Abstract
Duck circovirus was a newly discovered pathogen that causing ducks immunosuppression in recent years, but it can not been cultured in vitro that limited its depth study. In the present study, the Cap gene that defect the nuclear localization signal (NLS) of DuCV was amplified and connected to the express vector pET-32a (+), to express the recombinant Cap protein in the bacterium Escherichia coli Rosetta. The recombinant Cap protein was purified and an indirect ELISA method was developed based on the recombinant Cap protein. The results showed that the truncated Cap gene was 567 bp and cloned into pET-32a (+) vector successfully. The recombinant Cap protein was expressed as inclusion bodies. The results of optimization for indirect ELISA revealed that the optimal antigen and serum dilutions were selected to be 4 μg/well and 1:40, respectively; the coating condition was 37°C for 1 h and 4°C overnight; the blocking time in 1% BSA was 1 h at 37°C and the second antibody dilution was 1:800, the cut-off value was 0.352. Known anti-sera samples of other duck common pathogens were tested by the developed ELISA and the results showed it was specific for DuCV anti-sera detection. The sensitivity of indirect ELISA reached 1:2560, and the coefficient of variation between intra-assay and inter-assay were less than 10%. Compared the PCR and indirect ELISA methods, the positive compliance rate was 95.6% for detected 59 duck samples.
Keywords: Duck circovirus, cap gene, prokaryotic expression, indirect ELISA
Introduction
Duck circovirus (DuCV) was a new member of Circovirus families, it was first described in Germany in female 6-week-old Mallard ducks from a German farm [1,2]. Since then, DuCV was isolated in Hungary [3], Taiwan area [4], America [5] and China mainland [1]. Now the DuCV has been reported in many cities in China, such as Shandong [2] and Fujian [6]. DuCV infected birds showed feathering disorder, poor body condition and low weight for their age [6], it has caused great losses for duck farms. The characteristic of Circovirus infection is that it is commonly associated with damage of the lymphoreticular tissues, and it can lead to immunosuppression which could increase the chance of secondary infection [7].
DuCV is a small, nonenveloped and single-stranded DNA virus. The genome has three major open reading frames (ORFs), the ORFC1, encoded the virus Capsid protein (Cap), Cap protein is the major immunogenic protein and the only structure protein of DuCV, it plays a critical role in virus antigenic. Cap protein had a large number of arginine residues concentrated at the N-terminus, which can inhibits the mRNA translation, and most information of antigenic epitopes of circovirus suggested that the NLS region might not be the major domain for the conformational epitopes [8].
Due to lack of cell culture propagation system for DuCV, the most reliable serological diagnostic assays for antibody detection against DuCV consist of PCR and RT-PCR [1,3,5,9]. However, these assays not only asked the expensive experiments, but also carry the risk of virus contamination, and had a high time-consuming. Whereas commercially available enzyme-linked immunosorbent assays (ELISA) based on a recombinant Capsid protein expressed in baculovirus expression systems or bacterial expression systems are more convenient [10-12]. As a result, in this study, a recombinant NLS truncated Cap protein of DuCV expressed in Escherichia coli system was used as an antigen for indirect ELISA development.
Materials and methods
Plasmid construction
DuCV GH01strain (GenBank Accession No. JX499186) was isolated from spleen of sick Muscovy duck in Sichuan Province of China in 2012. A pair of primers were designed, Cap F (5’-GGCGAATTCCTAGAAGATGGGGCTCAGTACAC-3’) as the sense primer, Cap R (5’-ACGCGTCGACGCTAGAACCCGGTGAACTGACC-3’) as the antisense primer, containing two restriction sites (underlined) EcoR I and Sal I. The truncated Cap gene was 567 bp in length and digested with EcoR I and Sal I. then cloned into pMD18-T vector. The recombinant plasmid pMD18-Cap was verified by double enzymes digestion and DNA sequencing.
Prokaryotic expression and purification of Cap protein of DuCV
The recombinant expression plasmid transformed to E. coli Rosetta cells. After confirmed by restriction enzyme digestion and DNA sequencing, the positive colonies were inoculated into LB medium containing ampicillin (AMP) and the inoculum were induced with isopropy-β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.8 Mm. The bacterial lysates were analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Based on the His-tag at the N-terminal end, the Cap fusion protein was purified by Ni-NTA affinity chromatography His-Bind Resin according to the manufacturer’s instructions, and stored at -20°C.
Establishment of an indirect ELISA based on recombinant Cap protein
An indirect ELISA was established using the purified recombinant Cap protein for the serological surveillance. All of the assay parameters were optimized and standardized. The 96-well microtiter plates were coated with 100 μL Cap protein in 0.05 M carbonate buffer (PH9.6) and incubated overnight at 4°C. After washing three times with 0.01 M PBST, the plate was blocked with 100 μL/well of 1% BSA for 2 h at 37°C. Following three washes with PBST, the DuCV positive serum in PBST was added to the plate 100 μL/well and incubated for 2 h at 37°C. Then after washing for three times, 100 μL of goat anti-duck-HRP IgG diluted 1/1000 in 0.01 M PBST was added to each well, followed by 2 h incubation at 37°C and then washing for three times. Last, 100 μL TMB was added into every well, and incubated in 37°C. for 15 min in dark. The reaction was stopped by adding 50 μL 2 M H2SO4 in each well. The absorbance at 450 nm was determined by automatic ELISA plate reader.
Optimization of indirect ELISA working conditions
Based on the procedure described above, the optimal antigen concentration and sera dilution were determined through standard checkerboard titration procedures [13]. Briefly, the Cap protein was immobilized onto 96-well microtiter plates in serial dilutions from 1 μg/well to 64 μg/well. Correspondingly, the DuCV positive and negative serum dilutions were from 1:10 to 1:160, and the phalanx titration was used to determine the best working conditions. After the establishment of antigen concentration antigen and antisera dilutions, the HRP-labeled goat anti-duck IgG condition was optimized by using different dilutions, including 1:200, 1:400, 1:800, 1:1600, 1:3200 and 1:6400. The conditions that gave the highest OD450 ratio between positive and negative serum (P/N value) and the OD450 value of positive serum close to 1.0 were scored as optimal working conditions [12].
Determination of the cut-off value for the indirect ELISA
Thirty-five samples of DuCV negative serum were used to assess the cut-off value under the optimal conditions of indirect ELISA. The cut-off value was calculated by means of OD450 plus triple standard deviation (SD), which will be used as a standard to determine the positive and negative serum of DuCV.
Reproducibility and sensitivity of the indirect ELISA
Five positive serum samples of DuCV were used for the reproducibility experiment. For intra-assay reproducibility, three replicates of each serum sample were assigned in the same plate. For inter-assay, three replicates of each sample were run in different plates. Mean S/N ratio, SD and coefficient of variation (C.V) were calculated. Four positive serum samples of DuCV were used to test the sensitivity of indirect ELISA with twofold serial dilution from 1:40 to 1:5120. The dilution of serum was measured by the indirect ELISA.
Specificity of indirect ELISA
The positive serum of avian influenza virus (AIV), duck hepatitis virus (DHV), duck swollen head septicemia virus (DSHSV), Riemerella anatipestifer (RA), Escherichia coli (E. coli) and Salmonella enteritidis (S.E) (stored in our laboratory) were used in the established indirect ELISA method for the evaluation of the antigenic cross-reactivity of recombinant Cap protein. Each serum sample was replicated in triplicate, and the DuCV positive serum and PBS was served as a positive and negative control respectively.
Comparison of the indirect ELISA and PCR for DuCV detection
For the further diagnosis of DuCV using indirect ELISA, 59 clinical suspected DuCV-infection samples were tested by PCR and indirect ELISA. The PCR primers were as same as aforementioned. Meanwhile, all the positive and negative sera of DuCV were tested by indirect ELISA. Compared the results was obtained from PCR and indirect ELISA and evaluate the positive compliance rate for DuCV detection.
Results
Construction of recombinant plasmid
The truncated Cap gene of DuCV GH01 strain was amplified successfully by PCR (Figure 1), which was corresponding to the expected size (567 bp). The recombinant plasmid pMD-18-Cap was digested with the EcoR I and Sal I (Figure 2).
Figure 1.
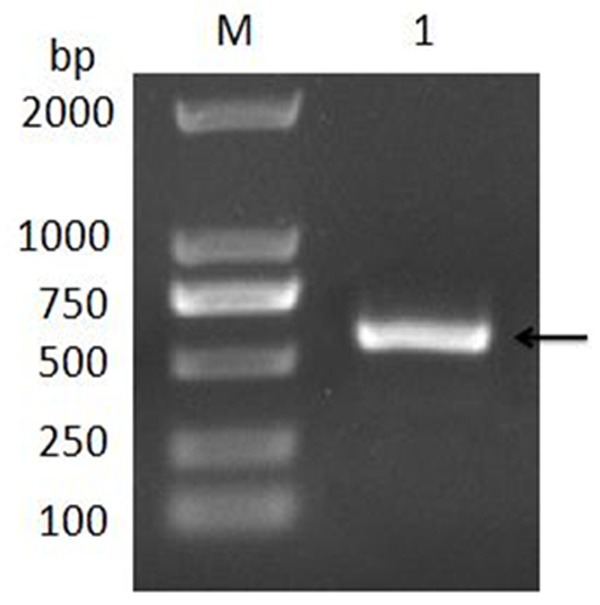
Identification of Cap gene amplified by PCR. M: DL2000 DNA Marker; 1: PCR product.
Figure 2.
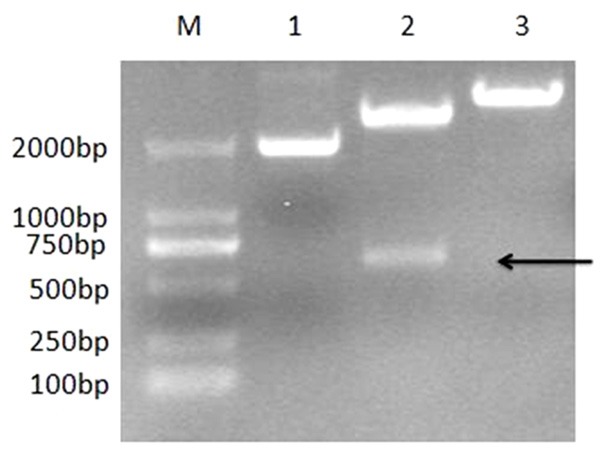
Restriction enzyme analysis of pMD18-Cap. M: DL2000 DNA Marker; 1: The T cloned products of pMD18-Cap; 2: The products digested from pMD18-Cap by EcoR I and Sal I; 3: The products digested from pMD18-Cap by EcoR I.
Expression the recombinant Cap protein
After optimization of induction conditions, the results showed that with the induction of 0.8 Mm IPTG for 16 h at 30°C, the recombinant Cap protein can be expressed at the highest level. When the expressed protein was analyzed by SDS-PAGE, demonstrated that Cap fusion protein was existed in the cell debris pellets in the form of inclusion (data not shown). The fusion protein exhibited a relatively high level of expression with the molecular weight approximately 39 KDa, which is corresponding to the expected molecular weight of His-tagged Cap protein expressed by Rosetta (Figure 3, lane 4). Obvious band of approximately 18 KDa that corresponding to the expected molecular weight of His tag, existed in the cell lysates of Rosetta harboring pET-32a (+) (Figure 3, lane 2).
Figure 3.
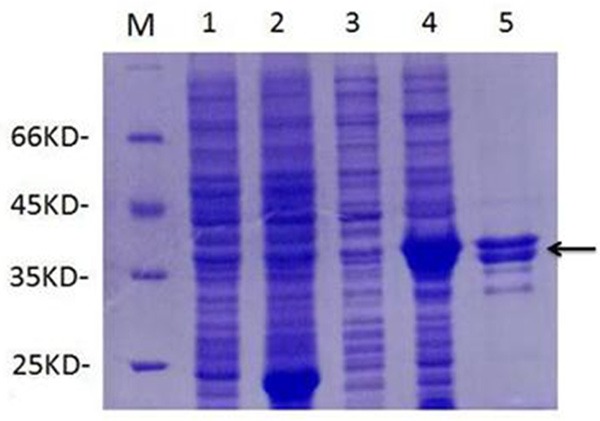
Products from recombinant Cap protein express by SDS-PAGE analysis. M: Protein Marker; 1: pET-32a (+) without IPTG; 2: pET-32a (+) with IPTG; 3: pET-32-Cap without IPTG; 4: pET-32a-Cap with IPTG; 5: protein purification products.
Purification of Cap fusion protein
Purification of the His-tagged Cap fusion protein was performed with a Ni-NTA resin column. The SDS-PAGE results showed that two clear bands corresponding to the expected molecular weight (39 KDa) of the target protein were detected after purification (Figure 3, lane 5).
Establishment of indirect ELISA
The indirect ELISA method was optimized using the checkerboard titration protocol (Table 1). The optimal antigen concentration and serum dilutions were 4 μg/well and 1:40 respectively, while the incubation condition was 37°C for 1 h followed by incubation at 4°C overnight. And the blocking time in 1% BSA was 1 h at 37°C and the second antibody dilution was 1:800. DuCV-negative serum specimens were analyzed to obtain the cut-off value for the indirect ELISA, and the mean was 0.223 with a SD of 0.0429. Thus, the cut-off value was 0.352.
Table 1.
Determination of the coating concentration of protein and serum dilution
| Antisera at different dilutions | OD values of antigen at different coatingconcentration | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 1 μg | 2 μg | 4 μg | 8 μg | 16 μg | 32 μg | 64 μg | |
| 1:10 (+) | 0.988 | 1.243 | 1.560 | 1.684 | 1.825 | 1.988 | 2.125 |
| 1:10 (-) | 0.203 | 0.233 | 0.224 | 0.255 | 0.273 | 0.289 | 0.304 |
| 1:20 (+) | 0.747 | 1.135 | 1.423 | 1.521 | 1.642 | 1.810 | 2.042 |
| 1:20 (-) | 0.195 | 0.217 | 0.204 | 0.243 | 0.264 | 0.277 | 0.291 |
| 1:40 (+) | 0.590 | 0.892 | 1.208 | 1.316 | 1.454 | 1.672 | 1.877 |
| 1:40 (-) | 0.183 | 0.198 | 0.206 | 0.227 | 0.257 | 0.262 | 0.270 |
| 1:80 (+) | 0.472 | 0.724 | 1.109 | 1.147 | 1.289 | 1.458 | 1.690 |
| 1:80 (-) | 0.174 | 0.186 | 0.200 | 0.225 | 0.211 | 0.248 | 0.259 |
| 1:160 (+) | 0.365 | 0.548 | 0.769 | 0.929 | 1.067 | 1.169 | 1.358 |
| 1:160 (-) | 0.155 | 0.136 | 0.120 | 0.189 | 0.223 | 0.197 | 0.236 |
Reproducibility and sensitivity of indirect ELISA
Three replicates of 5 specimens were analyzed for reproducibility. The results showed that the CVs were range from 2.66% to 6.05%, with the mean value of 3.92% in intra-assay. The results were different after each group was performed at different times, so the inter-assay showed that the CVs were range from 2.57% to 5.56% with a mean value of 3.96% (Table 2). All of the results showed that the coefficient were lower than 10%, which indicated that the indirect ELISA was highly reproducible and stable.
Table 2.
Repeatability and reproducibility analysis of indirect ELISA
| Sample No. | Intra-assay (OD450) | Inter-assay (OD450) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| 1 | 2 | 3 | Mean | SD | CV% | 1 | 2 | 3 | Mean | SD | CV% | |
| 1 | 0.847 | 0.78 | 0.755 | 0.794 | 0.048 | 6.05 | 0.861 | 0.899 | 0.822 | 0.861 | 0.039 | 4.53 |
| 2 | 0.692 | 0.689 | 0.733 | 0.705 | 0.025 | 3.55 | 0.72 | 0.757 | 0.738 | 0.738 | 0.019 | 2.57 |
| 3 | 0.84 | 0.802 | 0.841 | 0.828 | 0.022 | 2.66 | 0.839 | 0.801 | 0.781 | 0.807 | 0.029 | 3.59 |
| 4 | 1.083 | 1.085 | 1.163 | 1.110 | 0.046 | 4.14 | 1.077 | 1.034 | 1.109 | 1.073 | 0.038 | 3.54 |
| 5 | 0.757 | 0.782 | 0.807 | 0.782 | 0.025 | 3.20 | 0.822 | 0.742 | 0.755 | 0.773 | 0.043 | 5.56 |
Sensitivity analysis of ELISA was performed by detecting the DuCV positive serum. The minimum detection limit of the sera was 1:2560 according to the cut-off value 0.352 (data not shown).
The specificity of indirect ELISA
Six known positive sera samples of AIV, DHV, DSHSV, RA, E. coli and S.E were detected by indirect ELISA to evaluate the cross-reactivity of this assay. The results showed that the OD450 values of all the known sera and PBS control were lower than cut-off value (Figure 4).
Figure 4.
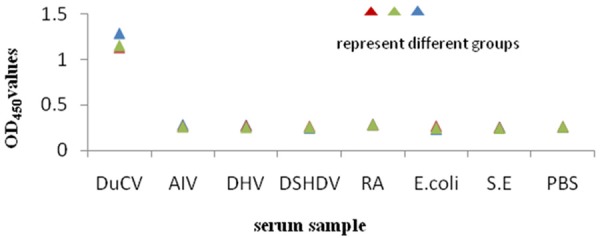
The specificity of indirect ELISA.
Comparison the PCR and indirect ELISA detection methods
Suspected DuCV-infection samples were detected by PCR and indirect ELISA. PCR results showed that 45 samples were detected as DuCV positive, while 43 positive samples were detected by indirect ELISA (Table 3). The coincident rate between indirect ELISA and PCR was 95.6% (43/45).
Table 3.
Comparison of the indirect ELISA and PCR assay for DuCV detection
| Indirect ELISA | PCR | ||
|---|---|---|---|
|
| |||
| Positive (+) | Negative (-) | Total | |
| Positive (+) | 43 | 0 | 43 |
| Negative (-) | 2 | 14 | 16 |
| Total | 45 | 14 | 59 |
Discussions
With the dramatic increase cases in duck farms, DuCV has become a potential threat to global duck industry. As DuCV is an immunosuppressive virus, detecting DuCV in ducks is essential to help control this virus-infection, and it can provide a basis material to learn other viruses in mix infection [14]. But so far, no cell culture method is available to maintain DuCV infect. Therefore, recombinant protein expression technologies have been utilized to analyzed the characteristics of DuCV.
The advantages of the E. coli expression system contain a well-known genetic background, amenability to high cell-density fermentation, high production capacity, relatively simple protein purification methods and inexpensive culture media [15-19]. But, however, E. coli is not suit for every foreign gene expression efficiently, because the existed codon usage bias is different between foreign and native E. coli genes [3]. It has been reported that rare codons, especially those encoding Arg (AGA, AGG, CGA and CGG), can greatly reduce expression levels of recombinant protein in E. coli because of translation termination [4,5]. Sequence analysis reveals that the N terminal of Cap gene contains 11 rare codons for E. coli, and it has a high proportion of arginine residues concentrated at the N terminus of the protein [20]. These may be the important factors that Cap protein with NLS fail to express in E. coli [21]. Therefore, in this study, with the deletion of gene fragment encoding arginine in Cap gene, Cap protein, Cap protein can be expressed well in E. coli.
The recombinant fusion protein, encoded by truncated Cap gene and expressed in E. coli, contains the tagged protein and Cap protein, with the molecular weight of about 18 kD and 21 kD, respectively. Therefore, the recombinant fusion protein is about 39 kD, which is in accordance with the expect value. After purification by Ni-NTA and SDS-PAGE analysis, showed that the recombinant protein was expressed at a high level as inclusion bodies, which would lay the material foundation for the subsequent investigation of DuCV.
The use of recombinant protein as antigens in serological diagnosis has major advantages because they are cheap, easy to produce, and their antigenicity could be carefully defined [22]. In this study, the indirect ELISA method for testing DuCV was established. The great immunoreactivity was observed by using the recombinant Cap protein as antigen, suggesting the functional antigenic sites were within the C-terminal region. The clinical comparison results of PCR and indirect ELISA test results showed that coincident rate between indirect ELISA and PCR was 95.6%.
In conclusion, DuCV Cap gene without nuclear localization signal can be expressed in E. coli successfully. The established indirect ELISA method has a good specificity, reproducibility and sensitivity. Therefore, this test will serve as a valuable tool for further diagnostic study and provide a basis for follow-up experiments.
Acknowledgements
The research was supported by China Agricultural Research System (CARS-43-8), the Ministry of Education Program (20125103110013), Sichuan Province Research Programs (2013HH0042/2013TD0015/11ZA084/12TD005/2011ZO0034/2011JO0040) and China 973 program (2011CB111606).
Disclosure of conflict of interest
None.
References
- 1.Jiang SJ, Zhang XX, Liu SN, Wang Y, Kong YB, Wei XL, Sun YN, Zhao Q. PCR detection and sequence analysis of duck circovirus in sick Muscovy ducks. Virologica Sinica. 2008;23:265–271. [Google Scholar]
- 2.Zhang X, Jiang S, Wu J, Zhao Q, Sun Y, Kong Y, Li X, Yao M, Chai T. An investigation of duck circovirus and co-infection in Cherry Valley ducks in Shandong Province, China. Vet Microbiol. 2009;133:252–256. doi: 10.1016/j.vetmic.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 3.Fringuelli E, Scott A, Beckett A, McKillen J, Smyth J, Palya V, Glavits R, Ivanics E, Mankertz A, Franciosini M. Diagnosis of duck circovirus infections by conventional and real-time polymerase chain reaction tests. Avian Pathol. 2005;34:495–500. doi: 10.1080/03079450500368334. [DOI] [PubMed] [Google Scholar]
- 4.Chen CL, Wang PX, Lee MS, Shien JH, Shieh HK, Ou SJ, Chen CH, Chang PC. Development of a polymerase chain reaction procedure for detection and differentiation of duck and goose circovirus. Avian Dis. 2006;50:92–95. doi: 10.1637/7435-090705R1.1. [DOI] [PubMed] [Google Scholar]
- 5.Banda A, Galloway-Haskins RI, Sandhu TS, Schat KA. Genetic analysis of a duck circovirus detected in commercial Pekin ducks in New York. Avian Dis. 2007;51:90–95. doi: 10.1637/0005-2086(2007)051[0090:GAOADC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 6.Wan CH, Fu GH, Shi SH, Cheng LF, Chen HM, Peng CX, Lin S, Huang Y. Epidemiological investigation and genome analysis of duck circovirus in Southern China. Virol Sin. 2011;26:289–296. doi: 10.1007/s12250-011-3192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Todd D, Gortazar C. Infectious Diseases of Wild Mammals and Birds in Europe. 2012. Circovirus Infections; pp. 67–72. [Google Scholar]
- 8.Jittimanee S, Ayudhya SNN, Kedkovid R, Teankum K, Suradhat S, Thanawongnuwech R. An indirect enzyme-linked immunosorbent assay using a recombinant truncated capsid protein of Porcine circovirus-2. J Vet Diagn Invest. 2012;24:1129–1132. doi: 10.1177/1040638712461251. [DOI] [PubMed] [Google Scholar]
- 9.Wan C, Huang Y, Cheng L, Fu G, Shi S, Chen H, Peng C, Lin F, Lin J. The development of a rapid SYBR Green I-based quantitative PCR for detection of Duck circovirus. Virol J. 2011;8:465. doi: 10.1186/1743-422X-8-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanchard P, Mahe D, Cariolet R, Truong C, Le Dimna M, Arnauld C, Rose N, Eveno E, Albina E, Madec F. An ORF2 protein-based ELISA for porcine circovirus type 2 antibodies in post-weaning multisystemic wasting syndrome. Vet Microbiol. 2003;94:183–194. doi: 10.1016/S0378-1135(03)00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcekova Z, Psikal I, Kosinova E, Benada O, Sebo P, Bumba L. Heterologous expression of full-length capsid protein of porcine circovirus 2 in Escherichia coli and its potential use for detection of antibodies. J Virol Methods. 2009;162:133–141. doi: 10.1016/j.jviromet.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang SB, Li YF, Guo JQ, Wang ZT, Chen QX, Shen HG, Zhou JY. Development and validation of a recombinant capsid protein-based ELISA for detection of antibody to porcine circovirus type 2. Res Vet Sci. 2008;84:150–157. doi: 10.1016/j.rvsc.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 13.Crowther JR, Walker JM. The ELISA guidebook. Springer; 2009. [Google Scholar]
- 14.Liu SN, Zhang XX, Zou JF, Xie ZJ, Zhu YL, Zhao Q, Zhou EM, Jiang SJ. Development of an indirect ELISA for the detection of duck circovirus infection in duck flocks. Vet Microbiol. 2010;145:41–46. doi: 10.1016/j.vetmic.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 15.Sahdev S, Khattar SK, Saini KS. Production of active eukaryotic proteins through bacterial expression systems: a review of the existing biotechnology strategies. Mol Cell Biochem. 2008;307:249–264. doi: 10.1007/s11010-007-9603-6. [DOI] [PubMed] [Google Scholar]
- 16.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 17.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sørensen HP, Mortensen KK. Advanced genetic strategies for recombinant protein expression in Escherichia coli. J Biotechnol. 2005;115:113–128. doi: 10.1016/j.jbiotec.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 19.Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72:211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- 20.Xiang QW, Zou JF, Wang X, Sun YN, Gao JM, Xie ZJ, Wang Y, Zhu YL, Jiang SJ. Identification of two functional nuclear localization signals in the capsid protein of duck circovirus. Virology. 2013;436:112–117. doi: 10.1016/j.virol.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 21.Zhou JY, Shang SB, Gong H, Chen QX, Wu JX, Shen HG, Chen TF, Guo JQ. In vitro expression, monoclonal antibody and bioactivity for capsid protein of porcine circovirus type II without nuclear localization signal. J Biotechnol. 2005;118:201–211. doi: 10.1016/j.jbiotec.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Wu PC, Chien MS, Tseng YY, Lin J, Lin WL, Yang CY, Huang C. Expression of the porcine circovirus type 2 capsid protein subunits and application to an indirect ELISA. J Biotechnol. 2008;133:58–64. doi: 10.1016/j.jbiotec.2007.09.015. [DOI] [PubMed] [Google Scholar]


