Abstract
Interleukin-33 (IL-33) is a dual-function protein that acts both as a secreted cytokine and as a nuclear factor regulating gene transcription. It has been demonstrated that IL-33 exerts its nuclear function in promoting the transcription of NF-κB p65. Here, we show that USP21-mediated deubiquitination of IL-33 affects the transcription of p65. IL-33 can be post-translationally modified by ubiquitination. Ubiquitin-specific protease 21 (USP21) interacts with IL-33 and also localizes in nucleus. The protein stability of IL-33 is maintained by USP21 through deubiquitination. Furthermore, depletion of USP21 reduces IL-33 protein levels and IL-33-mediated NF-κB p65 promoter activity. Our findings reveal the role of ubiquitination modification in regulating the protein stability and the nuclear function of IL-33.
Keywords: IL-33, USP21, deubiquitinase, stability
Introduction
IL-33 is a member of the IL-1 family which plays an important role in inflammatory and autoimmune diseases. It is mainly expressed by fibroblasts, epithelial cells and endothelial cells, particularly in high endothelial venules [1]. IL-33 is a crucial regulator of inflammation and is mainly involved in type 2 immunity and inflammation initiated by type 2 cells, including T helper 2 (Th2) cells and innate lymphoid cell-2 (ILC2) [2]. In addition, IL-33 also acts as a novel alarmin released after trauma or infection [3].
IL-33 functions as an extracellular cytokine by binding to its receptor ST2 [1]. ST2 contains two main isoforms: the soluble, secreted form (sST2) and the transmembrane, long form (ST2L) [4]. However, these two isoforms have diametrically opposite functions during inflammation. The sST2 exerts anti-inflammatory properties by inhibiting IL-33 signals [5], while the ST2L has a vital role in initiating inflammation through binding to IL-33 [6].
Besides its importance as a cytokine, IL-33 is simultaneously a chromatin-associated nuclear protein (also known as the nuclear factor from high endothelial venule, NF-HEV), which has been related to many diseases, including asthma, fibrosis, rheumatoid arthritis and so on [2]. The transcription factor nuclear factor κB (NF-κB) plays a critical role in orchestrating the expression of cytokines, survival factors and proinflammatory molecules in various immune responses [7]. Highly expressed IL-33 in the nuclei of endothelial cells participates in inflammation as a transcriptional regulator of NF-κB p65 [8]. Moreover, another study suggests that the binding of nuclear IL-33 to the NF-kB p65 subunit reduces p65-triggered gene expression to dampen the production of several proinflammatory cytokines [9].
It has been shown that the ubiquitination system is involved in the regulation of the IL-33/ST2 signaling pathway, thus influencing the inflammatory effects of IL-33. F-box protein FBXL19, an E3 ubiquitin ligase, abrogates the inflammatory effects of IL-33 by selectively regulating the polyubiquitination status of ST2L, finally targeting the IL-33 receptor for degradation [10]. Despite its recognized importance as a transcriptional regulator, little is known about the molecular regulation of nuclear IL-33 expression, particularly relating to ubiquitination or deubiquitination.
Human genome encodes almost 79 deubiquitinases (DUBs) with catalytic activity. These DUBs have been divided into five subfamilies: Ubiquitin-specific proteases (USPs), Ubiquitin C-terminal hydrolases (UCHs), ovarian tumor (OTU), JAB1/MPN/Mov34 metalloenzyme domain zinc-dependent metalloprotease (JAMM) family and Josephin domain [11,12]. USP21 is a deubiquitinase with a C-terminal catalytic DUB domain [13]. It has been found that USP21 negatively regulates the NF-κB signaling pathway through deubiquitinating receptor-interacting protein 1 (RIP1) [14]; USP21 also acts as a negative regulator in antiviral immune defense through its ability to deubiquitinate retinoic acid-inducible gene 1 (RIG-1) [15]; Furthermore, USP21 positively regulates the Th2 specific transcriptional factor GATA3, which is important for the function of regulatory T cells [16]. Collectively, USP21 is essential in innate and adaptive immune responses.
Here, we report USP21 as a deubiquitinase for IL-33. We found that USP21 regulates the stability of IL-33 through deubiquitination. Consistently, silencing of endogenous USP21 resulted in reduced IL-33 protein levels and decreased IL-33-mediated NF-κB p65 promoter activity. Our work thus identifies a novel mechanism of positive regulation of IL-33 by deubiquitination.
Materials and methods
Plasmid and antibodies
Expression plasmids encoding human IL-33, USP21 and ubiquitin, fused with Flag-, Myc-, HA- or His-tag, were constructed based on the pIRES vector by standard molecular biology techniques. The USP21C221A mutant was constructed with the Quickchange II site-directed mutagenesis kit (Stratagene). The antibodies used in this study are listed below: anti-Flag (M2, Sigma), anti-Myc (9E10, Santa Cruz), anti-HA (F-7, Santa Cruz), anti-USP21 (AP1069a, ABGENT), anti-Ubiquitin (sc-8017, Santa Cruz), anti-tubulin (DM1A, Sigma), anti-PARP (436400, Invitrogen), anti-β-actin (KM9001, Sungene Biotech), anti-mouse IgG HRP (85-18-8817-31, eBioscience) and anti-rabbit IgG HRP (85-18-8816-31, eBioscience).
Cell culture and transfection
Human embryonic kidney (HEK) 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Cells were transfected with appropriate plasmids by using polyethylenimine (PEI) reagent (Polysciences) according to the manufacturer’s instructions.
Ubiqutin pull-down assay
HEK293T cells transfected with Flag-tagged IL-33, Myc-USP21 and His-tagged ubiquitin were treated with 20 μM MG132 (CAS 133407-82-6, Merck) for 4 h before harvesting. Cells were washed with ice-cold PBS and lysed in a pH 8.0 urea buffer (10 mM Tris pH 8.0, 8 M urea, 100 mM Na2HPO4, 0.2% Triton X-100, 1 mM N-ethylmaleimide and 10 mM imidazole) for 30 min. Cell lysates were incubated with Ni-NTA beads (30210, Qiagen) for 3 h at room temperature after sonication (2×20 s, 30% power). Beads were washed three times in pH 8.0 urea buffer before incubation. After 3 h incubation, the beads were washed twice in pH 8.0 urea buffer, twice in pH 6.3 urea buffer (10 mM Tris pH 6.3, 8 M urea, 100 mM Na2HPO4, 0.2% Triton X-100 and 10 mM imidazole), and once in wash buffer (20 mM Tris pH 8.0, 100 mM NaCl, 20% glycerol, 1 mM dithiothreitol and 10 mM imidazole). Beads were boiled in 30 μl 2X loading buffer for 10 min and separated on SDS-PAGE gel, and then ubiquitination was determined by immune blot with specific antibodies.
Immunoblotting and co-immunoprecipitation
Cells were lysed in RIPA buffer consisting of 20 nM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% Glycerol, 0.5% Na-deoxycholate, 1 mM PMSF, 1 mM NaF, 1 mM Na3VO4 and 1% Protease Inhibitor Cocktail (P8340, Sigma). The cell lysates were used for immunoblotting and co-immunoprecipitation. For immunoprecipitation, cell lysates were immunoprecipitated with indicated antibodies for 2 h at 4°C and then incubated with protein A/G PLUS Agarose (sc-2003, Santa Cruz) for 1 h at 4°C. The immunoprecipitates were washed with RIPA buffer (containing 1 mM PMSF, 1 mM NaF and 1 mM Na3VO4) three times and then analyzed by immune blot with indicated antibodies.
Cytoplasmic and nuclear fractionation
Transfected HEK293T cells were harvested and washed with ice-cold PBS. Cells were lysed in Cytoplasm buffer (10 mM Tris-HCl pH 7.5, 10 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF) for 15 min and Cytoplasm buffer plus 10% NP-40 for another 15 min. After collecting the supernatant containing the cytoplasmic fraction, the pellet was further lysed in Nuclear Extract buffer (20 mM Tris-HCl pH 8.0, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF and 1% Protease Inhibitor Cocktail) for 30 min. After centrifugation, the supernatant, constituting the nuclear fraction, was collected for further analysis.
Knockdown assay
The shRNA sequences of USP21 were synthetized at Shanghai Sunny Biotechnology Co. Ltd. and the oligos were cloned into the shRNA lentiviral vectors PLKO.1 (Addgene). 293T cells were transfected with shUSP21 to silence USP21. The shRNA sequences used in this study were as follows: 5’-CAACAAGATGAAGAGCACCAA-3’ (shCK); 5’-CCACTTTGAGACGTAGCACTT-3’ (shUSP21-1); 5’-GACCCTCTGCAATATCACTTT-3’ (shUSP21-2).
Luciferase reporter assay
The 1.1-kb region upstream of the human NF-κB p65 transcriptional starting site (Location: Chromosome 11: 65, 421, 067-65, 430, 565) was amplified from human genomic DNA and cloned into the pGL3-Basic vector to generate the pGL3-p65-luc reporter construct. For luciferase, pGL3-p65-luc was co-transfected with the indicated plasmids and an internal control plasmid encoding β-gal into HEK293T cells. After 48 h, cells were lysed and luciferase assay was performed following the manufacturer’s instructions (Beyotime).
Results
USP21 deubiquitinates IL-33
To investigate whether IL-33 could be ubiquitinated, we co-transfected Flag-tagged IL-33 and His-tagged ubiquitin into 293T cells. The ubiquitinated proteins were bound to Ni-NTA beads and pulled down through the ubiquitin pull-down assay. As shown in Figure 1A, IL-33 could be ubiquitinated. Since ubiquitination can be reversed by a large family of deubiquitinases, we then tested a panel of potential deubiquitinases to see whether they could reduce the ubiquitination status of IL-33. Flag-tagged IL-33, HA-tagged ubiquitin and Myc-tagged USPs were co-transfected into 293T cells for a co-immunoprecipitation assay. Among these deubiquitinases, we found that USP21 could reduce the ubiquitination status of IL-33 (data not shown). To verify the screening results, we co-transfected Flag-IL-33 with HA-ubiquitin and Myc-USP21 or its catalytically inactive mutant into 293T cells, and found that USP21 wild type, but not its enzymatic inactive C221A mutant, could reduce the ubiquitination of IL-33 (Figure 1B). Furthermore, we confirmed the USP21-mediated deubiquitination of IL-33 through ubiquitin pull-down assay under denaturing condition. As expected, wild-type USP21, but not its mutant, reduced the ubiquitination of IL-33 (Figure 1C). Taken together, our results suggest that USP21 deubiquitinates IL-33.
Figure 1.
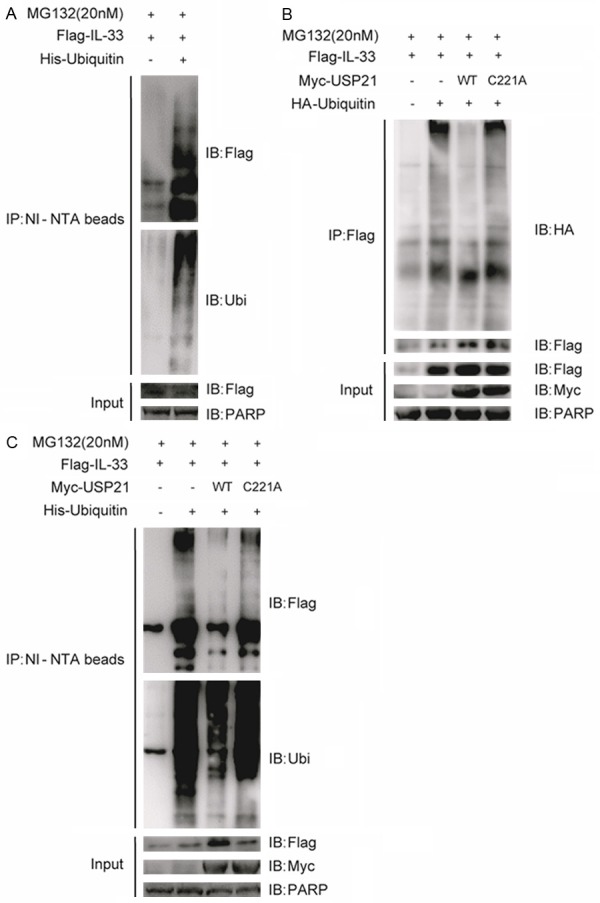
USP21 deubiquitinates IL-33. A. HEK293T cells were transfected with the plasmids encoding Flag-tagged IL-33 and His-tagged ubiquitin. Cells were treated with 20 μM MG132 for 4 h before harvesting. Ubiquitinated IL-33 was pulled down by Ni-NTA beads upon denaturing conditions as described in the Methods. Immune blot (IB) was performed with indicated antibodies. B. HEK293T cells were transfected with Flag-IL-33, HA-ubiquitin and Myc-USP21 or its catalytically inactive mutant C221A (USP21C221A). Cell lysates were immunoprecipitated with anti-Flag antibody and detected by immune blot as indicated. C. HEK293T cells were transfected with Flag-IL-33, His-ubiquitin and Myc-USP21 or its catalytically inactive mutant USP21C221A. The ubiquitin pull-down assay was performed as described in the Methods. Data are representative of at least three independent experiments.
USP21 stabilizes IL-33
Knowing that ubiquitination is a key mechanism in regulating the proteasome-mediated proteolysis of target proteins, we supposed that USP21 might affect the stability of IL-33. To verify our hypothesis, Flag-tagged IL-33, increasing concentrations of Myc-tagged USP21 or controlled vector were introduced into 293T cells and the protein levels were detected by immune blot. We found that the protein levels of IL-33 positively correlated with the increasing concentrations of co-expressed USP21, suggesting that USP21 could stabilize IL-33 in a dose-dependent manner (Figure 2A). We also treated the cells co-expressing IL-33 with or without USP21 or its catalytically inactive mutant USP21C221A in the presence of the protein synthesis inhibitor cycloheximide (CHX, C7698-5G, Sigma) at the indicated time points. In accordance with the previous result, overexpression of USP21 prolonged the half-life of IL-33, while this effect was lost in the presence of the USP21 mutant (Figure 2B, 2C). These data attest that USP21 stabilizes IL-33 through deubiquitination.
Figure 2.
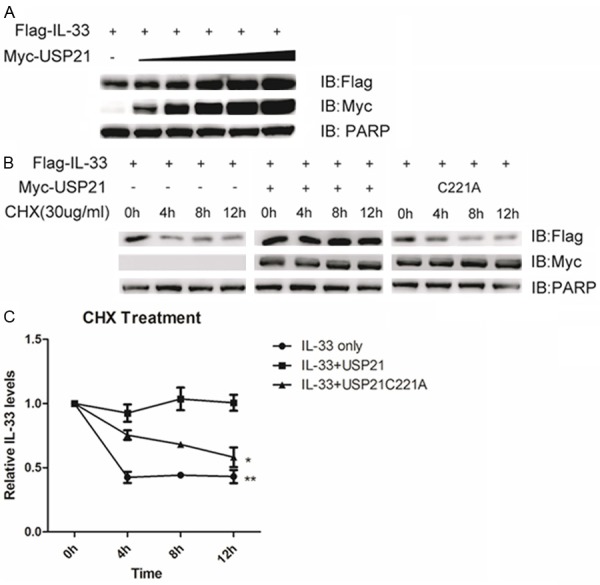
USP21 stabilizes IL-33. A. HEK293T cells were transfected with Flag-IL-33 and increasing concentrations of Myc-USP21. Immune blot was performed with the indicated antibodies. B. HEK293T cells were transfected with Flag-IL-33 and Myc-USP21 or its inactive mutant USP21C221A. Cells were treated with CHX (30 μg/ml) at the indicated time points. Immune blot was performed with the indicated antibodies. C. The protein levels of IL-33 were quantified by ImageJ software. All protein intensity values were normalized to 0h protein values. Data are representative of at least three independent experiments. **p < 0.01. *p < 0.05. Error bars represent mean ± SEM.
USP21 interacts with IL-33 and also localizes in nucleus
The finding that USP21 could maintain IL-33 protein stability prompted us to test their potential interaction and localization. Therefore, Myc-tagged USP21 and Flag-tagged IL-33 were co-transfected into 293T cells for a co-immunoprecipitation assay with anti-Flag antibody or anti-Myc antibody. We found that USP21 and IL-33 could interact reciprocally (Figure 3A, 3B). Moreover, 293T cells transfected with Myc-USP21 and Flag-IL-33 were used to perform nuclear/cytoplasmic fractionation in order to evaluate IL-33 and USP21 localization. We separated cytoplasmic and nuclear fractions and found that IL-33 and USP21 could be detected in both the cytoplasm and the nucleus (Figure 3C). These results show that USP21 can interact with IL-33 and also localize in nucleus.
Figure 3.
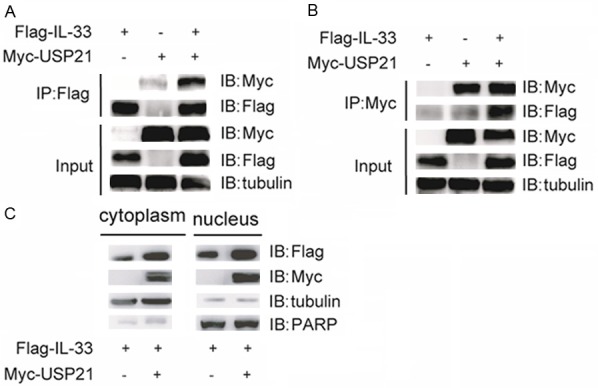
USP21 interacts with IL-33 and also localizes in nucleus. (A, B) HEK293T cells were transfected with Flag-IL-33 and/or Myc-USP21. Immunoprecipitation was performed with either anti-Flag antibody (A) or anti-Myc antibody (B). Immune blot was performed with indicated antibodies. (C) HEK293T cells were transfected with Flag-IL-33 and Myc-USP21. Nuclear/cytoplasmic fractionation was performed to separate cytoplasmic and nuclear fractions. Immune blot was performed with indicated antibodies. Data are representative of at least three independent experiments.
Knockdown of endogenous USP21 decreases IL-33 protein levels and IL-33-mediated NF-κB p65 promoter activity
To further validate the mechanism of USP21 in the regulation of IL-33 stability, we constructed specific shRNAs for USP21 to reduce the endogenous expression of USP21 in 293T cells. Although both shRNAs targeted USP21, shUSP21-1 had the better knockdown efficacy and was chosen for further functional assays (Figure 4A). After silencing USP21, Flag-IL33 was transfected into 293T cells. The results obtained showed that silencing of endogenous USP21 resulted in a decrease in IL-33 protein levels (Figure 4B).
Figure 4.
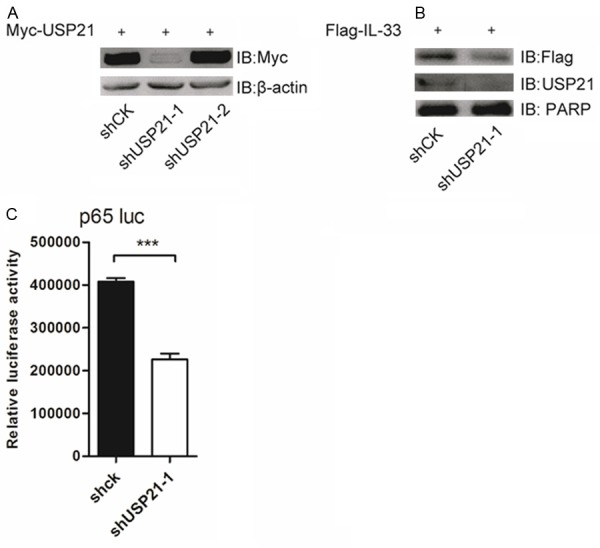
Knockdown of endogenous USP21 decreases IL-33 protein levels and IL-33-mediated NF-κB p65 promoter activity. A. HEK293T cells were transfected with Myc-USP21 and shCK or shUSP21-1 or shUSP21-2. Immune blot was performed with indicated antibodies. B. HEK293T cells were transfected with Flag-IL-33 and shCK or shUSP21-1. Immune blot was performed with indicated antibodies. C. HEK293T cells were transfected with Flag-IL-33, pGL3-p65-luc, β-gal and shCK or shUSP21-1. The luciferase activity was detected 48 hours post-transfection. Data are representative of at least three independent experiments. ***p < 0.001. Error bars represent mean ± SEM.
It has been reported that IL-33 can bind to the NF-κB p65 promoter region in the nucleus, inducing endothelial cell activation [8]. Based on this, we constructed p65 promoter reporter plasmids to which nuclear IL-33 could directly bind. Then, p65 promoter luciferase reporter plasmids and Flag-IL-33 were introduced into the USP21-deficient 293T cells. We found that the silencing of USP21 could significantly decrease IL-33-mediated p65 promoter activity (Figure 4C). These evidences suggest that USP21 stabilizes IL-33 and promotes IL-33-mediated NF-κB p65 promoter activity.
Discussion
Post-translational modifications (PTMs) are processes that change the properties of a protein by proteolytic cleavage, degradation of the entire protein or by covalent addition of modifying groups to one or more amino acids [17]. These modifications include phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation and proteolysis [18]. PTMs occur on nearly all eukaryotic proteins and control their functions through regulating their stability, localization, interactions with other cellular molecules and their activation status [19]. Post-translational modifications of IL-33 mainly lead to its proteolytic cleavage [20]. Indeed, previous studies have found that IL-33, which is active in its full-length form, can be inactivated by caspase-1, 3 and 7 during apoptosis in order to avoid any unwanted inflammatory response [21,22], while proteases from neutrophils (elastase, cathepsin G and proteinase 3) can cleave the full-length IL-33 generating highly active forms [23,24]. All these modifications affect the function of IL-33, particularly as a cytokine. However, IL-33 is also known to be a nuclear factor involved in the regulation of transcription [8], and the molecular mechanisms controlling its activity are still unclear. For this reason, we were interested in understanding whether other forms of PTMs could affect the function and the activity of IL-33.
Modulation of the IL-33-ST2L axis by ubiquitination might serve as an important regulatory mechanism for inflammation. For example, one study found that the E3 ubiquitin ligase FBXL19 selectively bound to ST2L to mediate its polyubiquitination and proteasomal degradation, finally alleviating the severity of lung injury in mouse models of pneumonia [10]. Since ubiquitination plays critical roles in regulating a variety of cellular processes [25], we attempted to detect whether IL-33 could directly be subjected to ubiquitination. Interestingly, ubiquitin pull-down assay showed that ubiquitin chains could be linked to IL-33. As IL-33 is constitutively expressed in the nucleus under normal conditions [20], we will examine the subcellular localization of ubiquitination modification for IL-33 further in the future.
Deubiquitinases reverse protein ubiquitination by removing ubiquitin chains from the modified proteins, leading to reversal of ubiquitin signalling or to protein stabilization by rescue from degradation [12]. Our study figured out one of the deubiquitinases for IL-33 through immunoprecipitation and ubiquitin pull-down assay. Among the different USPs tested, USP21 was shown to interact with IL-33 and could also localize in nucleus. Moreover, USP21 could stabilize the protein levels of IL-33 by deubiquitination. Taken together, our results have revealed one of the mechanisms involved in the regulation of IL-33 protein stability.
Besides being associated with proteasomal degradation, ubiquitination is also involved in regulatory events in a proteasome-independent manner. Until recently, ubiquitination modification has emerged as an important regulator of diverse cellular processes, such as activation of protein kinases, control of gene transcription, DNA repair and replication, intracellular trafficking, virus budding and so on [25-27]. Having established that IL-33 stability can be regulated by ubiquitination and deubiquitination, we next examined whether this kind of modification could affect the function of IL-33. It has been shown that IL-33 participates in the inflammatory process through an up-regulation of the transcription of NF-κB p65 [8]. We found that silencing of USP21 decreased IL-33-mediated p65 promoter activity, highlighting a previous unknown mechanism of regulation of IL-33 in inflammation. Collectively, our data reveal that ubiquitination modification participates in regulating the stability and the nuclear function of IL-33.
Increasing evidences suggest that nuclear IL-33 is involved in chronic inflammation [28]. We hypothesized and demonstrated that post-translational modifications of IL-33 can be crucial in regulating its role in inflammation. Our study identified ubiquitination as a key post-translational modification for IL-33, and USP21 as a deubiquitinase involved in its regulation. Our results also established ubiquitination as a novel pathway in the regulation of IL-33 nuclear function. Future studies are still needed to explore the role of ubiquitinated or deubiquitinated IL-33 in inflammation and to broaden other hidden nuclear functions of IL-33.
Acknowledgements
We thank members of the Pulmonary Medicine Department of Ruijin Hospital and the Key Laboratory of Molecular Virology and Immunology from Institut Pasteur of Shanghai for supervision, attentive instruction and technical help. Our work was supported by National Basic Research Program of China (973 Program) 2014CB541803, NSFC 81270083, 81330072, 31370863, 31170825, 31350110505; The 2014 Shanghai Municipal Education Commission Research and Innovation Projects (Natural Science) 14ZZ107; and The Chinese Academy of Sciences Fellowships for Young International Scientists 2013Y1SB0005.
Abbreviations
- DUB
deubiquitinase
- IL-33
interleukin-33
- Ubi
ubiquitin
- USP
ubiquitin-specific protease
- USP21
ubiquitin-specific protease 21
References
- 1.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moussion C, Ortega N, Girard JP. The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: a novel ‘alarmin’? PLoS One. 2008;3:e3331. doi: 10.1371/journal.pone.0003331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trajkovic V, Sweet MJ, Xu D. T1/ST2--an IL-1 receptor-like modulator of immune responses. Cytokine Growth Factor Rev. 2004;15:87–95. doi: 10.1016/j.cytogfr.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Li XY, Yuan BH, Zhang BB, Hu SL, Gu HB, Jin XB, Zhu JY. Adenovirus-mediated overexpression of soluble ST2 provides a protective effect on lipopolysaccharide-induced acute lung injury in mice. Clin Exp Immunol. 2011;164:248–255. doi: 10.1111/j.1365-2249.2011.04326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi YS, Choi HJ, Min JK, Pyun BJ, Maeng YS, Park H, Kim J, Kim YM, Kwon YG. Interleukin-33 induces angiogenesis and vascular permeability through ST2/TRAF6-mediated endothelial nitric oxide production. Blood. 2009;114:3117–3126. doi: 10.1182/blood-2009-02-203372. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 8.Choi YS, Park JA, Kim J, Rho SS, Park H, Kim YM, Kwon YG. Nuclear IL-33 is a transcriptional regulator of NF-kappaB p65 and induces endothelial cell activation. Biochem Biophys Res Commun. 2012;421:305–311. doi: 10.1016/j.bbrc.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, Martin MU. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol. 2011;187:1609–1616. doi: 10.4049/jimmunol.1003080. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, Zou C, Mallampalli RK, Zhao Y. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–658. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 13.Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu G, Tan X, Wang H, Sun W, Shi Y, Burlingame S, Gu X, Cao G, Zhang T, Qin J, Yang J. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–978. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, Zhang H, An L, Zhao Y, Xu X, Chen Z, Kogiso M, Zhang D, Zhang H, Zhang P, Jung JU, Li X, Xu G, Yang J. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211:313–328. doi: 10.1084/jem.20122844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Chen C, Hou X, Gao Y, Lin F, Yang J, Gao Z, Pan L, Tao L, Wen C, Yao Z, Tsun A, Shi G, Li B. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J Biol Chem. 2013;288:9373–9382. doi: 10.1074/jbc.M112.374744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 18.Lothrop AP, Torres MP, Fuchs SM. Deciphering post-translational modification codes. FEBS Lett. 2013;587:1247–1257. doi: 10.1016/j.febslet.2013.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YC, Peterson SE, Loring JF. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 2014;24:143–160. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lefrancais E, Cayrol C. Mechanisms of IL-33 processing and secretion: differences and similarities between IL-1 family members. Eur Cytokine Netw. 2012;23:120–127. doi: 10.1684/ecn.2012.0320. [DOI] [PubMed] [Google Scholar]
- 21.Cayrol C, Girard JP. The IL-1-like cytokine IL-33 is inactivated after maturation by caspase-1. Proc Natl Acad Sci U S A. 2009;106:9021–9026. doi: 10.1073/pnas.0812690106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luthi AU, Cullen SP, McNeela EA, Duriez PJ, Afonina IS, Sheridan C, Brumatti G, Taylor RC, Kersse K, Vandenabeele P, Lavelle EC, Martin SJ. Suppression of interleukin-33 bioactivity through proteolysis by apoptotic caspases. Immunity. 2009;31:84–98. doi: 10.1016/j.immuni.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Lefrancais E, Roga S, Gautier V, Gonzalez-de-Peredo A, Monsarrat B, Girard JP, Cayrol C. IL-33 is processed into mature bioactive forms by neutrophil elastase and cathepsin G. Proc Natl Acad Sci U S A. 2012;109:1673–1678. doi: 10.1073/pnas.1115884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae S, Kang T, Hong J, Lee S, Choi J, Jhun H, Kwak A, Hong K, Kim E, Jo S, Kim S. Contradictory functions (activation/termination) of neutrophil proteinase 3 enzyme (PR3) in interleukin-33 biological activity. J Biol Chem. 2012;287:8205–8213. doi: 10.1074/jbc.M111.295055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond-Martel I, Yu H, Affar el B. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24:410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278:35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 27.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 28.Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 2010;10:103–10. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]


