Abstract
Accumulating studies revealed that the expression levels of several miRNAs are up or down-regulated in osteosarcoma (OS). The aim of this study was to investigate the functional significance and molecular of the let-7g in OS cells. The expression levels of let-7g was significantly down-regulated in OS cell lines U2-OS and HOS cell compared to osteoblast cell lines HOB cell. Moreover, bioinformatic prediction suggested that Aurora-B, which is overexpressed and functions as an oncogene in OS cells, is a putative target gene of let-7g. Using mRNA and protein expression analysis and luciferase assays, we further identified let-7g directly regulated Aurora-B expression in OS cells. Functional investigation revealed both restoration of let-7g and silencing Aurora-B induce cell apoptosis and suppressed cell viability, migratory and invasive ability in OS cells. Finally, we found that silencing Aurora-B in OS cells could partly dampen anti-let-7g mediated tumor promotion. Thus, our findings suggested that let-7g inhibits OS cell malignant phenotype at least partly through targeting Aurora-B. Targeting of let-7g and Aurora-B may be a novel therapeutic strategy for treating OS.
Keywords: Osteosarcoma, let-7, Aurora-B, metastasis
Introduction
Osteosarcoma (OS) is the most common primary malignant tumor of bone occurred in young adolescent and children. This disease has a poor prognosis because of the metastases in the period of tumor progression, which are usually developed previous to the clinical diagnosis. Clearly, the impact of identifying factors that govern proliferation and metastasis is essential in the management of osteosarco- ma.
Aurora-B is one of the major protein kinases that ensures the proper execution and fidelity of mitosis. Increasing evidences showed that up-regulated Aurora-B expression involved in growth, invasion and metastasis in various malignant tumors, which make it believed to be an important anti-tumor target [1,2]. Li Y et al. showed that down-regulation of Aurora-B could inhibit proliferation and metastasis, induce G2/M phase arrest in clear cell renal cell carcinoma cells and exert antitumor activity in an SN12C xenograft model [3]. Our previous studies revealed that Aurora-B was elevated expression in OS tissues and knockdown of Aurora-B suppressed OS cells proliferation, migratory and invasion in vitro [4]. However, the molecular mechanisms on overexpression of Aurora-B in OS are still uncertain.
Numerous studies showed that MicroRNAs (miRNAs) plays an important role in regulating expression of oncogenes. MiRNAs are a category of small non-coding RNA molecule approximately 22 nucleotides in length. Mature miRNAs are involved in the transcriptional and post-transcriptional regulation of gene expression, through binding to complementary sequences in the 3’ untranslated regions (3’ UTRs) of target mRNA transcripts [5]. Accumulating studies have demonstrated that miRNAs played critical role in cell proliferation, apoptosis and metastasis [6-9]. Let-7 cluster has been shown to be significantly correlated with the occurrence and development of cancer, suggesting it is involved in the regulation of oncogenic pathways in numerous types of tumors [10-13]. However, whether let-7 cluster involved in the development of osteosarcoma and the potential molecular mechanisms are remain unexplored.
In this study, we found that let-7g, one member of let-7 cluster, mediated Aurora-B overexpression and promoted cell proliferation, migration and invasion in OS cells.
Results
Aurora-B may be a target for let-7 cluster
To identify potential miRNAs that may regulate Aurora-B expression, the bioinformatic research analysis (Pictar, Targetscan) was performed. Result showed that Aurora-B is included in the conserved targets of let-7 cluster as shown in Figure 1. Furthermore, the 3’UTR of Aurora-B was cloned and subjected to a luciferase reporter assay in U2-OS cell along with let-7 cluster mimics. We found that 8 members of let-7 cluster (let-7a/b/c/d/e/f/g/i) can significantly inhibit the reporter activity of the Aurora-B 3’UTR (Figure 2). To Further explore whether let-7a/g/i/d/b/c/e regulates Aurora-B, its wild-type 3’UTR was mutated by deleting the 7-bp sequence (D-Mut) complementary to let-7a/g/i/d/b/c/e or by introducing point mutations (Point-Mut) (Figure 2). We found that the inhibitory effect of let-7a/b/c/d/e/f/g/i on AuroraB reporter activity was significantly restrained (Figure 2). It indicated that Aurora-B may be negatively targeted by let-7a/b/c/d/e/f/g/i in OS.
Figure 1.
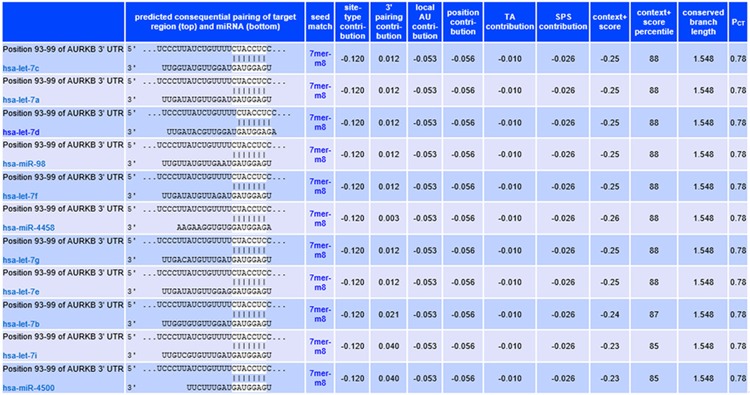
Results of bioinformatic research about microRNAs targeting Aurora-B. There are 11 membesr of let-7 cluster may target Aurora-B at the position of 93-99 bp.
Figure 2.
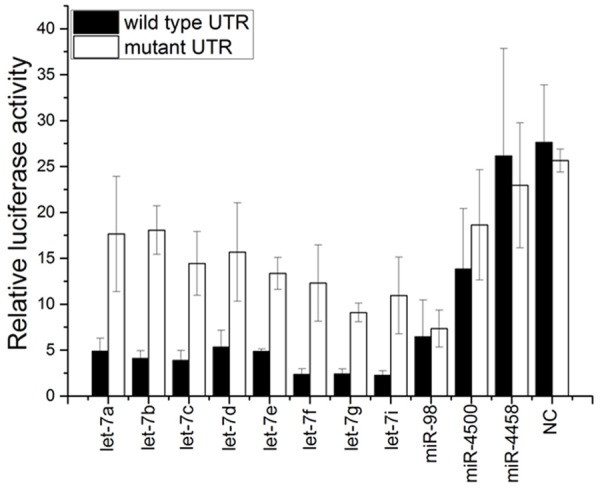
Luciferase reporter assay. Results indicated that Aurora-B may be negatively targeted by let-7a/b/c/d/e/f/g/i in OS.
Let-7a/b/c/d/e/f/g/i were downregulated expression in OS cells versus osteoblast cells
Previously, let-7 cluster has been reported as a tumor suppressor in colon,breast, lung endometrial and cancers [14,15]. Whereas the correlation between let-7 cluster dysregulation and OS remains unknown. The expression level of let-7a/b/c/d/e/f/g/i in OS cell (U2-OS and HOS) and osteoblast cell (HOB cell) was measured by qRT-PCR. The results revealed that the expression of let-7a/b/c/d/e/f/g/i was significantly lower in OS cells than those in osteoblast cells, espesially let-7g (Figure 3). It suggested that the let-7 microRNA family may play an essential role in OS malignant phenotype. Let-7g was the most significantly changed let-7 miRNA. Thus, let-7g was chosen as the candidate miRNA to conduct functional investigation in OS.
Figure 3.
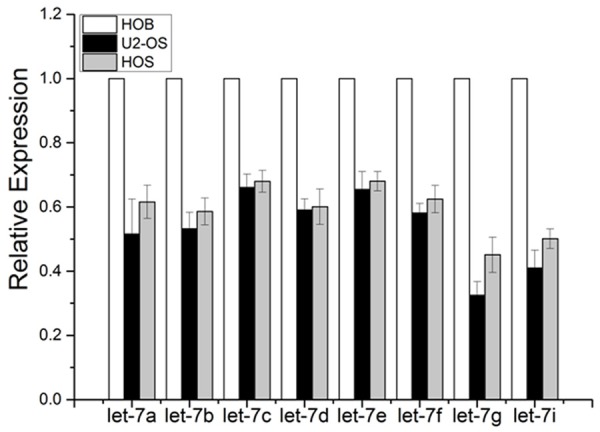
The expression level of let-7a/b/c/d/e/f/g/i in OS cell (U2-OS and HOS) and osteoblast cell (HOB cell) was evaluated by qRT -PCR. The results revealed that expression of let-7i was significantly down-regulated in OS cells compare to that in osteoblast cells, suggesting that let-7a/b/c/d/e/f/g/i may play an essential role in OS. Let-7g was the most significantly changed let-7 miRNA. So it was chosen as the candidate miRNA to conduct functional investigation in OS.
Let-7g inhibit Aurora-B expression in OS cells
To determine whether Aurora-B was regulated by let-7g in OS, the OS cell lines U2-OS and HOS cells that overexpression Aurora-B were treated with let-7g mimic and anti-let-7g, respectively. The expression of Aurora-B was measured by western blot and qRT-PCR. The results showed that the expression levels of Aurora-B mRNA and protein were significant lower in cells treated with let-7g mimic than those cells treated with anti-let-7g inhibitor and untreated (Figure 4). It indicated that let-7g inhibit Aurora-B expression in OS cells.
Figure 4.
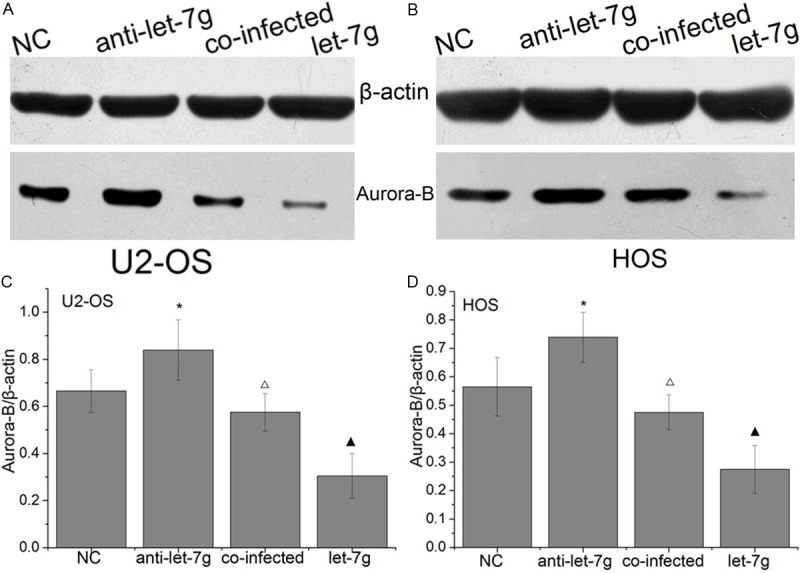
Let-7g inhibits Aurora-B expression in OS cells. Aurora-B mRNA (C, D) and protein (A, B) were significant lower in cells treated with let-7g mimic than those cells treated with anti-let-7g inhibitor and untreated. (Columns, mean (n=6); bars, S.D. *P < 0.05, ΔP < 0.05 VS NC, ▲P < 0.05 VS NC and anti-let-7g. NC, Negative control. anti-let-7g, anti-let-7g inhibitor (OS cells transfected by the reverse complementary sequence of let-7g). Co-infected, OS cells transfected by LV-shAurora-B combined with anti-let-7g. Let-7g, OS cells transfected by let-7g mimics).
Let-7g changes the malignant phenotype of OS cells by targeting Aurora-B
To explore the functional relationship between let-7g and Aurora-B in OS, the U2-OS and HOS cells were treated with let-7g mimic or anti-let-7g inhibitor and the ability of cells proliferation, migration and invasion was measured by MTT, wound healing and transwell invasion assays. It was found that the cell proliferation, migratory and invasive ability in elevated let-7g cells was significantly reduced compared to lower let-7g cells (Figures 5, 6 and 7), suggesting that let-7g has anti-malignant phenotype effects in OS cells.
Figure 5.
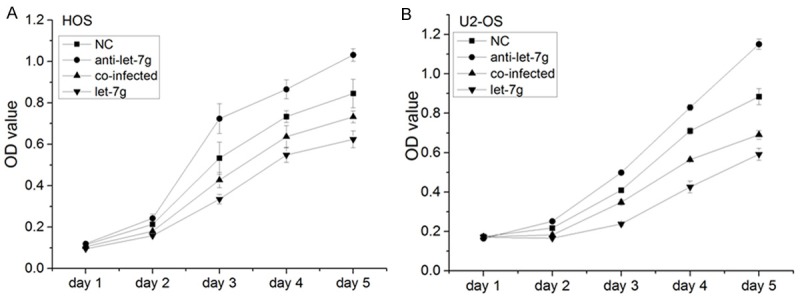
The OS cell proliferation was evaluated by MTT assays. The results revealed that the viability of OS cells was inhibited by restoration expression of let-7g in OS cells, which indicated that let-7g could inhibit OS cells viability in vitro. The tumor proliferation promotion mediated by anti-let-7g was partly inhibited by silencing Aurora-B in OS cells. (Columns, mean (n=6); bars, S.D. *P < 0.05, ΔP < 0.05 VS NC, ▲P < 0.05 VS NC and anti-let-7g.).
Figure 6.
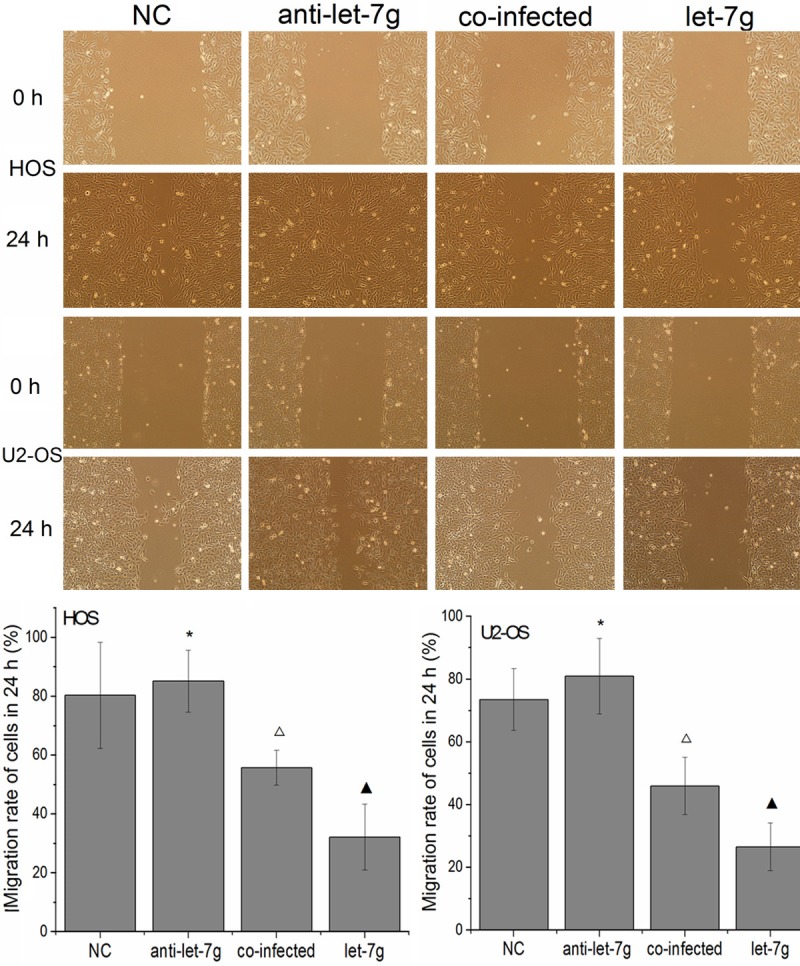
The migratory ability of OS cells was measured by wound healing assays. The migratory rate was significantly lower in cells infected with let-7g mimics than Let-7g changes malignant phenotype of OS cells that in cells in Let-7g changes malignant phenotype of OS cells infected with negative mimics, suggesting that enhanced expression of let-7g could inhibit OS cells migration in vitro. The tumor migratory promotion mediated by anti-let-7g was partly inhibited by silencing Aurora-B in OS cells. (Columns, mean (n=6); bars, S.D. *P < 0.05, ΔP < 0.05 VS NC, ▲P < 0.05 VS NC and anti-let-7g.).
Figure 7.
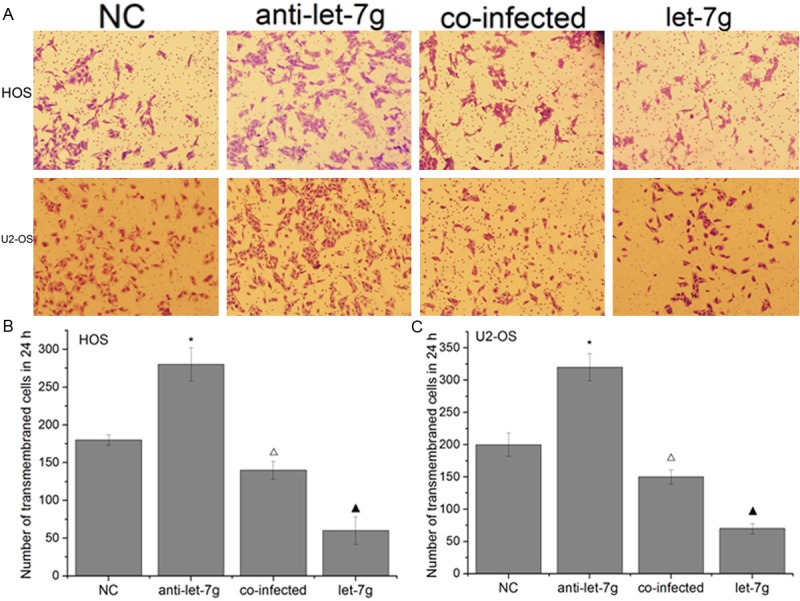
The invasive ability of cells was measured by transwell assays. The number of invasive cells was significantly lower in cells infected with let-7g mimics than that in cells infected with negative mimics, suggesting that enhanced expression of let-7g could inhibit OS cells invasion in vitro. The tumor invasive promotion mediated by anti-let-7g was partly inhibited by silencing Aurora-B in OS cells. (Columns, mean (n=6); bars, S.D. *P < 0.05, ΔP < 0.05 VS NC, ▲P < 0.05 VS NC and anti-let-7g.).
Furthermore, to investigate whether let-7g inhibits OS cells malignant phenotype by targeting Aurora-B, the OS cells were infected with let-7g mimic, anti-let-7g inhibitor and LV-sh Aurora-B combined with anti-let-7g (co-infected), respectively. In western blot assays, the results revealed that the Aurora-B protein level was significantly inhibited in cells infected with let-7g mimic. However, partially down-regulated Aurora-B protein level in co-transfected cells was observed (Figures 5, 6 and 7). The malignant phenotype of cell was investigated by measures the proliferation, migrator and invasive ability. The data showed the tumor promotion mediated by anti-let-7g was partly inhibited by silencing Aurora-B in OS cells (Figures 5, 6 and 7). These data suggested that let-7g alters OS cells malignant phenotype at least partly by targeting Aurora-B in vitro.
Discussion
In this study, we firstly found that let-7g expression is decreased in OS cells and restoring let-7g expression inhibits cell proliferation, migration and invasion by targeting Aurora-B in vitro.
A larger number of evidences revealed that Aurora-B involved in malignant tumor cells growth and metastasis [16]. In our previous study, we found that Aurora-B was overexpressed in OS tissues and cells and inhibition of Aurora-B by shRNA and small molecular inhibitor could suppress U2-OS cell proliferation, migration and invasion. In this study, to explore the potential molecular mechanisms on up-regulated Aurora-B expression in OS, we performed the bioinformatic research analysis to identify potential miRNAs that may interact with Aurora-B. The results showed that 11 members of let-7 cluster may be target Aurora-B. Furthermore, the luciferase reporter assay was performed to clarify whether Aurora-B is a potential target. The data indicated that eight mature miRNAs of let-7 cluster, including let-7a/b/c/d/e/f/g/i, may negatively target Aurora-B gene in OS cell.
One member of let-7 cluster was the first identified human miRNA in 2000 by Reinhart [17]. Numerous members of the let-7 cluster have been identified in various species [18]. So far, 11 mature subtypes of the let-7 cluster have been found in humans, including let-7a, -7b, -7c, -7d, -7e, -7f, -7g, -7i, miR-98, miR-4500 and miR-4458. Increasing studies have reported that members of let-7 cluster are down-regulated in various types of cancer, including lung cancer, gastric tumors, colon cancer, nasopharyngeal carcinoma, endometrial carcinoma and Bur-kitt’s lymphoma [19-21]. Decreased expression levels of let-7 cluster have been associated with the clinical postoperative survival of patients with malignant tumor [22,23]. The let-7 cluster is involved in the proliferation, apoptosis and metastasis of cancer cells [24]. Interesting, the upregulated expression of let-7 cluster was also observed in head and neck squamous cell carcinoma [25]. In present study, we measured the expression levels of members of let-7 cluster (including let-7a, -7b, -7c, -7d, -7e, -7f, -7g, -7i) in OS cells lines (U2-OS and HOS cell) and osteoblast cells (HOB) by qRT-PCR. We found that those members of let-7 cluster were lower expression in OS cells compared to HOB cells and let-7g was the most significant dysregulated miRNAs. It suggested that those mRNAs may play an important role in regulating cellular processes in osteosarcoma.
Decreased expression of MicroRNAs promoted tumorgenesis in various malignant tumors [24]. In this study, of the most significantly dysregulated miRNAs, let-7g, was highlighted for further functional analysis to identify its role in OS. The OS cells were treated with let-7g mimic and anti-let-7g respectively and the cell malignant phenotype was evaluated by investigating the cell viability, migratory and invasive ability. We found that let-7g prevented OS cells proliferation, migration and invasion in vitro.
To further elucidate whether let-7g alters OS cells malignant phenotype via directly targeting Aurora-B in OS, the correlation between let-7g and the Aurora-B expression was measured using qRT-PCR in OS cell lines. The data revealed that let-7g negative regulates Aurora-B expression at the post-transcriptinal level. In addition, the functional relationship between let-7g and Aurora-B in OS cells was analyzed. The results showed that up-regulated let-7g inhibit cell viability, migratory and invasive ability, and the tumor promotion mediated by anti-let-7g inhibitor was partly inhibited by silencing Aurora-B. These data suggested that let-7g alters OS cells malignant phenotype at least partly by targeting Aurora-B in vitro.
In conclusion, we findings suggested that down-regulated expression of let-7 microRNA family member is closely related to OS and let-7g alters the malignant of OS cells by negative regulating Aurora-B expression. Let-7g and Aurora-B corporately provide novel diagnostic biomarkers for OS, and targeting of let-7g and Aurora-B may aid in the development of novel therapeutic strategy for OS management. However, the tumor microenvironment played a key role in tumor development, progression and metastasis, therefore, further experiments in vivo are required to elucidate the potential of let-7g and Aurora-B as a target for the treatment of OS metastasis and a predictor of prognosis.
Materials and methods
Bioinformatic prediction
To predict miRNA targeting potentially at Aurora-B, we used the following bioinformatic algorithms: PicTar [26], TargetScan6.2 [27].
Luciferase reporter assay
Primers were designed in accordance with the human Aurora-B (AURKB) gene (NM_004217.3) sequences. And a fragment of the 3’-UTR of AURKB was amplified from U2-OS cells by PCR using the forward primer 5’-CCCCTCGAGATGGTCCCTGTCATTCACT-3’ and the reverse primer 5’-CTCGCGGCCGCTGAGTACAAAAAGCTTCAGCCTT-3’. Following digestion of the PCR product by XhoI and NotI, the AURKB 3’-UTR was cloned into pSiCHECK2 (Promega, Madison, WI, USA) at the XhoI and NotI sites. All PCR products were verified by DNA sequencing. U2-OS cells were co-transfected with the pSiCHECK2 vectors containing the 3’-UTR of AURKB (or variants) and let-7 or the negative control miRNA. Luciferase activity was measured 48 h after transfection. The firefly luciferase activity was then normalized to the Renilla luciferase activity.
Cell culture
The human OS cell line, U2-OS and HOS, and HOB was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA), and cells were routinely cultured in DMEM medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, USA) in a humidified 37°C incubator containing 5% CO2.
qRT-PCR
The total RNA was isolated from cells with Trizol reagent (Invitrogen, USA). Reverse transcription was performed with 2 mg of total RNA using PrimeScript Rtreagent Kit (Takara, Co, Japan). Then each sample was analyzed by quantitative real-time PCR (qPCR) (Bio-Rad, Hercules, CA, USA) under the conditions described in the SYBR Premix Ex Tap II (Invitrogen, USA): 50°C for 2 min, 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s. Primer sequences information was in summary as Table 1. Relative expression was calculated using the 2-ΔΔCt method. All experiments were repeated by six times over multiple days.
Table 1.
Primers information
| Primers | Sequences 5’ to 3’ |
|---|---|
| let-7a-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACTATA |
| Let-7a-5p-Q-F | GCCTGAGGTAGTAGGTTG |
| let-7b-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACCACA |
| Let-7b-5p-Q-F | GCGTGAGGTAGTAGGTTG |
| let-7c-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACCATA |
| Let-7c-5p-Q-F | GCCTGAGGTAGTAGGTTG |
| let-7d-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACTATG |
| Let-7d-5p-Q-F | GCGAGAGGTAGTAGGTTG |
| let-7e-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACTAT |
| Let-7e-5p-Q-F | GCTGAGGTAGGAGGTTGT |
| let-7f-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACTATA |
| Let-7f-5p-Q-F | GCGTGAGGTAGTAGATTG |
| let-7g-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACTGTA |
| Let-7g-5p-Q-F | GCGTGAGGTAGTAGTTTG |
| let-7i-5p-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAACAGCA |
| Let-7i-5p-Q-F | GCGTGAGGTAGTAGTTTG |
| U6-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAAAT |
| U6-Q-F | GCACTGGACTTGGAGTCA |
| Q-miR- 5p-R | CAGTGCAGGGTCCGAGGT |
| psi-AURKB-F | CCCCTCGAGATGGTCCCTGTCATTCACT |
| psi-AURKB-R | CTCGCGGCCGCTGAGTACAAAAAGCTTCAGCCTT |
| mut-AURKB-F | GAGCTCCTTTGTTTAATAAAGGCTGA |
| mut-AURKB-R | CATGAAAACAGATAAGGGAACAGTTA |
Transfection
The miRNA mimics for let-7g (hsa-let-7g, 5‘-UGAGGUAGUAGUUU GUACAGUU-3’), and the miRNA inhibitors for let-7g (The reverse complementary sequence of let-7g, 5‘-AACUGUACAAACU ACUACCUCA-3’), and miRNA negative control (NC) were obtained from GenePharma (Shuzhou). U2-OS or HOS cells were transfected with 10 nM of tested miRNA mimics or inhibitors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. OS cells were infected with Lentivirus-Vectors of down-regulating Aurora-B (LV-Aurora-B) and negative Lentivirus-Vectors (LV-Neg) at MOI of 12.5 (U2-OS) or 25 (HOS). After 48 h transfection, cells were harvested for appropriate subsequent assays.
Western blot
Total protein from cells was extracted using RIPA lysis buffer containing 60 μg/ml PMSF. Protein concentration was determined by Bradford assay. Western blot analysis was conducted using antibodies against Aurora-B (Abcam, UK) and β-actin (Santa Cruz). The immune complexes were detected with pro-light HRP Kit (pierce). The determination of the grayscale value was processed using ImageJ sofware. All experiments were repeated six times over multiple days.
Cell proliferation assay
Cells were cultured in 96-well tissue culture plates at a cell density of 5000 cells per well in culture medium. 3- (4, 5-dimethylthiazol-2-yl) -2, 5-diphenyltetrazolium bromide (MTT) assays were carried out at 490 nm wave length. All experiments were repeated six times over multiple days.
Transwell invasion assays
Cell invasion was measured in 24-well plates by Transwell assay using a chamber containing the polyethylene terephthalate filter membrane with 8-μm pores (BD Biosciences) according to the manufacturer’s protocol. The medium in the lower chamber contained 5% fetal calf serum as a source of chemoattractants. Cells were suspended in serum-free medium and added to the upper chambers at the same time. Cells that passed through the Matrigel-coated membrane were stained with Diff-Quik (Sysmex, Kobe, Japan) and photographed (×400). Photographs were taken at 24 h, and cell counting was measured by Image J (NCBI). The values for invasion were obtained by counting three fields per memberane and represented as the average of six independent experiments done over multiple days.
Wound healing assays
Cell migration was assessed by determining the ability of the cells to move into a cellular space in a two-dimensional in vitro ‘wound healing assay’. In brief, the cells were grown to confluence in 6-well tissue culture plastic dishes to a density of 5×106 cells/well. The cells were denuded by dragging a rubber policeman (Fisher Scientific, Hampton, NH, USA) through the center of the plate. The cultures were rinsed with PBS and replaced with fresh DMEM alone or containing 10% FBS, following which the cells were incubated at 37°C for 24 h. Images were captured at 0 and 24 h and the migrated distance was measured using ImageJ software (NIH, Bethesda, MD, USA). All experiments were repeated six times over multiple days.
Statistical analysis
Statistical comparisons were performed using SPSS software version 13.0 (SPSS Inc, Chicago, IL, USA). All measurement data are presented as the means±SD, and the one-way ANOVA with a post-hoc test (Student-Newman-Keuls test) was performed for statistical analysis. A P-value < 0.05 was considered to indicate a statistically significant difference.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (No. 81260400), the Natural Science Foundation of Jiangxi Province (No. 20114BAB205093) and Jiangxi Province Education Department of Science and Technology (No. GJJ12097, GJJ11316).
Disclosure of conflict of interest
None.
References
- 1.Bonet C, Giuliano S, Ohanna M, Bille K, Allegra M, Lacour JP, Bahadoran P, Rocchi S, Ballotti R, Bertolotto C. Aurora B is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem. 2012;287:29887–98. doi: 10.1074/jbc.M112.371682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuncel H, Shimamoto F, Kaneko Guangying Qi H, Aoki E, Jikihara H, Nakai S, Takata T, Tatsuka M. Nuclear Aurora B and cytoplasmic Survivin expression is involved in lymph node metastasis of colorectal cancer. Oncol Lett. 2012;3:1109–14. doi: 10.3892/ol.2012.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Zhou W, Wei L, Jin J, Tang K, Li C, Teh BT, Chen X. The effect of Aurora kinases on cell proliferation, cell cycle regulation and metastasis in renal cell carcinoma. Int J Oncol. 2012;41:2139–49. doi: 10.3892/ijo.2012.1633. [DOI] [PubMed] [Google Scholar]
- 4.Zhu XP, Liu ZL, Peng AF, Zhou YF, Long XH, Luo QF, Huang SH, Shu Y. Inhibition of Aurora-B suppresses osteosarcoma cell migration and invasion. Exp Ther Med. 2014;7:560–4. doi: 10.3892/etm.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilsen TW. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–9. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, Xie D, Shen Z, Sze J, Li K, Lu G, Chan DT, Poon WS, Kung HF, Lin MC. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. 2011;286:39703–14. doi: 10.1074/jbc.M111.293126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615–25. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S, Callegari E, Cescon M, Ravaioli M, Croce CM, Bolondi L, Gramantieri L. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–85. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 9.Zhang LY, Liu M, Li X, Tang H. miR-490-3p modulates cell growth and epithelial to mesenchymal transition of hepatocellular carcinoma cells by targeting endoplasmic reticulum-Golgi intermediate compartment protein 3 (ERGIC3) J Biol Chem. 2013;288:4035–47. doi: 10.1074/jbc.M112.410506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Chen X, Chin LJ, Paranjape T, Speed WC, Kidd KK, Zhao H, Weidhaas JB, Slack FJ. Extensive sequence variation in the 3’ untranslated region of the KRAS gene in lung and ovarian cancer cases. Cell Cycle. 2014;13:1030–40. doi: 10.4161/cc.27941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tay Y, Karreth FA, Pandolfi PP. Aberrant ceRNA activity drives lung cancer. Cell Res. 2014;24:259–60. doi: 10.1038/cr.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi YN, Xin XY, Ye HM. Effects of HMGA2 on malignant degree, invasion, metastasis, proliferation and cellular morphology of ovarian cancer cells. Asian Pac J Trop Med. 2014;7:289–92. doi: 10.1016/S1995-7645(14)60040-7. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi E, Satow R, Ono M, Masuda M, Honda K, Sakuma T, Kawai A, Morioka H, Toyama Y, Yamada T. MicroRNA expression and functional profiles of osteosarcoma. Oncology. 2014;86:94–103. doi: 10.1159/000357408. [DOI] [PubMed] [Google Scholar]
- 14.Haselmann V, Kurz A, Bertsch U, Hubner S, Olempska-Muller M, Fritsch J, Hasler R, Pickl A, Fritsche H, Annewanter F, Engler C, Fleig B, Bernt A, Roder C, Schmidt H, Gelhaus C, Hauser C, Egberts JH, Heneweer C, Rohde AM, Boger C, Knippschild U, Rocken C, Adam D, Walczak H, Schütze S, Janssen O, Wulczyn FG, Wajant H, Kalthoff H, Trauzold A. Nuclear death receptor TRAIL-R2 inhibits maturation of let-7 and promotes proliferation of pancreatic and other tumor cells. Gastroenterology. 2014;146:278–90. doi: 10.1053/j.gastro.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Paz EA, LaFleur B, Gerner EW. Polyamines are oncometabolites that regulate the LIN28/let-7 pathway in colorectal cancer cells. Mol Carcinog. 2014;53(Suppl 1):E96–106. doi: 10.1002/mc.22051. [DOI] [PubMed] [Google Scholar]
- 16.Jha HC, Lu J, Saha A, Cai Q, Banerjee S, Prasad MA, Robertson ES. EBNA3C-mediated regulation of aurora kinase B contributes to Epstein-Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases. J Virol. 2013;87:12121–38. doi: 10.1128/JVI.02379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 18.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–14. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan XM, Jia J, Guo XM, Li ZH, Zhang Z, Qin HJ, Xu GH, Gao LB. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer. 2014;13:93–7. doi: 10.1007/s10689-013-9681-4. [DOI] [PubMed] [Google Scholar]
- 20.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K, Mochizuki T. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 22.Farazi TA, Horlings HM, Ten Hoeve JJ, Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F, van Kouwenhove M, Kreike B, Sie D, Hovestadt V, Wessels LF, van de Vijver MJ, Tuschl T. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443–53. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Vito C, Riggi N, Suva ML, Janiszewska M, Horlbeck J, Baumer K, Provero P, Stamenkovic I. Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS One. 2011;6:e23592. doi: 10.1371/journal.pone.0023592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123:2791–7. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 27.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]


