CH_2 monomer: 14.1 kDa), St.: PageRuler Prestained Protein Ladder.
The enzymes were investigated for their ability to catalyse the synthesis of mandelonitrile from benzaldehyde and HCN (Table 1). Only CH_6 did not show any conversion of benzaldehyde after 24 h. Three candidates, CH_1, CH_3 and CH_4 were slightly better than GtHNL and reached about 97% conversion with enantiomeric excesses (ee) between 90.2 and 93.1%. Remarkably, CH_2 showed nearly full conversion (99.5%) after 24 h with a very high ee (R) of 96.7%. Furthermore, a high conversion of benzaldehyde (94%) and an ee of 96.6% were achieved already after 8 h reaction time. Importantly, the ee was stable during the whole course of the reaction.
Table 1.
Stereoselective conversion of benzaldehyde to (R)-mandelonitrile in the presence of CH_1–10.
| 8 h |
24 h |
|||
|---|---|---|---|---|
| Enzymea | Conv. [%] | ee [%] | Conv. [%] | ee [%] |
| CH_1 | 77.3 | 93.3 | 97.6 | 93.1 |
| CH_2 | 94.0 | 96.6 | 99.5 | 96.7 |
| CH_3 | 76.2 | 92.3 | 97 | 92.3 |
| CH_4 | 77.5 | 90.8 | 97 | 90.2 |
| CH_5 | 36.9 | 79.2 | 75.7 | 78.5 |
| CH_6 | ND | ND | ND | ND |
| CH_7 | 53.9 | 86.7 | 69.6 | 81.8 |
| CH_9 | 75.3 | 81.7 | 96.1 | 81.2 |
| CH_10 | 5.2 | 16.7 | 18.4 | 11.9 |
| GtHNL | 57.3 | 89.0 | 91.9 | 88.9 |
| BpHNL | 83.1 | 95.0 | 96.6 | 94.9 |
| pET26b(+) | ND | ND | 10.4 | 0.1 |
Reaction conditions: Two-phase system comprising 2 mL MTBE containing 2 M HCN and 0.5 M benzaldehyde and 1 mL concentrated cleared lysates (45 mg/mL, pH 4), 1000 rpm, 5 °C.
ND: not determined due to the low conversion.
100 μM MnCl2 added to expression medium.
The structure of GtHNL, which was solved recently [10,18], indicates that the active site of the enzyme is located in a large cavity, which should provide enough space to bind bulky substrates. Thus, the new cupin-HNLs were examined in the synthesis of 3-phenoxybenzaldehyde cyanohydrin. Seven out of nine enzymes showed weak or no activity in the synthesis of 3-phenoxybenzaldehyde cyanohydrin (data not shown). But, very interestingly, two enzymes exhibited notable activity (Table 2). Although the conversion was higher with CH_7, excellent enantioselectivity was only obtained with CH_2 (> 99.9% after 1 and 4 h).
Table 2.
Conversion of 3-phenoxybenzaldehyde.
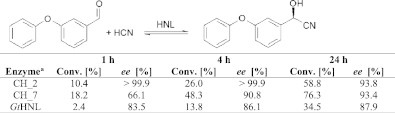
Reaction conditions: Two-phase system comprising 1 mL MTBE containing 2 M HCN and 0.5 M 3-phenoxy-benzaldehyde and 0.5 mL concentrated cleared lysates (45 mg/mL, pH 4), 1000 rpm, 5 °C.
a 100 μM MnCl2 added to expression medium.
As for none of the chosen target proteins, 3D structures are available, models were build using the SWISS-MODEL protein structure homology-modelling server [19] and the structure of GtHNL as template [10] to be able to identify differences in the active site. Structural alignment in Pymol and cavity analysis using parts of the programme VASCo [20] and a LIGSITE algorithm [21] revealed that there are hardly any significant differences in the size and the shape of the cavities in the different target proteins. This is not unexpected as many of the active site amino acids (V42, F44, L61, H96, H106 and W120, numbering refers to GtHNL) are identical (Fig. 1, and S2A). The positions A40 and Q110 are less conserved. Moreover, CH_2 contains a valine at position 25, where an isoleucine is found in most other cupin HNLs creating a little bit more space for bulky substrates. In CH_7 a leucine is found at the respective position (Figure S2B).
Considering the results obtained so far, it was decided to choose CH_2 from A. capsulatum ATCC 51196 (subsequently renamed AcHNL) for a more detailed analysis concerning its substrate scope. For this purpose the industrially relevant substrates 2-chlorobenzaldehyde, 3-phenylpropanal and 3-phenylprop-2-enal were tested. However, only low conversion (10–20% above the background reaction after 24 h) and no or very low ee (< 20%) were observed (data not shown).
For a more detailed analysis, AcHNL (expressed in the presence or absence of MnCl2, subsequently named AcHNL+Mn and AcHNL-Mn) was purified by anion-exchange chromatography and subsequent size-exclusion chromatography. The elution profile of AcHNL is identical to GtHNL (Figure S3). Thus, we can assume that AcHNL is a tetramer as it was observed in the GtHNL structure [10]. The metal contents of the purified proteins were determined by inductively coupled plasma/optical emission spectroscopy. Without addition of MnCl2 to the medium 0 mol% manganese and only traces of iron (6.3 mol%) and zinc (4.6 mol%), which are generally present in LB medium, were found per metal-binding site. In contrast, the expression of AcHNL in LB medium supplemented with 100 μM MnCl2 led to a loading of 70 mol% manganese per metal-binding site. Both enzyme preparations were tested for mandelonitrile synthesis. Purified AcHNL+Mn showed 97.3% conversion and 94.7% ee (R) after 24 h (Fig. 4). This is significantly higher than values obtained previously for purified GtHNL, for which a large difference in the activity of lysate and purified enzyme was observed (50% conversion with 89.3% ee after 24 h using 18 mg/mL of protein in otherwise identical reaction conditions), which was attributed to the stabilizing effect of the lysate [10]. The only slightly decreased conversion and enantiomeric excess obtained with purified AcHNL in comparison to the concentrated lysate (99.5% conversion and 96.7% ee (R) after 24 h) can be also explained by the lower amount of enzyme present in the reaction (~ 22.5 mg/mL of cupin in cleared lysate, compared to 11.8 mg/mL of purified enzyme). As expected, AcHNL expressed without manganese addition, exhibited only 35.3% conversion and a strongly decreased ee (R) of 35.8% after 24 h confirming the manganese-dependence of cupin HNLs.
Fig. 4.
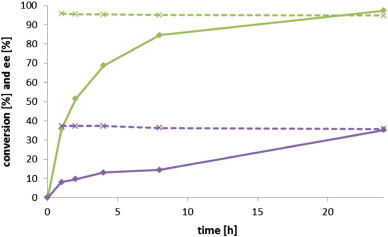
Conversion of benzaldehyde by purified AcHNL. Reaction conditions: Two-phase system comprising 1 mL MTBE containing 2 M HCN and 0.5 M benzaldehyde and 0.5 mL purified AcHNL expressed ± MnCl2 (11.8 mg/mL, pH 4), 1000 rpm, 5 °C. Green: AcHNL+Mn, purple: AcHNL-Mn, solid lines: conversion, dashed lines: ee.
The activity of purified AcHNL+Mn in the cyanogenesis of (R)-mandelonitrile was examined spectrophotometrically at pH 4.0, pH 4.5, pH 5.0 (sodium oxalate buffer) and pH 5.5 (MES oxalate buffer) by following the formation of the product benzaldehyde at 280 nm (see Supplemental information and Table S2). The enzyme displayed a specific activity of 0.69 ± 0.02 U/mg at pH 5.5. At pH 5.0, AcHNL exhibited a slightly decreased specific activity of 0.63 ± 0.06 U/mg, whereas a further drop of the pH to 4.5 led to a significant decrease of the specific activity with 0.12 ± 0.02 U/mg which can be attributed to a partial denaturation of the enzyme which was visible as faint cloudiness after the measurement in the wells of the microtitre plate. At pH 4.0 the enzyme was clearly precipitated during the measurement and thus, reliable data evaluation was not possible. Kinetic measurements of purified AcHNL+Mn (0.5 mg/mL) were performed in sodium oxalate buffer, pH 5.0, in a substrate concentration range between 3 and 30 mM, which revealed a Km of 4.2 ± 0.8 mM and a turnover number kcat of 9.5 s− 1, corresponding to a specific activity of 0.71 ± 0.035 U/mg at vmax.
In summary, a novel manganese-dependent bacterial HNL from A. capsulatum ATCC 51196 was discovered by database mining. Compared to other cupin HNLs it shows higher conversion and an excellent enantioselectivity in the synthesis of (R)-mandelonitrile.
For additional experimental procedures refer to supplemental information.
Acknowledgements
This work was supported by the Austrian Bundesministerium für Wirtschaft, Familie und Jugend, the Austrian Bundesministerium für Verkehr, Innovation und Technologie, the SFG, the Standortagentur Tirol, and the Technology Agency of the City of Vienna through the COMET Funding Program managed by the Austrian Forschungsförderungsgesellschaft. The research leading to these results has received funding from the European Union's Seventh Framework Programme for Research, Technological Development and Demonstration under grant agreement no. 289646 (Kyrobio). We thank Helmar Wiltsche, Institute of Analytical Chemistry and Food Chemistry, TU Graz for the ICP-OES analysis, and Karin Reicher and Elisa Lanfranchi for their help with cyanohydrin synthesis reactions.
Appendix A. Supplementary data
Supplementary material.
References
- 1.Gruber-Khadjawi M., Fechter M.H., Griengl H. Cleavage and formation of cyanohydrins. In: Drauz K., Gröger H., May O., editors. Enzyme catalysis in organic synthesis. Wiley-VCH Verlag GmbH & Co; Weinheim: 2012. pp. 947–990. [Google Scholar]
- 2.Lanfranchi E., Steiner K., Glieder A., Hajnal I., Sheldon R.A., van Pelt S. Recent developments in hydroxynitrile lyases for industrial biotechnology. Recent Pat Biotechnol. 2013;7:197–206. doi: 10.2174/18722083113076660010. [DOI] [PubMed] [Google Scholar]
- 3.Purkarthofer T., Skranc W., Schuster C., Griengl H. Potential and capabilities of hydroxynitrile lyases as biocatalysts in the chemical industry. Appl Microbiol Biotechnol. 2007;76:309–320. doi: 10.1007/s00253-007-1025-6. [DOI] [PubMed] [Google Scholar]
- 4.Gruber K., Kratky C. Biopolymers for biocatalysis: structure and catalytic mechanism of hydroxynitrile lyases. J Polym Sci Polym Chem. 2004;42:479–486. [Google Scholar]
- 5.Dadashipour M., Asano Y. Hydroxynitrile lyases: insights into biochemistry, discovery, and engineering. ACS Catal. 2011;1:1121–1149. [Google Scholar]
- 6.Winkler M., Glieder A., Steiner K. Hydroxynitrile lyases: from nature to application. In: Carreira E.M., Yamamoto H., editors. Comprehensive chirality. Elsevier; Amsterdam: 2012. pp. 350–371. [Google Scholar]
- 7.Andexer J.N., Langermann J.V., Kragl U., Pohl M. How to overcome limitations in biotechnological processes — examples from hydroxynitrile lyase applications. Trends Biotechnol. 2009;27:599–607. doi: 10.1016/j.tibtech.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Hanefeld U. Immobilisation of hydroxynitrile lyases. Chem Soc Rev. 2013;42:6308–6321. doi: 10.1039/c3cs35491a. [DOI] [PubMed] [Google Scholar]
- 9.Avi M., Wiedner R.M., Griengl H., Schwab H. Improvement of a stereoselective biocatalytic synthesis by substrate and enzyme engineering: 2-hydroxy-(4′-oxocyclohexyl)acetonitrile as the model. Chem Eur J. 2008;14:11415–11422. doi: 10.1002/chem.200800609. [DOI] [PubMed] [Google Scholar]
- 10.Hajnal I., Łyskowski A., Hanefeld U., Gruber K., Schwab H., Steiner K. Biochemical and structural characterization of a novel bacterial manganese-dependent hydroxynitrile lyase. FEBS J. 2013;280:5815–5828. doi: 10.1111/febs.12501. [DOI] [PubMed] [Google Scholar]
- 11.Hussain Z., Wiedner R., Steiner K., Hajek T., Avi M., Hecher B. Characterization of two bacterial hydroxynitrile lyases with high similarity to cupin superfamily proteins. Appl Environ Biotechnol. 2012;78:2053–2055. doi: 10.1128/AEM.06899-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunwell J.M., Purvis A., Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Chavez F.A., Banerjee A., Sljivic B. Modeling the metal binding site in cupin proteins. In: Pramatarova L., editor. On biomimetics. In tech. 2011. pp. 3–28. [Google Scholar]
- 14.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 16.Gouet P., Courcelle E., Stuart D.I., Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 17.Krammer B., Rumbold K., Tschemmernegg M., Pöchlauer P., Schwab H. A novel screening assay for hydroxynitrile lyases suitable for high-throughput screening. J Biotechnol. 2007;129:151–161. doi: 10.1016/j.jbiotec.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 18.Łyskowski A., Steiner K., Hajnal I., Steinkellner G., Schwab H., Gruber K. Crystallization of a novel metal-containing cupin from Acidobacterium sp. and preliminary diffraction data analysis. Acta Crystallogr F. 2012;68:451–454. doi: 10.1107/S1744309112006550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Steinkellner G., Rader R., Thallinger G.G., Kratky C., Gruber K. VASCo: computation and visualization of annotated protein surface contacts. BMC Bioinformatics. 2009;10:32. doi: 10.1186/1471-2105-10-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendlich M., Rippmann F., Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–363. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.


