Abstract
Human prothymosin-α (PTMA) plays an important role in tumorigenesis, and its overexpression triggers a TP53 response. In this study, we identified that PTMA expression was up-regulated at both the transcriptional and translational level in tumor tissue compared to that in adjacent normal tissue. PTMA overexpression was significantly associated with the depth of tumor invasion, lymph node metastasis (LNM), distant metastasis, advanced AJCC stage, and tumor differentiation. There was also a significant association between PTMA over-expression and mutant TP53 expression (r=0.515, P < 0.001). Survival analysis revealed that the disease-free survival (DFS) and overall survival (OS) rates were significantly lower among patients with PTMA- and TP53-positive tumors. Hence, PTMA might play an important role in the progression of CRC, and the assessment of both PTMA and mutant TP53 expression can help predict colon cancer prognosis.
Keywords: PTMA, TP53, colorectal cancer, prognosis, biomarker
Introduction
Colorectal cancer (CRC) is the third most common cancer and the third leading cause of cancer death both in men and women worldwide [1]. Colorectal adenocarcinoma is the most common form of colonic cancer affecting approximately 112,000 new patients every year [2]. Although many advanced methods of diagnosis and treatment have been employed over the last few decades, the overall survival (OS) rate of CRC patients has not markedly improved [3]. Sensitive biomarkers are crucial for early diagnosis and predicting prognosis, but none has been incorporated into routine clinical practice. Therefore, the identification of novel factors that can accurately predict postoperative tumor recurrence will greatly improve CRC management.
Human prothymosin-α (PTMA) is a member of the α-thymosin family comprising 110 amino acids, and its sequence is highly conserved in mammals [4]. To the best of our knowledge, PTMA plays an important role in cell biology, including cell cycle regulation, proliferation, transcription, and apoptosis [5-7]. Over-expression of PTMA has been reported in various malignancies including breast, lung, bladder, and head and neck cancer [8-11], and both PTMA and c-myc were over-expressed at the mRNA level in human CRCs compared with adjacent normal tissues, and there was a significant correlation between them [12]. However, there have been no reports concerning PTMA protein expression in CRC and its association with clinical outcome.
TP53 is one of the best characterized tumor suppressor genes and is the most frequently altered gene in human cancers, being mutated in more than 50% of carcinomas [13]. The wild-type TP53 protein is usually undetectable by standard immunohistochemistry; however, mutant TP53 protein is frequently detected at a high level in many primary tumors and tumor cell lines [14]. Analysis of the TP53 gene in a large cohort of CRC patients revealed that its mutation had prognostic significance [15]. Similar to Myc, Ras, E2F, and β-catenin, over-expression of PTMA results in the activation of TP53, which is now generally accepted as an innate tumor suppressive mechanism [16], and a critical cellular response to various stress stimuli [17]. However, PTMA does not increase the transcriptional activity of mutant TP53, negating this tumor suppressive mechanism [16]. At present, there is no agreement on whether mutant TP53 is associated with colon cancer prognosis [18]. Moreover, the relationship between mutant TP53 and PTMA, and especially the prognostic value of their combined expression, has not been evaluated.
In this study, we examined the PTMA and mutant TP53 expression patterns, evaluated their association with clinicopathologic features in CRC, and assessed whether the combination of PTMA and mutant TP53 could be an effective predictive marker for CRC.
Materials and methods
Patients and tissue specimens
Specimens were collected from 185 patients who had undergone radical colectomy at the General Surgery Department of Shanghai Jiaotong University affiliated Shanghai First People’s Hospital Medical Center between January 2001 and December 2003. None of the patients had undergone preoperative chemotherapy or radiotherapy. At least two pathologists confirmed the diagnosis. Staging was based on pathological findings according to the American Joint Committee on Cancer (AJCC) guidelines. There were 79 men and 106 women with a mean age of 65.82 years (range, 22-95 years).
Thirty pairs of fresh CRC tumors and adjacent normal mucosa (10 cm from the primary CRC) were obtained from patients who had undergone tumor resection without preoperative therapy. Tissues were put immediately into RNA Keeper Tissue Stabilizer (Vazyme Biotech Co., Ltd, Jiangsu, China) during the operation, stored at 4°C overnight, and then transferred to -80°C for long-term storage. The study was approved by the institutional review boards of Shanghai Jiaotong University Affiliated Shanghai First People’s Hospital Medical Center. Every patient enrolled in this study had provided written, informed consent.
Immunohistochemistry
Tissue microarray (TMA) slides were prepared as previously described [19]. Citrate buffer (0.01 M, pH 6.0) was used for antigen retrieval of the paraffin-embedded sections. The slides were then incubated with rabbit polyclonal antibody against PTMA (1:700, ABGENT, San Diego, CA) and TP53 (1:100, Abcam, Cambridge, UK) for 16 h at 4°C. The primary antibody was detected using the anti-mouse or anti-rabbit EnVision™ two-step Visualization System (Gene Tech, Shanghai, China) for 30 min at room temperature. Finally, the slides were counterstained with Mayer’s hematoxylin and mounted with a coverslip.
Evaluation of immunohistochemistry staining and scoring
Immunoreactivity was evaluated by a scoring system for both staining intensity and extent. Staining intensity for PTMA was scored as 0 for negative, 1 for mild, 2 for moderate, and 3 for intense. Staining extent scoring was based on the percentage of the positively immunostained cells as follows: 0, 0%; 1, 1-25%; 2, 26-50%; 3, 51-75%; and 4, 76-100%. Based on the overall score, which was calculated by adding the scores for staining intensity and extent, the specimens were divided into 3 groups as follows: 0-2, negative expression; 3-4, weak positive expression; and 5-7, strong positive expression. Based on the TP53 index, samples were divided into 2 groups: negative (< 10% of cells with positive nuclei) and positive (> 10% of cells with positive nuclei). All slides were evaluated independently by two researchers who were blinded to patient information.
Western blot analysis
Total protein was extracted from 4 pairs of colon tumors and their adjacent normal tissue, using RIPA lysis buffer (Beyotime Biotechnology, Jiangsu, China). The concentration of the protein was measured using the BCA protein assay kit (Beyotime Biotechnology, Jiangsu, China). Equal amounts of protein (30 μg) were electrophoresed on a 10% sodium dodecyl sulfate -polyacrylamide gel for 2 h, and then transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA) following standard protocols. The membranes were blocked using 5% non-fat milk with 0.1% Tween-20 at room temperature for 1 h, followed by incubation with the appropriate primary antibodies, anti-PTMA (1:1000, ABGENT, San Diego, CA) and anti-β-actin (1:1000, Abcam, Cambridge, UK), at 4°C overnight. After washing with TBST, membranes were incubated with goat anti-rabbit IgG-HRP (1:2000, Santa Cruz Biotechnology, USA). Protein was visualized using ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) according to the manufacturer’s instructions.
RNA extraction and quantitative real-time polymerase chain reaction (RT-PCR)
Total RNA was extracted using a TrizolTM reagent (Invitrogen Life Technologies, Carlsbad, CA), and 500 ng of total RNA was reverse-transcribed into first strand cDNA using the PrimeScriptTM RT reagent kit (Takara, Shiga, Japan) following manufacturer’s instructions. RT-PCR was performed using the SYBR Premix Ex Taq II (Takara, Shiga, Japan) reaction system on a MastercyclerTM ep Realplex (Eppendorf, Germany) under the following cycling conditions: initial denaturation (30 s at 95°C), followed by 40 cycles of denaturation (5 s at 95°C), and annealing and extension (30 s at 60°C). The human PTMA gene was amplified with forward primer: 5’-TGAGGAAGAGGATGGAGATGA-3’, and reverse primer: 5’-GGGAAGTGGAGGGTGAATAG-3’, and the GAPDH gene with forward primer: 5’-AGAAGGCTGGGGCTCATTTG-3’ and reverse primer: 5’-AGGGGCCATCCACAGTCTTC-3’; the latter was used as an internal control. All reactions were repeated in triplicate. The relative PTMA quantification was based on the 2-ΔΔCt values, and it was calculated using the formulas:
Statistical analysis
The two-tailed χ 2 test and Fisher exact test were used to determine the statistical significance of differences between experimental groups. The association between PTMA and TP53 protein expression was assessed using Spearman’s test, and the survival rate was analyzed using the Kaplan-Meier method. A log-rank test was used to compare survival curves. A Cox proportional hazards model was used to calculate univariate and multivariate hazard ratios. All analyses were performed using the SPSS 19.0 software (SPSS Inc., Chicago, IL). A P value < 0.05 was considered statistically significant.
Results
PTMA expression in colon tissues
Among the 30 pairs of fresh-frozen tissues used to estimate the mRNA level of PTMA, 20 (66.7%) colon cancers showed at least a 2-fold increase in PTMA mRNA level compared with that in the adjacent normal mucosa (Figure 1A). The mean PTMA quantification (-ΔCt value) in the colon tumor group (2.17±0.23; 0.80-1.93) was significantly higher than that in the normal tissue group (0.80±0.16; 0.80-2.00; P < 0.001). Likewise, Western blot analysis showed a significant up-regulation of PTMA protein in tumors compared with that in the corresponding normal tissue (Figure 1B), confirming that PTMA expression was elevated at the both transcriptional and translational level.
Figure 1.
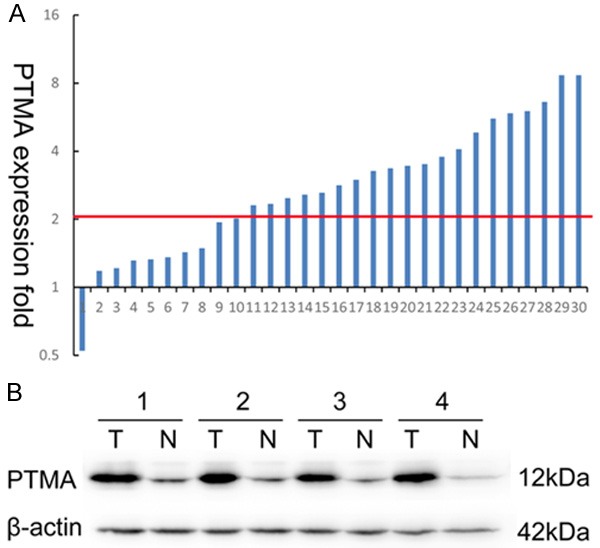
Expression of prothymosin-α (PTMA) in colon cancer tumors and adjacent normal mucosa. A. Relative PTMA mRNA levels in 30 matched colorectal tumors compared with that in normal mucosa specimens. A logarithmic scale of 2-ΔΔCT was used to represent the fold change in quantitative real-time polymerase chain reaction detection; B. Western blotting analysis of PTMA protein expression in 4 representative paired colon tumor/normal tissue pairings, β-actin was used as the loading control.
Association of PTMA and mutant TP53 expression in colon cancer with clinicopathologic parameters
Among the 185 samples on the paired TMA, 136 (73.5%) showed negative staining in normal mucosa. In contrast, up-regulated PTMA expression was apparent in colon tumors, with weak staining in 82 (44.3%) specimens, strong staining in 51 (27.6%) specimens, and negative staining in 52 (28.1%) specimens (Table 1). It was noteworthy that 54 of the 63 (85.7%) LNM samples also exhibited PTMA over-expression. Positive staining was prominent in the nuclei of colonic epithelial and tumor cells, but was only rarely present in the cytoplasm (Figure 2). The association between PTMA expression and a range of clinicopathologic parameters is summarized in Table 2. PTMA over-expression was significantly associated with the depth of tumor invasion (pT stage), LNM (pN stage), distant metastasis (M stage), advanced AJCC stage, and tumor differentiation. No associations were found between PTMA expression and age, sex, location, or vascular invasion. Moreover, PTMA expression was more frequently detected in patients with positive mutant TP53 staining than in those with negative mutant TP53 staining (Figure 3), with a significant correlation between them (r=0.515, P < 0.001). Positive TP53 staining was significantly associated with pT stage, pN stage, M stage, AJCC stage, and differentiation.
Table 1.
PTMA and TP53 immunohistochemical staining in normal colonic mucosa, tumors, and lymph node metastases
| Tissue sample | n | PTMA nuclei expression | P value | TP53 expression | P value | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Negative (%) | Weak (%) | Strong (%) | Negative (%) | Positive (%) | ||||
| Normal mucosa | 185 | 136 (73.5) | 35 (18.9) | 14 (7.6) | < 0.001* | 183 (98.9) | 2 (1.1) | < 0.001* |
| Tumor | 185 | 52 (28.1) | 82 (44.3) | 51 (27.6) | 90 (48.6) | 95 (51.4) | ||
| LNM | 63 | 9 ( 14.3) | 14 (22.2) | 40 (63.5) | 11 (17.5) | 52 (82.5) | ||
P value is based on the chi-square test.
Figure 2.
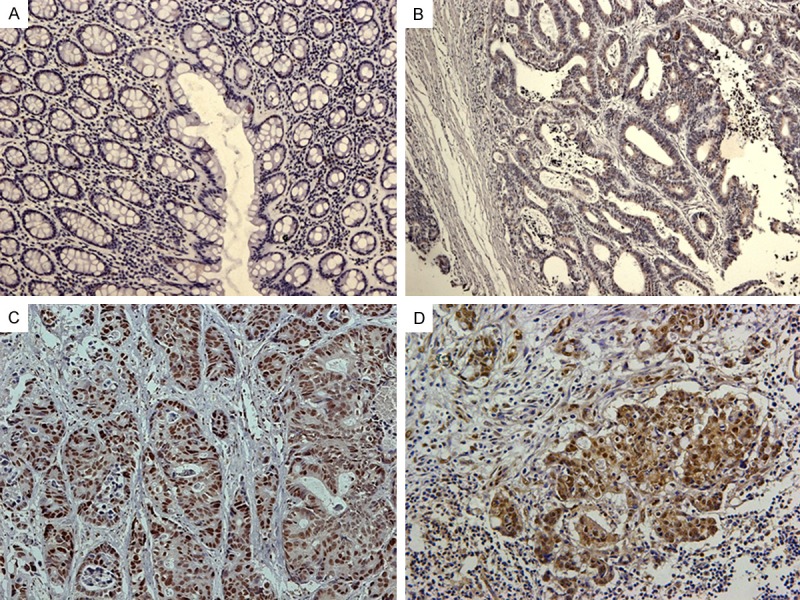
Immunohistochemical staining for prothymosin-α (PTMA) in normal and malignant colon tissue. A. Negative PTMA expression in normal colonic epithelium; B. Weak PTMA staining in a well-differentiated colorectal tumor; C. Diffuse, intense PTMA staining in a moderately to poorly differentiated colorectal tumor; D. Strong PTMA staining in a colon cancer lymph node metastasis sample. Original magnification ×200.
Table 2.
Association between clinicopathologic features and PTMA or TP53 protein expression
| PTMA expression | TP53 expression | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Negative | Weak | Strong | P value | Negative | Positive | P value | |
| Age (years) | |||||||
| < 65 | 18 | 31 | 23 | 0.539 | 34 | 38 | 0.765 |
| ≥ 65 | 34 | 51 | 28 | 56 | 57 | ||
| Sex | |||||||
| Male | 24 | 34 | 21 | 0.861 | 42 | 37 | 0.302 |
| Female | 28 | 48 | 30 | 48 | 58 | ||
| Location | |||||||
| Right | 16 | 34 | 24 | 0.552 | 34 | 40 | 0.865 |
| Transverse | 6 | 8 | 4 | 9 | 9 | ||
| Left | 30 | 40 | 23 | 47 | 46 | ||
| T stage | |||||||
| T1 | 5 | 2 | 0 | 0.002* | 3 | 4 | 0.003* |
| T2 | 10 | 9 | 2 | 15 | 6 | ||
| T3 | 21 | 36 | 15 | 42 | 30 | ||
| T4 | 16 | 35 | 34 | 30 | 55 | ||
| N stage | |||||||
| N0 | 36 | 44 | 16 | < 0.001* | 54 | 42 | 0.012* |
| N1 | 15 | 25 | 18 | 28 | 30 | ||
| N2 | 1 | 13 | 17 | 8 | 23 | ||
| M stage | |||||||
| M0 | 50 | 77 | 41 | 0.015* | 87 | 81 | 0.010* |
| M1 | 2 | 5 | 10 | 3 | 14 | ||
| AJCC stage | |||||||
| I | 14 | 6 | 2 | < 0.001* | 16 | 6 | 0.004* |
| II | 20 | 37 | 14 | 38 | 33 | ||
| III | 16 | 34 | 25 | 33 | 42 | ||
| IV | 2 | 5 | 10 | 3 | 14 | ||
| Differentiation | |||||||
| High | 38 | 37 | 15 | < 0.001* | 54 | 36 | 0.006* |
| Moderate | 10 | 35 | 23 | 28 | 40 | ||
| Low | 4 | 10 | 13 | 8 | 19 | ||
| Vascular invasion | |||||||
| Yes | 2 | 6 | 4 | 0.698 | 4 | 8 | 0.374 |
| No | 50 | 76 | 47 | 86 | 87 | ||
| TP53 expression | |||||||
| Negative | 43 | 40 | 7 | < 0.001* | |||
| Positive | 9 | 42 | 44 | ||||
P < 0.05 indicates a significant association among the variables.
Figure 3.
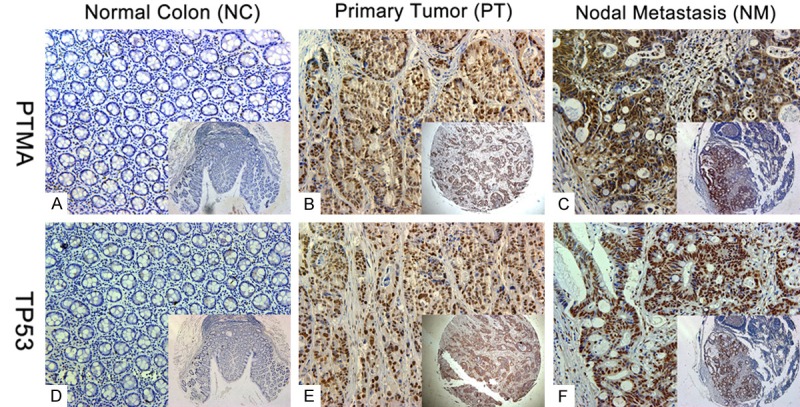
Expression of prothymosin-α (PTMA) and TP53. Representative images of PTMA and TP53 expression in normal colon (A and D), primary tumor (B and E), and nodal metastasis specimens (C and F). PTMA expression was more frequently present in specimens that stained positive for mutant TP53. Original magnification ×200 (×50 for inset images).
Over-expression of PTMA alone or combined with mutant TP53 predicts poor prognosis
Survival analysis was performed on 177 patients who had undergone curative operations, excluding 8 patients with stage disease and who had undergone non-curative surgery to avoid the potential confounding influence of unresectable metastatic tumors. At the end of the study, 50 of 177 patients (28.2%) had died of their disease, and 127 patients were still alive. Of the 177 patients, 64 (36.2%) experienced disease relapse. The Kaplan-Meier plots showed that patients with negative tumor PTMA expression had a better disease-free survival (DFS) and OS rate than those with PTMA tumor over-expression (P < 0.01; Figure 4A). Mutant TP53 expression was not related to OS but was associated with DFS (P=-0.027; Figure 4B). We also divided the patients into 3 groups depending on the concomitant expression of PTMA and mutant TP53: group 1, tumors with no PTMA or TP53 expression (43 cases); group 2, overexpression of one protein (56 cases); group 3, abnormal expression of both proteins (78 cases). Notably, patients in group 1 with both PTMA- and TP53-negative expression had a significantly better DFS and OS rates (Figure 4C) than those in group 3 with both PTMA- and TP53-positive expression.
Figure 4.
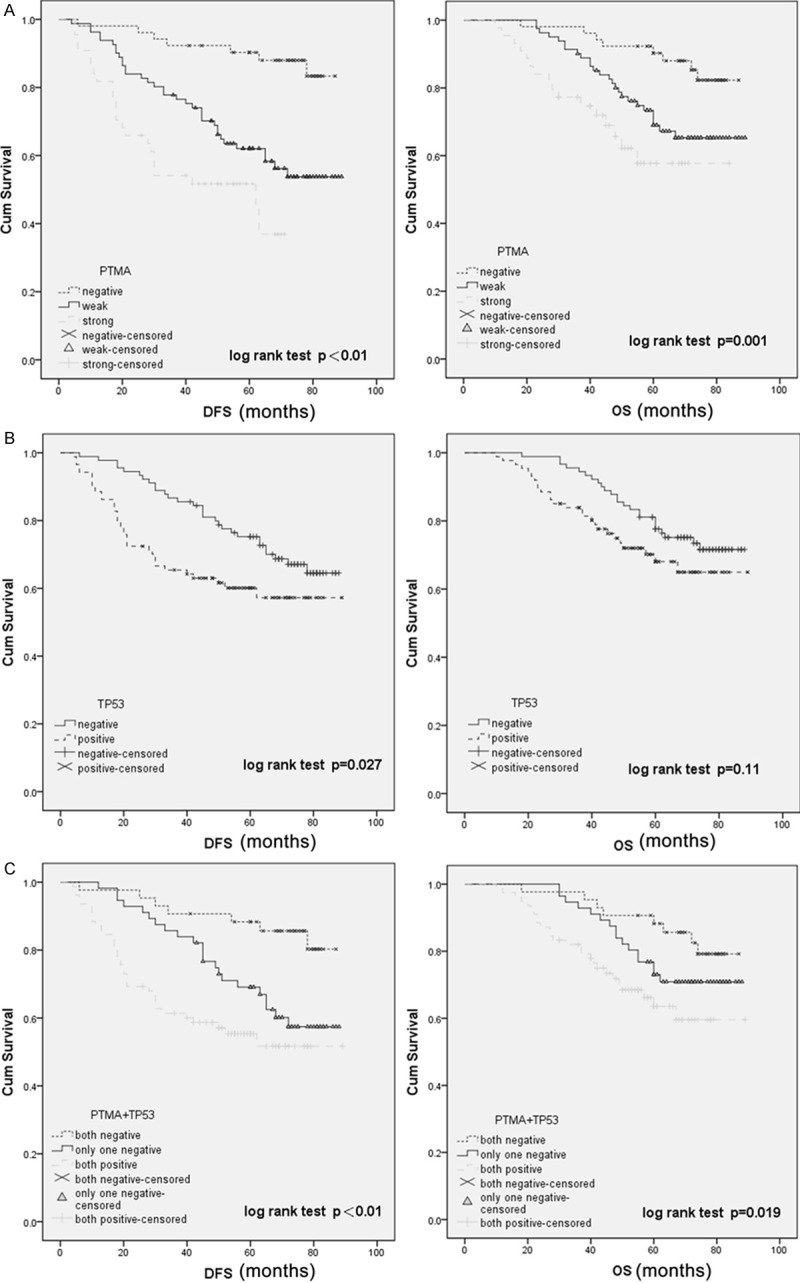
Kaplan-Meier analysis with a log-rank test of survival. A. The disease-free survival (DFS) and overall survival (OS) of patients were associated with prothymosin-α (PTMA) expression that was determined by immunohistochemical staining; B. DFS was significantly longer in patients with TP53-negative tumors than in those with TP53-positive tumors (P=0.027); C. DFS and OS were significantly shorter amongst patients with PTMA- and TP53-positive tumors than in those with PTMA- or TP53-negative tumors.
We conducted a multivariate analysis using the Cox proportional hazards model for all the significant variables in the univariate analysis. The results demonstrated that positive PTMA expression was a significant independent prognostic factor for disease recurrence and shorter survival (Table 3). Although mutant TP53 expression alone was not a prognostic indicator, expression of both PTMA and mutant TP53 was found to be a significant prognostic factor for DFS (hazard ratio [HR] 2.094; 95% confidence interval [CI], 1.457-3.051; P < 0.001) and OS (HR 2.348; 95% CI, 1.493-3.692; P < 0.001).
Table 3.
Multivariate analysis of the disease-free survival (DFS) and overall survival (OS) of 177 colon cancer patients
| Variable | Disease-free survival (DFS) | Overall survival (OS) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| P value | HR | 95% CI | P value | HR | 95% CI | |
| AJCC stage (I/II vs. III/IV) | 0.0144* | 11.372 | 1.623-79.654 | < 0.001* | 50.709 | 8.634-297.828 |
| pN stage | 0.024* | 2.179 | 1.110-4.274 | 0.045* | 1.733 | 1.013-2.968 |
| PTMA (negative vs. positive) | 0.019* | 4.631 | 1.287-16.664 | 0.010* | 3.624 | 1.354-9.700 |
| TP53 (negative vs. positive) | 0.164 | 1.659 | 0.814-3.382 | 0.118 | 1.663 | 0.880-3.142 |
| PTMA+TP53 (both negative vs. at least one positive) | < 0.001* | 2.348 | 1.493-3.692 | < 0.001* | 2.094 | 1.457-3.051 |
NS, not significant; HR, hazard ratio; CI, confidence interval.
P < 0.05 indicates that the lower limit of the 95% CI of HR is > 1.
Discussion
Previous studies have shown that the deregulation of PTMA results in increased cell proliferation and inhibits apoptosis by preventing formation of the apoptosome [6,20]. Over-expressed PTMA was associated with aggressive tumors and a poor prognosis in breast, hepatocellular, pituitary, and head and neck cancer [21]. Using an Expression Difference Mapping analysis, Mieko Shiwa et al. discovered that PTMA expression in colon cancer cells lines was significantly higher than that in normal colon cells [22]. In our study, up-regulated PTMA expression was found to be associated with CRC progression and was an independent prognostic marker for the disease. We also found a positive association between PTMA expression and advanced tumor stage, suggesting that the over-expression of PTMA may contribute to CRC progression. These data indicate that PTMA might therefore also be a prognostic marker for CRC patients after surgery.
In the absence of cellular stress, TP53 protein expression is maintained at low steady state and exerts very little, if any, effect on cell fate. However, TP53 becomes activated in response to various types of stress, including oncogene activation and DNA damage. This is reflected in elevated protein levels, as well as augmented biochemical capabilities [23]. Activated TP53 suppresses cellular transformation mainly by inducing apoptosis, inhibiting cell cycle progression, senescence, and differentiation, and accelerating DNA repair in damaged cells [18]. Accordingly, TP53 function is almost always compromised in tumor cells, usually as a result of somatic mutations, which occur in approximately half of all human cancers and constitute a cornerstone in tumorigenesis [24,25]. Mdm2 is a negative regulator of TP53, which can bind to TP53 and act as a TP53-specific E3 ubiquitin ligase. Elevated levels of Mdm2 will interfere with TP53 activity, even under conditions where TP53 is normally expected to be functional [23]. However, the mutant TP53 protein falls outside of this negative feedback loop [26].
In this study, we evaluated PTMA and TP53 expressions, and found that they were both over-expressed in tumors compared with the expressions in the normal epithelium, with a strong positive association between them. The mechanism that underlies the co-expression of these proteins is unclear. It is noteworthy that over-expressed PTMA, as an inappropriate growth stimulus, triggers a TP53 response, resulting in increased mRNA and protein levels of the endogenous TP53 target genes Mdm2 and p21 [16]. This is probably because PTMA over-expression induces wild-type TP53 protein degradation through Mdm2. However, the TP53 mutant protein is more stable due to its altered conformation, and is thus less readily degraded [27]. Mutant TP53 protein can therefore accumulate and promote tumorigenesis. On the other hand, in the absence of a functional TP53 pathway, PTMA is free to exert its oncogenic effects and promote the development of a malignant phenotype, like β-catenin and other oncogenes [28]. The combination of PTMA and TP53 might completely block tumor suppression. Further studies are needed to elucidate the role of PTMA and mutant TP53 in CRC progression.
In this study, we also found that PTMA staining was notably higher in lymph node metastatic CRC cells than in the primary tumors. PTMA expression was linked to unfavorable survival outcomes, indicating that increased PTMA expression was associated with invasive behavior and metastasis of CRC. This is supported by the previously identified role of PTMA in ovarian cancer cell adhesion, migration, and proliferation [29]. Multivariate analysis showed that PTMA expression alone or combined with mutant TP53 expression was an independent predictive factor for OS and DFS in CRC. However, mutant TP53 expression alone was not related to cancer prognosis, which concurs with the findings of previous studies [30].
In summary, increased PTMA expression was found in CRC tumors and was associated with multiple clinicopathologic factors as well as OS and DFS. These findings highlight the potential of PTMA as a therapeutic target in CRC, and justify the further study of the role of PTMA and mutant TP53 in this malignancy. These preliminary results need to be confirmed in a larger, prospective, controlled clinical study.
Acknowledgements
This project was supported by the funds: National High Technology Research and Development Program (SS2014AA020803), National Natural Science Foundation of China (81220108021, 81072008), Project of Shanghai Science and Technology Commission (11431921000), Joint Research Projects of Shanghai Municipal Hospital (SHDC12012105), Project of Shanghai Industrial Technology Institute (12DZ942500), Project of Shanghai JiaoTong University (YG2012ZD01).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Henderson-Jackson EB, Helm J, Ghayouri M, Hakam A, Nasir A, Leon M, Bui M, Yeatman T, Coppola D. Correlation between Mcl-1 and pAKT protein expression in colorectal cancer. Int J Clin Exp Pathol. 2010;3:768–774. [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan XS, Zhang Y, Guan XY, Dong B, Zhao M, Mao LL, Lu YY, Tian XY, Hao CY. p42.3: a promising biomarker for the progression and prognosis of human colorectal cancer. J Cancer Res Clin Oncol. 2013;139:1211–1220. doi: 10.1007/s00432-013-1434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haritos AA, Goodall GJ, Horecker BL. Prothymosin alpha: isolation and properties of the major immunoreactive form of thymosin alpha 1 in rat thymus. Proc Natl Acad Sci U S A. 1984;81:1008–1011. doi: 10.1073/pnas.81.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eilers M, Schirm S, Bishop JM. The MYC protein activates transcription of the alpha-prothymosin gene. Embo J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Kim HE, Shu H, Zhao Y, Zhang H, Kofron J, Donnelly J, Burns D, Ng SC, Rosenberg S, Wang X. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science. 2003;299:223–226. doi: 10.1126/science.1076807. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez P, Vinuela JE, Alvarez-Fernandez L, Buceta M, Vidal A, Dominguez F, Gomez-Marquez J. Overexpression of prothymosin alpha accelerates proliferation and retards differentiation in HL-60 cells. Biochem J. 1998;331:753–761. doi: 10.1042/bj3310753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magdalena C, Dominguez F, Loidi L, Puente JL. Tumour prothymosin alpha content, a potential prognostic marker for primary breast cancer. Br J Cancer. 2000;82:584–590. doi: 10.1054/bjoc.1999.0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki H, Nonaka M, Fujii Y, Yamakawa Y, Fukai I, Kiriyama M, Sasaki M. Expression of the prothymosin-a gene as a prognostic factor in lung cancer. Surg Today. 2001;31:936–938. doi: 10.1007/s005950170040. [DOI] [PubMed] [Google Scholar]
- 10.Tripathi SC, Matta A, Kaur J, Grigull J, Chauhan SS, Thakar A, Shukla NK, Duggal R, Choudhary AR, Dattagupta S, Sharma MC, Ralhan R, Siu KW. Overexpression of prothymosin alpha predicts poor disease outcome in head and neck cancer. PLoS One. 2011;6:e19213. doi: 10.1371/journal.pone.0019213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai YS, Jou YC, Lee GF, Chen YC, Shiau AL, Tsai HT, Wu CL, Tzai TS. Aberrant prothymosin-alpha expression in human bladder cancer. Urology. 2009;73:188–192. doi: 10.1016/j.urology.2008.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Mori M, Barnard GF, Staniunas RJ, Jessup JM, Steele GD Jr, Chen LB. Prothymosin-alpha mRNA expression correlates with that of c-myc in human colon cancer. Oncogene. 1993;8:2821–2826. [PubMed] [Google Scholar]
- 13.Strehl JD, Hoegel J, Hornicek I, Hartmann A, Riener MO. Immunohistochemical expression of IMP3 and p53 in inflammatory lesions and neoplastic lesions of the gastric mucosa. Int J Clin Exp Pathol. 2014;7:2091–2101. [PMC free article] [PubMed] [Google Scholar]
- 14.Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–1189. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- 15.Russo A, Bazan V, Iacopetta B, Kerr D, Soussi T, Gebbia N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005;23:7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T, Wang T, Maezawa M, Kobayashi M, Ohnishi S, Hatanaka K, Hige S, Shimizu Y, Kato M, Asaka M, Tanaka J, Imamura M, Hasegawa K, Tanaka Y, Brachmann RK. Overexpression of the oncoprotein prothymosin alpha triggers a p53 response that involves p53 acetylation. Cancer Res. 2006;66:3137–3144. doi: 10.1158/0008-5472.CAN-05-2112. [DOI] [PubMed] [Google Scholar]
- 17.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 18.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 19.Yan DW, Li DW, Yang YX, Xia J, Wang XL, Zhou CZ, Fan JW, Wen YG, Sun HC, Wang Q, Qiu GQ, Tang HM, Peng ZH. Ubiquitin D is correlated with colon cancer progression and predicts recurrence for stage II-III disease after curative surgery. Br J Cancer. 2010;103:961–969. doi: 10.1038/sj.bjc.6605870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orre RS, Cotter MA 2nd, Subramanian C, Robertson ES. Prothymosin alpha functions as a cellular oncoprotein by inducing transformation of rodent fibroblasts in vitro. J Biol Chem. 2001;276:1794–1799. doi: 10.1074/jbc.M008560200. [DOI] [PubMed] [Google Scholar]
- 21.Ioannou K, Samara P, Livaniou E, Derhovanessian E, Tsitsilonis OE. Prothymosin alpha: a ubiquitous polypeptide with potential use in cancer diagnosis and therapy. Cancer Immunol Immunother. 2012;61:599–614. doi: 10.1007/s00262-012-1222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiwa M, Nishimura Y, Wakatabe R, Fukawa A, Arikuni H, Ota H, Kato Y, Yamori T. Rapid discovery and identification of a tissue-specific tumor biomarker from 39 human cancer cell lines using the SELDI ProteinChip platform. Biochem Biophys Res Commun. 2003;309:18–25. doi: 10.1016/s0006-291x(03)01520-1. [DOI] [PubMed] [Google Scholar]
- 23.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 24.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 25.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 26.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji F, Jin X, Jiao CH, Xu QW, Wang ZW, Chen YL. FAT10 level in human gastric cancer and its relation with mutant p53 level, lymph node metastasis and TNM staging. World J Gastroenterol. 2009;15:2228–2233. doi: 10.3748/wjg.15.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damalas A, Kahan S, Shtutman M, Ben-Ze’ev A, Oren M. Deregulated beta-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. Embo J. 2001;20:4912–4922. doi: 10.1093/emboj/20.17.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hapke S, Kessler H, Luber B, Benge A, Hutzler P, Hofler H, Schmitt M, Reuning U. Ovarian cancer cell proliferation and motility is induced by engagement of integrin alpha(v)beta3/Vitronectin interaction. Biol Chem. 2003;384:1073–1083. doi: 10.1515/BC.2003.120. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Tang H, Wu Z, Zhou C, Jiang T, Xue Y, Huang G, Yan D, Peng Z. Overexpression of RBBP6, alone or combined with mutant TP53, is predictive of poor prognosis in colon cancer. PLoS One. 2013;8:e66524. doi: 10.1371/journal.pone.0066524. [DOI] [PMC free article] [PubMed] [Google Scholar]


