Abstract
Background: Colorectal cancer (CRC) is one of the leading causes of cancer-related death all over the world. Ribosomal s6 kinase4 (RSK4), an X-linked gene, firstly was found as to be a potential tumor suppressive gene in a variety of cancers and is widely participated in signaling pathway. However its role in CRC is unclear. This study is to explore the correlation between the protein expression of RSK4 and clinical pathologic characteristics in colorectal tumors, which might serve as a prognostic determinant of colorectal cancers. Methods: Biopsies of 103 colorectal cancer and 46 matched adjacent noncancerous tissues were collected for analysis of RSK4 protein by immunohistochemistry. The correlation between RSK4 protein expression and the clinical pathological features of colorectal cancers were evaluated by Chi-square test and Fisher’s exact test. The survival rates were analyzed by the Kaplan-Meier method, and the relationship between prognostic factors and patient survival was analyzed by the Cox proportional hazard models. Results: RSK4 was conversely correlated with some pathological classifications (P<0.05 for N, G and clinical staging), and there were no statistically significant differences in age, CEA expression in blood, CA199 and tumors t-staging (x2 test, P>0.05 for all categories) respectively. Furthermore, patients with high protein level of RSK4 showed prolonged overall survivals (P<0.05). Moreover, multivariate analysis showed that low expression level of RSK is an independent risk factor for high mortality in colorectal cancer. Conclusions: Low RSK4 expression is correlated with advanced clinical pathologic classifications and is a poor overall survival in colorectal cancer patients. These findings suggest that RSK4 may serve as a useful marker in prognostic evaluation for patients with colorectal cancer.
Keywords: Colorectal cancer, RSK4, immunohistochemistry, survival analysis
Introduction
Colorectal cancer remains a major cause of cancer-related death worldwide, even though survival for patients with metastatic colorectal cancer is improved dramatically over the past decade. In the recent years, molecularly targeted agents become a popular and promising way to treat colorectal cancers. One way to reduce the staggering mortality rate is to predict outcome based on the aggressiveness of the tumor biology in order to treat patients accordingly to their risk profile. Although advance has beendramatic in the treatment for colorectal cancer, the prognosis of the cancer remains a challenge clinically.Worse still, there are few tumor markers useful in the monitoring of colorectal cancer such as CEA, CA199 et al. As such, it comes as no surprise that prognostic biomarker discovery is a hot topic in colorectal cancer research. Therefore, it is necessary to develop novel markers that may show increased specificity and/or sensitivity in evaluating the prognosis of colorectal cancer clinically.
Recently, mounting evidence has proved that the Ras-mitogen-activated protein kinase (MAPK) pathway is a key signalling pathway involved in proliferation, survival, growth and differentiation [1]. The 90 kDa ribosomal S6 kinases (RSKs) are a family of Ser/Thr kinases downstream of the Ras-MAPK cascade. This family consists of four human isoforms (RSK1-4) and two structurally related homologues. RSK1-4 possess 73-80% amino acid identity and have the same general structure [2]. The most striking feature of the RSK family is the two functional and non-identical phosphortransferase domains within the same polypeptide [3,4]. It is thought that, during evolution, the genes for two distinct protein kinases have fused, generating a single protein kinase capable of receiving an upstream activating signal from ERK1/2 to its CTKD (C-terminal kinase domain) and transmitting, with high efficiency and fidelity, an activating input to its NTKD (Nterminal kinase domain). There are many studies clearly showed that RSK1-2 [5-7] is associated with the regulation of cell cycle, proliferation and differentiation in normal human cells. Recently, it is shown that RSK3 is abnormally expressed in human cancers, such as ovarian cancer, which suggested that it may function as a tumor suppressive gene [8]. RSK4, ribosomal protein S6 kinas 4, is primitively found as an X-linked gene in patients with mental retardation and abundantly expressed in brain and kidney [9]. RSK4, as the last member of this kinase family identified, is different from other RSK proteins presenting a tumor inhibitory role in cell proliferation. RSK4 is associated with the cell signaling pathway in cellular proliferation and differentiation [10,11].
However, up to now, the real role of RSK4 in colorectal carcinoma is not clear. The purpose of our present study is to explore the expression pattern of RSK4 in human colorectal cancer samples compared to the normal benign tissues and to investigate the prognostic value of RSK4 in clinical colorectal cancer patients.
Materials and methods
Patients and tissue specimens
This study complied with the Helsinki Declaration and was approved by the Human Ethics the Research Ethics committees of the First Affiliated Hospital of Yangtze University of China. Through the surgery consent form, patients were informed that the resected specimens were kept in our hospital and might be used for scientific research, and that their privacy would be maintained. Follow-up survival data were collected retrospectively through medical-record analyses. The formalin-fixed, paraffin-embedded pathology specimens of 103 colorectal cancer tissue samples, 46 matched adjacent noncancerous tissues were used as control from the above-mentioned patients. Pathological parameters, including age, gender, grade, nodal metastasis, clinical stage and survival data were carefully reviewed in all cases. None of the patients received radiotherapy or chemotherapy before surgery. Informed consent was obtained from patients prior to investigation. The diagnosis was microscopically confirmed by two pathologists between 2006 and 2008. Conventional clinical features were evaluated including age, presence of CEA and CA199 in blood. Clinical staging was assessed according to the 2009 TNM staging of International Union Against Cancer. The observation period was from 2006 to 2013.
Immunohistochemical staining and evaluation
All tissue samples were fixed in 4% formalin for 24 h, and then embedded routinely into paraffin for hematoxylin and eosin (H and E) and immunohistochemistry (IHC) staining. Five-micrometer sections were cut from the paraffin blocks and placed on glass slides. The slides were dried in an incubator at 60°C for 60 min, deparaffinized in xylene, and then rehydrated in an ethanol series. After washing in water, antigen retrieval with citrate buffer was performed at high temperature and pressure. The sections were cooled for 20 min, washed twice with phosphate buffered solution (PBS) for 5 min each, and then incubated in serum for 10 min. The primary antibody (RSK4 rabbit anti-person monoclonal antibody, USA) was diluted in 1% PBS and incubated for 45 min after the serum was tipped. The slides were then washed twice with PBS and incubated with the anti-rabbit secondary antibody (GTVisionTM I, China) for 30 min. After an additionally two times of wash in PBS, the slides were incubated with diaminobenzidine (DAB, Invitrogen, United States) for 10 min to visualize immunoglobulin. After washing, the sections were counterstained with hematoxylin (Invitrogen, United States). PBS was used as the negative control instead of the primary antibody on each slide for RSK4. H and E-stained samples were individually examined microscopically by two independent pathologists. Clinic pathological characteristics of tumors are provided in Table 1.
Table 1.
Expression RSK4 of colorectal cancer tissue, adjacent noncancerous tissues and normal tissues
| Tissue Type | Case number | Positive number | positive rate (%) | X2 | P |
|---|---|---|---|---|---|
| Colorectal cancerous tissue | 46 | 7 | 15.0 | 13.16 | 0.001 |
| Noncancerous Issues | 46 | 19 | 41.3 | ||
| Normal colorectal tissues | 46 | 23 | 50 |
Appropriate positive and negative controls were included for each run. Cytoplasm staining was considered positive. The staining intensity was scored as weak, moderate, or strong. The percentage of positively stained tumor cells was scored in 4 grades: 0 (0%-10%), 1 (10%-25%), 2 (25%-50%) and 3 (50%-100%). To achieve objectivity, the intensity of positive staining was also used in a four scoring system: 0 (negative staining), 1 (weak staining exhibited as light yellow), 2 (moderate staining exhibited as yellow brown), and 3 (strong staining exhibited as brown). The intensity and percentage of positively stained tumor cells were scored after counting at least 10 high-power fields at 200 ×. RSK4 expression index = (intensity score) (positive score). The cut-off value for high and low levels of expression was based on a measurement of heterogeneity with a log-rank test statistical analysis with respect to overall survival. Using this assessment system, optimal cut-off values were identified: expression index scores of ≥4 were used to define high expression of RSK4 and expression index scores of <4 were indicative of low expression of RSK4.
Statistical analyses
All statistics data were analyzed by SPSS16.0 statistical software package. Because of the non-normal distribution of protein expression, statistical evaluation was performed using nonparametric tests. The Chi-square test was employed to evaluate differences in the expression level of RSK4 among three categories of tissues. The correlation between RSK4 expression and clinical pathological characteristics were evaluated by Chi-square test and Fisher’s exact tests. Relative risks (RR) of death associated with RSK4 expression and other variables were estimated using unvaried and multivariate Cox proportional hazards model. In our analyses, we defined a RR of 1.000 as baseline for factors including age (≤60 years), T1, N0, M0, clinical stage I, high level of RSK4 expression [12]. Survival curves were plotted using Kaplan-Meier survival analysis and compared by log-rank test. Further analyses of survival curves were computed based on stratifying TNM classifications and clinical stages. Multivariate survival analysis was performed on all the significant characteristics measured by univariate survival analysis (gender, age, tumor size, differentiation, lymph node metastasis, tumor stage, the expression of CEA, CA199 and RSK4 through the Cox proportional hazard regression model. In all statistical analyses, P<0.05 was considered significant.
Progression free survival (PFS) was calculated as the time lapsed between the date of treatment and the date of relapse or progressive disease. Patients with no signs of relapse were censored at the time of last follow-up or death. Overall survival (OS) was analyzed from the day of diagnosis until death or the last follow-up.
Results
Decreased expression of RSK4 in colorectal cancer tissues
The protein expression level of RSK4 was evaluated in 103 CRC tissues, 46 noncancerous tissues and normal colorectal tissues. We found that RSK4 dispersedly expressed in the cytoplasm as brown granules. The expression levels of RSK4 in cancerous, noncancerous and normal tissues are shown in Figure 1. Immunohistochemical assay showed that the positive rate of RSK4 protein expression is gradually reduced from normal colorectal tissues (50%, 23/46) to noncancerous tissues (41.3%, 19/46) to cancerous tissues (15.0%, 7/46). The protein expression levels of RSK4 in the same patient were significantly lower in primary tumor tissues than that in corresponding matched adjacent noncancerous tissues and normal tissues (x2=13.16, P=0.001; Table 1).
Figure 1.

Immunohistochemical photomicrographs of RSK4 in colorectal cancer tissues and colorectal noncancerous tissues (200 ×). A, B. RSK 4 displayed negative and weak positive cytoplasm staining in colorectal cancer (CRC) tissues; C, D. RSK 4 displayed weak positive and strongest positive staining in normal tissues (200 ×).
Correlation between decreased protein expression level of RSK4 and clinical aggressiveness of colorectal cancer
Baseline characteristics of the 103 patients with colorectal, including gender, age, primary clinical stage, tumor TNM, histological grade, CEA and CA199. The correlation between RSK4 protein expression and clinic pathological factors are shown in Table 2.
Table 2.
Correlation between RSK4 protein expression level and clinic pathologic characteristics
| Patient characteristics | RSK4 | P value | |
|---|---|---|---|
|
| |||
| Low expression (n) | High expression (n) | ||
| Age (years) | |||
| ≤60 | 38 | 10 | 0.734 |
| >60 | 45 | 10 | |
| T classification | |||
| T1 | 4 | 2 | 0.123 |
| T2 | 10 | 6 | |
| T3 | 37 | 8 | |
| T4 | 32 | 4 | |
| N classification | |||
| N0 | 17 | 10 | 0.017 |
| N1 | 27 | 6 | |
| N2 | 39 | 4 | |
| M classification | |||
| M0 | 71 | 18 | 0.874 |
| M1 | 12 | 2 | |
| G classification | |||
| G1 | 25 | 13 | 0.005 |
| G2 | 20 | 5 | |
| G3 | 38 | 2 | |
| Clinical stage | |||
| I | 1 | 2 | 0.007 |
| II | 12 | 8 | |
| III | 58 | 8 | |
| IV | 12 | 2 | |
| CEA | |||
| Negative | 59 | 14 | 0.924 |
| Positive | 24 | 6 | |
| CA199 | |||
| Negative | 58 | 15 | 0.651 |
| Positive | 25 | 5 | |
To evaluate the relationship between RSK4 protein and colorectal cancer progression, we further analyzed the correlation between RSK4 protein expression and clinic pathological features of colorectal cancers. As shown in Table 1, RSK4 staining was relatively higher in the 20 clinical cases and lower in the 83 clinical cases. Through analyzing the clinical characteristics of the 103 colorectal cancer cases, it indicated that the expression of RSK4 protein was significantly reduced in colorectal cancer cases with lymph node metastasis (13.16%, 10/76) than that without metastasis (37.04%, 10/27) (P=0.017). Its expression was also significantly lower in the poorly and moderately differentiated (G2, G3) colorectal cancers (11.11%, 7/63) than in well-differentiated (G1) cases (34.21%, 13/38) (P=0.005). Similarly, for the TNM clinical stages, the positive rate of RSK4 protein was 14.29% (10/70) in the late stage (III-IV) colorectal cancers, but 43.48% (10/23) in early stage cases (I-II) (P=0.007). There were no statistically significant association between RSK4 expression and other clinical characteristics in terms of age, blood CEA, CA199 and tumors t-staging (x2 test, P>0.05 for all categories). These results dramatically indicated that the protein expression profiles of RSK4 were strongly correlated with tumor differentiation degree (G), lymph node metastasis (N) and clinical staging (x2 test, P<0.05 for all categories), respectively. These observations suggested a correlation between the decreased expression of RSK4 and clinical progression in patients with colorectal cancer.
Correlation between decreased protein expression level of RSK4 and prognosis of colorectal cancer
As we known, clinical pathologic classifications such as T, N, M and clinical stages were important indicators in the progressiveness of colorectal cancer [13]. Blood CEA and CA199 status also appeared to have important prognostic value clinically [14,15]. As we have demonstrated the correlation of RSK4 expression with clinical progressive pathologic features, we proceeded to investigate the relationship between RSK4 expression and patients’ survival and prognosis. As shown in Figure 2A, CRC patients with low RSK4 expression levels had lower survival rates than those with high RSK4 expression levels as determined using the Kaplan-Meier method (P<0.001). To explore the RSK4 role in the development of colorectal cancer, we further analyzed the patients’ survival and RSK4 expression in adjacent noncancerous tissues. As indicated in Figure 2B, the cases with higher expression levels of RSK4 in adjacent noncancerous tissues possess longer overall survival time than cases with lower expression levels of RSK4 (log-rank test, P<0.001). We found that colorectal cancer cases in the stage of T1 + T2 and T3 + T4 concomitant with lower RSK4 expression level had a significantly reduced 5-year survival rate (Figure 3A and 3B, P<0.05). Consistently, we also found that the low RSK4 expression level and poor 5-year survival rate were positively related in colorectal cancer cases with lymph node metastases. (Figure 3C and 3D, P<0.05). The similar observation were also existed in the colorectal cancer patients with M0 or M1 (P<0.001 and P=0.244, respectively).
Figure 2.
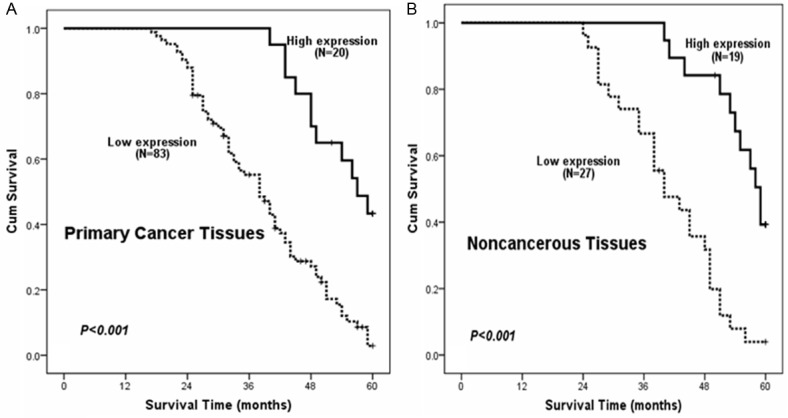
A. Kaplan-Meier curves show that patients with low RSK4 expression have poor overall survival by analyzing 103 primary colorectal cancer tissues (P<0.001). B. Patients show better overall survival with high RSK4 expression by analyzing 46 matched adjacent noncancerous colorectal tissues (P<0.001).
Figure 3.
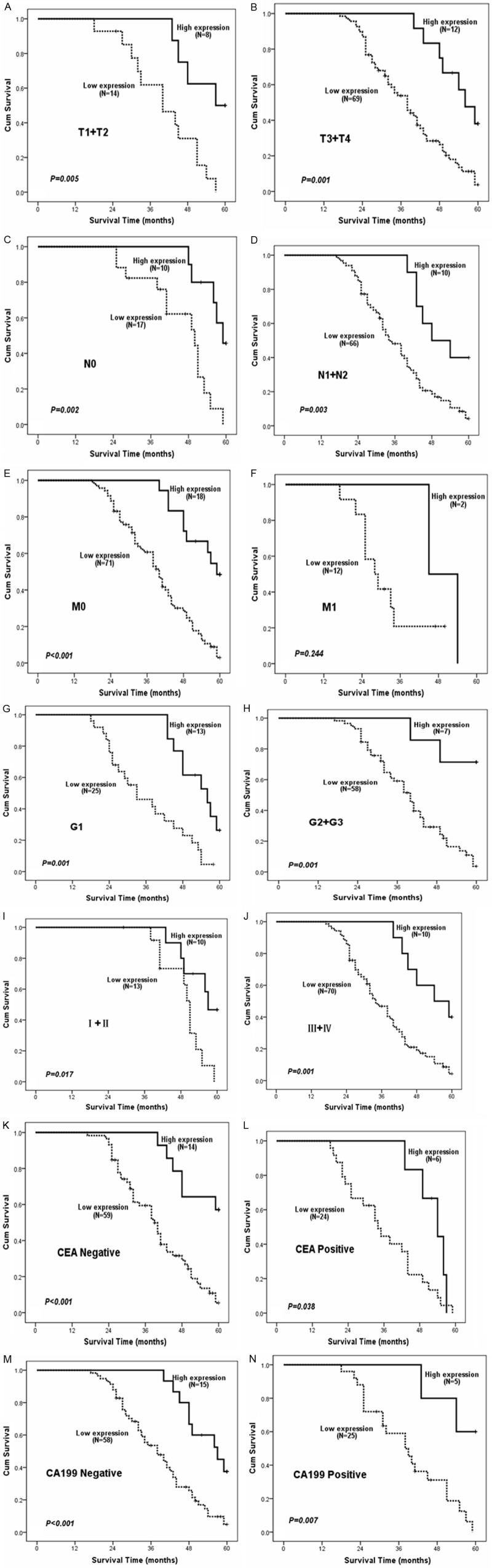
Overall survival curves stratified by RSK4 protein levels according to T, N, M classifications, clinical staging, and CEA, CA199. A, B. The differences of survival curves according to RSK4 protein expression were seen in T1 + T2 classification panel (P=0.005) and T3 + T4 classification panel (P=0.001). C, D. In N0 classification panel, patients with high RSK4 protein expression show much better overall survival (P=0.002), and N1 + N2 classification panel (P=0.003). E, F. Survival time is longer in patients with high RSK4 protein expression in M0 (P<0.001). G, H. Patients with high RSK4 protein expression show better overall survival in G1 and G2 + G3 (P=0.001). I, J. Patients with high RSK4 protein expression show better overall survival in I + II and III + IV (P<0.05). K-N. Patients with high RSK4 expression have longer survival time regardless of CEA, CA199 status.
Most importantly, stage (I-II and III-IV) colorectal cancers concomitant with low RSK4 expression had the lowest 5-year survival rate, which was significantly lower than those high RSK4 expression (Figure 3I and 3J, P=0.004). However, analyzing corresponding tumor grade (G0 and G1 + G2) suggested that levels of RSK4 seem to also have an impact on patients’ survival (Figure 3G and 3H, log-rank test, P<0.05). Interestingly, high RSK4 level revealed the favorable impact on outcome irrelevant of the presence of CEA and CA199 by subset analysis. Furthermore, it remained true that the expression levels of RSK4 are strongly correlated with patients’ survival even after stratifying the patients based upon their clinic pathologic classifications (Figure 3A-J).
Low expression of RSK4 is an independent prognostic factor in colorectal cancers by Cox proportional hazard regression model
To determine if RSK4 could serve as a risk factor for clinical application, we examined overall survival using Cox proportional hazard regression analyses. We found that colorectal cancer patients with high expression of RSK4 had significantly higher survival rates than those with low expressing of RSK4. Additionally, RSK4 expression was also positively associated with cancer survival rates in patients with lymph node metastasis (Table 3). These data suggest that RSK4 may be a favorable prognostic factor for colorectal cancer. Therefore, multivariate analysis was performed using the Cox proportional hazards model for all of the significant variables examined in the univariate analysis. The RRs showed no obvious differences when including clinical parameters, such as T classification, lymph node metastasis, tumor grade or clinical stage with the multivariate Cox regression model. Importantly, we found that low expression of RSK4 (RR: 3.468, 95% CI: 1.805-6.666) proved to be independent prognostic factors for survival in colorectal cancer (Table 4).
Table 3.
Univariate Cox regression analysis of potential prognostic factors for colorectal cancer patients
| Patient characteristics | RR (95% CI) | P value |
|---|---|---|
| Age (years) | ||
| ≤60 | 1.000 | |
| >60 | 1.185 (0.762-1.844) | 0.451 |
| T classification | ||
| T1 | 1.000 | |
| T2 | 1.554 (0.445-5.425) | 0.489 |
| T3 | 1.922 (0.588-6.285) | 0.280 |
| T4 | 2.179 (0.663-7.161) | 0.199 |
| N classification | ||
| N0 | 1.000 | |
| N1 | 2.007 (1.105-3.648) | 0.022 |
| N2 | 2.065 (1.178-3.619) | 0.011 |
| M classification | ||
| M0 | 1.000 | |
| M1 | 1.787 (0.938-3.408) | 0.078 |
| G classification | ||
| G1 | 1.000 | |
| G2 | 0.843 (0.467-1.520) | 0.569 |
| G3 | 1.110 (0.678-1.817) | 0.679 |
| Clinical stage | ||
| I | 1.000 | |
| II | 1.506 (0.342-6.642) | 0.588 |
| III | 2.964 (0.719-12.216) | 0.133 |
| IV | 4.303 (0.942-19.659) | 0.060 |
| RSK4 expression | ||
| High | 1.000 | |
| Low | 3.726(1.978-7.020) | <0.001 |
| CEA | ||
| Negative | 1.000 | |
| Positive | 1.522 (0.963-2.406) | 0.072 |
| CA199 | ||
| Negative | 1.000 | |
| Positive | 0.968 (0.597-1.570) | 0.895 |
CI, confidence interval; RR, relative risk.
Table 4.
Multivariate Cox regression analysis of potential prognostic factors for colorectal cancer patients
| Patient characteristics | RR (95% CI) | P value |
|---|---|---|
| N classification | ||
| N0 | 1.000 | |
| N1 | 1.356 (0.418-4.400) | 0.612 |
| N2 | 1.061 (0.307-3.660) | 0.926 |
| M classification | ||
| M0 | 1.000 | |
| M1 | 2.458 (0.415-14.558) | 0.322 |
| Clinical stage | ||
| I | 1.000 | |
| II | 1.101 (0.247-4.909) | 0.900 |
| III | 1.536 (0.241-9.794) | 0.650 |
| IV | 2.458 (0.415-14.558) | 0.322 |
| RSK4 expression | ||
| High | 1.000 | |
| Low | 3.468 (1.805-6.666) | <0.001 |
| CEA | ||
| Negative | 1.000 | |
| Positive | 1.556 (0.963-2.515) | 0.071 |
CI, confidence interval; RR, relative risk.
Discussion
Colorectal cancer is the third most common cancer and the fourth most common cancer cause of death globally. Even though much progress of treatment has been made in the past decades, there were still 1.2 million of new cases were diagnosed with and 600 thousand cases were died of colorectal cancer per year [16].
The slowly and steadily improved prognosis may drag the rear of CRC during the past decades. The heterogeneous molecular mechanisms underlying the development of CRC are clinically important due to their important role in prognosis and treatment of the patient [17,18]. Major advances have been achieved in the chemo therapeutic treatment of colorectal cancer, including the development of substances that inhibit the effect of vascular endothelial growth factor (bevacizumab and aflibercept), monoclonal antibodies of epidermal growth factor receptor (cetuximab and panitumumab) and kinase inhibition (regorafenib). Cetuximab and panitumumab should be used only for patients without mutations in the RAS gene (wildtype) in the tumor and are generally used as part of a combination therapy. Although therapeutic strategies have been improved in recent years, the fact is that 25% of the patients present with advanced colorectal cancer (ACRC) at diagnosis and an additional 25% eventually progress to advanced stage which makes it especially challenging to treat. Therefore, a number of new targets are being explored and appear promising in the treatment of colorectal cancer.
p90 Ribosomal S6 kinase (RSK) 4 is a serine-threonine kinase that belongs to the p90RSK family. RSK4 has been proposed as a tumor suppressor gene, related with anti-invasive activity, inhibition of the RAS-mitogen-activated protein kinase (MAPK) pathway and induction of senescence [19]. RSK4 is an unusual RSK in that it has very low expression levels and appears to be fully phosphorylated and activated in unstimulated cells [10]. In humans, RSK4 is down regulated in some tumors, such as colon and renal carcinomas, as well as some benign lesions, such as colon adenomas and benign breast papillomas [20,21]. An expression screen with mouse mRNA in Xenopus laevis embryos showed that RSK4 can disrupt mesoderm formation induced by the RAS-ERK pathway and was therefore proposed as an inhibitor of growth factor signal transduction [22]. Despite the related findings, little is known about RSK4 effectors. At present, the only characterization performed with RSK4 is its demonstration in various human and fetal tissues. RSK4 is a cytosolic protein and predominantly expressed in the human brain, while lowly expressed in kidney, heart, and skeletal muscle, and undetectable expression in lung, liver, spleen and pancreas [10]. In this study, the expression of RSK4 was evaluated in all 103 CRC tissues, 46 Noncancerous issues and normal colorectal tissue. Immunohistochemical assay showed that RSK4 expression is decreased in primary colorectal cancer tissues (15.0%) compared with their adjacent noncancerous tissues (41.3%), there was a statistically significant difference between two groups. This observations were similar with a previous report [23]. These observations presented here indicate that RSK4 acts as a tumor suppressive gene in human colorectal cancer, in keeping with a number of previous reports [19,23]. To evaluate the relationship between RSK4 protein and colorectal cancer progression, we further analyzed the correlation between RSK4 protein expression and clinical pathological features of colorectal cancers. The results demonstrated that there were no statistically significant differences in the age, blood CEA, CA199 and tumors t-staging. Unfortunately, we have not been able to identify a correlation between RSK4 expression and M classification in patients, possibly due to the limited sample size. Interestingly and for the first time, we have found that the expression profiles of RSK4 were strongly correlated with clinical pathologic classifications tumor differentiation degree (G), lymph node metastasis (N) and clinical staging, respectively. These observations suggested a correlation between decreased expression of RSK4 and clinical progression in colorectal cancer. RSK4 expression is reversely correlated with clinical pathologic classifications of colorectal cancer, which, to the best of our knowledge, has not been reported thus far. Indeed, further analyses of primary cancer tissues have revealed patients with colorectal cancer with lower RSK4 expression had lower survival rates than that high expression of RSK4. In light of the involvement of RSK4 in the development of colorectal cancer, we further analyzed the patients’ survival in terms of RSK4 expression in adjacent noncancerous tissues. As shown in Figure 2B, examining adjacent noncancerous tissues demonstrated that the overall survival time in patients expressing higher levels of RSK4 was much longer than patients expressing lower levels of RSK4. These indicated that there was significant clinical value in assessing the prognosis of colorectal cancer patients with RSK4 expression. Furthermore, we have probed the Overall survival curves stratified by RSK4 levels according to T, N, M classifications, clinical staging, and CEA, CA199. We found that colorectal cancer with T1 + T2 and T3 + T4 concomitant with RSK4 low expression had a significantly lower 5-year survival rate than RSK4 high expression. Similarly, colorectal cancer with or no lymph node metastasis and RSK4 low expression, had a significantly lower 5-year survival rate than colorectal cancer with or no lymph node metastasis in the high expression of RSK4. In M0 classification panel, patients with high RSK4 expression show much better overall survival. In M1 classification panel, the patients’ overall survival is not significantly different between the two groups. Most importantly, stage (I-II and III-IV) colorectal cancers concomitant with low RSK4 expression had the lowest 5-year survival rate, which was significantly lower than those high RSK4 expression. However, analyzing corresponding tumor grade (G0 and G1 + G2) suggested that expressed levels of RSK4 seem to also have an impact on patients’ survival. Interestingly, high RSK4 expression revealed the favorable impact on outcome irrelevant of the presence of CEA and CA199 by subset analysis. Overall, it remained true that the expression levels of RSK4 are strongly correlated with patients’ survival even after stratifying the patients based upon their clinic pathologic classifications. It should be emphasized that poor survival in patients with lower expression of RSK4 in colorectal cancer. Relevant to survival, we have also demonstrated a significantly increased risk of cancer-related death in patients with low expression of RSK4 using univariate and multivariate Cox regression analysis. Our findings, strongly suggest that RSK4 may be a novel, yet important, prognostic marker for colorectal cancer patients.
In summary, taking our clinical data and data from basic studies together we have been able to demonstrate that (1) the decreased expression of RSK4 is incremental depending upon the magnitude of cancer progression; (2) the expression patterns of RSK4 are correlated with clinic pathologic classifications of N, tumor grade and clinical staging, and (3) patients with diminished expression of RSK4 show poor overall survival. These findings strongly suggest that insufficient expression of RSK4 may serve as a risk factor predictive of prognosis of colorectal cancer patients following appropriate therapy.
Disclosure of conflict of interest
None.
References
- 1.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 2.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–44. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SW, Erikson E, Blenis J, Maller JL, Erikson RL. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc Natl Acad Sci U S A. 1988;85:3377–81. doi: 10.1073/pnas.85.10.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher TL, Blenis J. Evidence for two catalytically active kinase domains in pp90rsk. Mol Cell Biol. 1996;16:1212–9. doi: 10.1128/mcb.16.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012;441:553–69. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 6.Lara R, Mauri FA, Taylor H, Derua R, Shia A, Gray C, Nicols A, Shiner RJ, Schofield E, Bates PA, Waelkens E, Dallman M, Lamb J, Zicha D, Downward J, Seckl MJ, Pardo OE. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene. 2011;30:3513–21. doi: 10.1038/onc.2011.61. [DOI] [PubMed] [Google Scholar]
- 7.Saha M, Carriere A, Cheerathodi M, Zhang X, Lavoie G, Rush J, Roux PP, Ballif BA. RSK phosphorylates SOS1 creating 14-3-3-docking sites and negatively regulating MAPK activation. Biochem J. 2012;447:159–66. doi: 10.1042/BJ20120938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Dodds P, Emilion G, Mungall AJ, Dunham I, Beck S, Wells RS, Charnock FM, Ganesan TS. The human homologue of unc-93 maps to chromosome 6q27 - characterisation and analysis in sporadic epithelial ovarian cancer. BMC Genet. 2002;3:20. doi: 10.1186/1471-2156-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yntema HG, van den Helm B, Kissing J, van Duijnhoven G, Poppelaars F, Chelly J, Moraine C, Fryns JP, Hamel BC, Heilbronner H, Pander HJ, Brunner HG, Ropers HH, Cremers FP, van Bokhoven H. A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics. 1999;62:332–43. doi: 10.1006/geno.1999.6004. [DOI] [PubMed] [Google Scholar]
- 10.Dummler BA, Hauge C, Silber J, Yntema HG, Kruse LS, Kofoed B, Hemmings BA, Alessi DR, Frödin M. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J Biol Chem. 2005;280:13304–14. doi: 10.1074/jbc.M408194200. [DOI] [PubMed] [Google Scholar]
- 11.ME LL, Vidal F, Gallardo D, Diaz-Fuertes M, Rojo F, Cuatrecasas M, López-Vicente L, Kondoh H, Blanco C, Carnero A, Ramón y Cajal S. New p53 related genes in human tumors: significant downregulation in colon and lung carcinomas. Oncol Rep. 2006;16:603–8. doi: 10.3892/or.16.3.603. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, Jin H, To KF, Chan FK, Sung JJ. Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology. 2009;136:640–51. e1. doi: 10.1053/j.gastro.2008.10.050. [DOI] [PubMed] [Google Scholar]
- 13.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 14.Huang CS, Lin JK, Wang LW, Liang WY, Lin CC, Lan YT, Wang HS, Yang SH, Jiang JK, Chen WS, Lin TC, Chang SC. Assessment of the value of carcinoembryonic antigen reduction ratio as a prognosis factor in rectal cancer. Am J Surg. 2014;208:99–105. doi: 10.1016/j.amjsurg.2013.08.054. [DOI] [PubMed] [Google Scholar]
- 15.Pengjun Z, Xinyu W, Feng G, Xinxin D, Yulan L, Juan L, Xingwang J, Zhennan D, Yaping T. Multiplexed cytokine profiling of serum for detection of colorectal cancer. Future Oncol. 2013;9:1017–27. doi: 10.2217/fon.13.71. [DOI] [PubMed] [Google Scholar]
- 16.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 17.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, Lhermitte B, Olshen AB, Wiedenmann B, Cantley LC, Gray JW, Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, Rodermond H, van der Heijden M, van Noesel CJ, Tuynman JB, Dekker E, Markowetz F, Medema JP, Vermeulen L. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19:614–8. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Vicente L, Pons B, Coch L, Teixidó C, Hernández-Losa J, Armengol G, Ramon Y Cajal S. RSK4 inhibition results in bypass of stress-induced and oncogene-induced senescence. Carcinogenesis. 2011;32:470–6. doi: 10.1093/carcin/bgr003. [DOI] [PubMed] [Google Scholar]
- 20.Thakur A, Rahman KW, Wu J, Bollig A, Biliran H, Lin X, Nassar H, Grignon DJ, Sarkar FH, Liao JD. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res. 2007;5:171–81. doi: 10.1158/1541-7786.MCR-06-0071. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Vicente L, Armengol G, Pons B, Coch L, Argelaguet E, Lleonart M, Hernández-Losa J, de Torres I, Ramon y Cajal S. Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clin Cancer Res. 2009;15:4546–53. doi: 10.1158/1078-0432.CCR-08-3159. [DOI] [PubMed] [Google Scholar]
- 22.Myers AP, Corson LB, Rossant J, Baker JC. Characterization of mouse Rsk4 as an inhibitor of fibroblast growth factor-RAS-extracellular signal-regulated kinase signaling. Mol Cell Biol. 2004;24:4255–66. doi: 10.1128/MCB.24.10.4255-4266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thakur A, Sun Y, Bollig A, Wu J, Biliran H, Banerjee S, Sarkar FH, Liao DJ. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res. 2008;14:4427–36. doi: 10.1158/1078-0432.CCR-08-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]


