Abstract
Background: Manual evaluation of somatostatin receptor (SSTR) immunohistochemistry (IHC) is a time-consuming and cost-intensive procedure. Aim of the study was to compare manual evaluation of SSTR subtype IHC to an automated software-based analysis, and to in-vivo imaging by SSTR-based PET/CT. Methods: We examined 25 gastroenteropancreatic neuroendocrine tumor (GEP-NET) patients and correlated their in-vivo SSTR-PET/CT data (determined by the standardized uptake values SUVmax,-mean) with the corresponding ex-vivo IHC data of SSTR subtype (1, 2A, 4, 5) expression. Exactly the same lesions were imaged by PET/CT, resected and analyzed by IHC in each patient. After manual evaluation, the IHC slides were digitized and automatically evaluated for SSTR expression by Definiens XD software. A virtual IHC score “BB1” was created for comparing the manual and automated analysis of SSTR expression. Results: BB1 showed a significant correlation with the corresponding conventionally determined Her2/neu score of the SSTR-subtypes 2A (rs: 0.57), 4 (rs: 0.44) and 5 (rs: 0.43). BB1 of SSTR2A also significantly correlated with the SUVmax (rs: 0.41) and the SUVmean (rs: 0.50). Likewise, a significant correlation was seen between the conventionally evaluated SSTR2A status and the SUVmax (rs: 0.42) and SUVmean (rs: 0.62).Conclusion: Our data demonstrate that the evaluation of the SSTR status by automated analysis (BB1 score), using digitized histopathology slides (“virtual microscopy”), corresponds well with the SSTR2A, 4 and 5 expression as determined by conventional manual histopathology. The BB1 score also exhibited a significant association to the SSTR-PET/CT data in accordance with the high affinity profile of the SSTR analogues used for imaging.
Keywords: Somatostatin receptor, molecular imaging, immunohistochemistry, neuroendocrine tumor, PET/CT
Introduction
Neuroendocrine tumors (NET) are an extremely heterogeneous group of neoplasms. Therefore, the diagnostics and therapy are significantly influenced by various biological properties, such as degree of differentiation (grading), tumor proliferation (e.g., Ki-67 index) and staging [1-3].
Somatostatin receptors (SSTR) are expressed in nearly all neuroendocrine tumors (NET), especially in gastroenteropancreatic NET (GEP-NET). There are 5 human SSTR subtypes known (SSTR1, 2A, 3, 4, 5), which in a variable pattern and density are all expressed in GEP-NET. In GEP-NET, the diagnostics and treatment of each individual patient is based on the SSTR expression profile of the respective tumor [4]. SSTR serve as the molecular basis for high sensitive molecular imaging procedures (PET/CT) as well as therapy targets for long-acting somatostatin receptor analogues and peptide receptor-radionuclide therapy (PRRT) [5,6]. The maximum - and mean standardized uptake values (SUVmax, SUVmean) in SSTR-based PET are directly associated to the used peptide. These synthetic SSTR-analogues are characterized by a different SSTR-affinity profile to each SSTR-subtype [7]. As shown by Ocak et al. the clinical images with different peptides gave comparable results but the SUVvalues differ significantly [8]. The SSTR subtypes distribution and frequency of each tumor lesion are directly connected to uptake in PET and further medical treatment.
Immunohistochemistry is currently the routine standard method for assessing the extent of SSTR subtypes expression in NET cells. Evaluation of the amount of expression is done visually by means of different semiquantitative scoring systems, as e.g. the human epidermal growth factor receptor 2 (HER2/neu) score and the immunoreactive score (IRS). However, still none of this scoring systems has become established as shown by our group in previous studies, making it difficult to compare different immunohistochemical studies [9]. Immunohistochemical evaluation is a time-consuming, personnel- and cost-intensive process. Furthermore, the evaluation is semiquantitative, poorly standardised and inter-observer biased. Since few years, automated cellular imaging systems are available to improve histopathological investigations [10]. Automatic measurement of cell proliferation and immunohistochemical markers, automated vessel identification in immunohistochemical sections, automated in-situ hybridization (ISH) and semiautomated image analysis of tissue microarrays (TMAs) are performed in workflows already [11-15]. Studies have shown that automated procedures are objective, fast and reproducible with high levels of accuracy and strong correlations of results between manually and automated analysis procedures [16]. Particularly in large clinical trials they have a proven precision, are less observer dependent and have shown a better reproducibility of data than manual methods [17,18].
Especially for the diagnosis, therapy and prognosis of NET, it is essential to generate objective and quantitative, but also concordant data with the different methods used. However, the extent of correlation between the different modalities used for patient diagnosis and individual therapy has not been quantified so far. In order to achieve such quantifications, we correlated different data sets from different modalities (PET/CT and histopathological data) from the same patient. We quantified tissue morphology, staining distribution and intensity of staining, using both automated image analysis and manually performed Her2/neu and IRS scoring. In a next step, we computed correlations between image analysis results and manual Her2/neu and IRS scoring. In the present study, for the first time, we present correlations between molecular imaging of SSTR using PET/CT data (SUV max and SUVmean) and immunohistochemical data of SSTR (1, 2A, 4, 5) expression in comparison to an automated image analysis of digitized slides by Definiens Tissue Studio software.
Patients and methods
Twenty-five neuroendocrine tumor patients (Table 1) were routinely advised to a surgical procedure by an interdisciplinary tumor board.
Table 1.
Patients characteristic
| Number | Age | Sex | Primary tumor | Grading | Origin of lesions |
|---|---|---|---|---|---|
| 1 | 69 | male | CUP | 2 | I Liver - MTS |
| 2 | 71 | female | Duodenum | 2 | I Peritoneal - MTS |
| 3 | 43 | male | Appendix | 3 | I Appendix - PT |
| II Peritoneal - MTS | |||||
| 4 | 70 | male | Ileum | 2 | I Liver - MTS |
| 5 | 43 | female | Ileum | 2 | I Lymph node - MTS |
| 6 | 73 | male | Ileum | 2 | I Liver - MTS |
| 7 | 51 | male | Ileum | 2 | I Ileum - PT |
| 8 | 33 | female | Pancreas | 2 | I Pancreas - PT |
| 9 | 51 | male | Pancreas | 2 | I Meso - MTS |
| 10 | 57 | female | Pancreas | 2 | I Liver - MTS |
| 11 | 48 | female | Pancreas | 2 | I Liver - MTS |
| 12 | 82 | male | Ileum | 2 | I Meso - MTS |
| II Meso - MTS | |||||
| III Ileum - PT | |||||
| IV Peritoneal - MTS | |||||
| 13 | 49 | male | Stomach | 2 | I Liver - MTS |
| 14 | 65 | female | Pancreas | 2 | I Liver - MTS |
| 15 | 59 | female | Pancreas | 2 | I Pancreas - PT |
| 16 | 54 | male | Ileum | 1 | I Liver - MTS |
| II Ileum - PT | |||||
| 17 | 71 | male | Pancreas | 2 | I Small-intestine - MTS |
| 18 | 52 | female | Ileum | 2 | I Peritoneal - MTS |
| 19 | 77 | male | Pancreas | 2 | I Stomach - MTS |
| 20 | 50 | male | Ileum | 1 | I Meso - MTS |
| II Lymph node - MTS | |||||
| 21 | 53 | female | Ileum | 1 | I Ileum - PT |
| 22 | 73 | male | Pancreas | 2 | I Peritoneal - MTS |
| 23 | 59 | male | Pancreas | 1 | I Thyroid - MTS |
| 24 | 45 | male | Stomach | 3 | I Stomach - PT |
| 25 | 68 | female | Ileum | 2 | I Liver - MTS |
Abbreviations: CUP - carcinoma of unknown primary; PT - primary tumor; MTS - metastases.
All patients signed an informed consent and the study was approved by a local ethics committee. Retrospectively, we obtained PET/CT imaging data from these 25 randomly selected neuroendocrine tumor patients. The patients had received an injection of Ga-68 DOTANOC (n = 17) or DOTATATE (n = 8), which are Gallium-68-radiolabeled analogues of somatostatin and are used in conjunction with PET to image neuroendocrine tumors and their metastases (Figure 1). The PET/CTs were routinely performed during the staging process, and Standardized Uptake Values (SUV) as SUVmax and SUVmean of each single tumor lesion were calculated. All patients were examined using a dual-modality PET/CT tomograph (Biograph LSO Duo; Siemens Medical Solutions, U.S.A), as described previously [19]. Patients were then operated and each single tumor lesion (n = 31) was removed, marked and correlated to the lesions, that had already been detected on Ga-68 SSTR PET/CT. Then, immunohistochemical analysis of tumor tissue was performed with specific polyclonal and monoclonal antibodies. The detection of SSTR-subtypes was performed using the labeled streptavidin-biotin-method (LSAB) and counterstaining was done with haematoxylin. The monoclonal rabbit antibody used for detection of SSTR2A was custom produced by Epitomics, Burlingame, CA (USA), and the polyclonal rabbit antibodies for detection of SSTR1, 4 and 5 by Gramsch Laboratories, Schwabhausen (Germany). The rabbit antibodies were generated against the respective carboxyl-terminal tail of each human SSTR-subtype.
Figure 1.
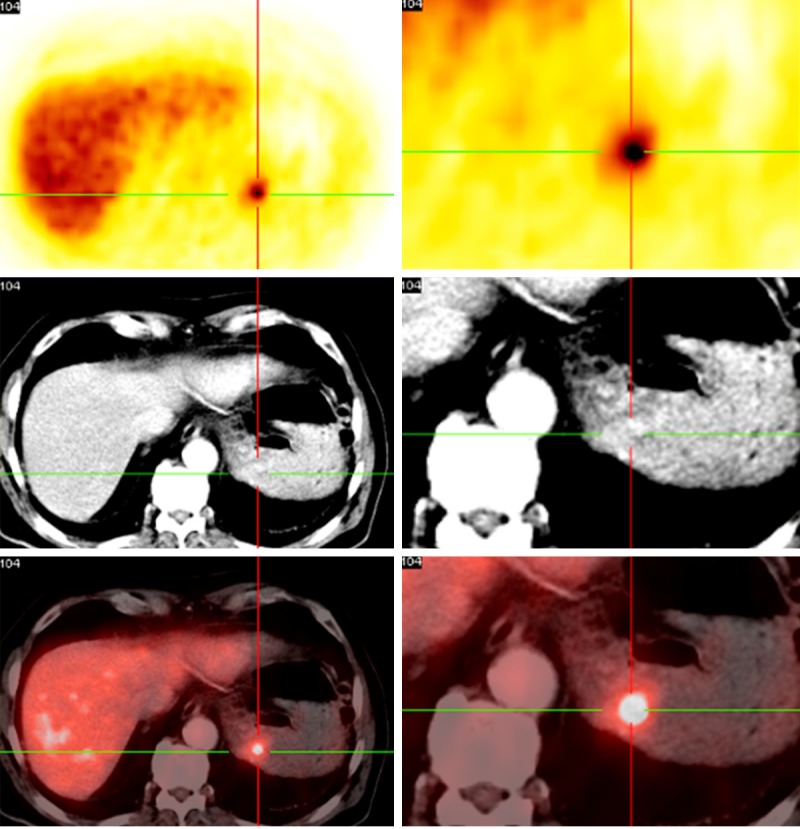
68Ga-DOTA-NOC PET/CT demonstrates a gastric metastasis of a neuroendocrine pancreatic tumor. The metastasis corresponds with the immunohistochemical image in Figure 2.
The analysis of the stained sections was done with light microscopy according to the immunoreactive score (IRS) by Remmele and Stegner and to the Her2/neu DAKO scoring system as previously described by our group [19,20] (Table 2).
Table 2.
IRS and HER2-scoring system
| Percentage of positive cells | X Intensity of Staining | = IRS (0-12) |
|
| ||
| 0 = no positive cells | 0 = no colour reaction | 0-1 = negative |
| 1 ≤ 10% of positive cells | 1 = mild reaction | 2-3 = mild |
| 2 = 10-50% positive cells | 2 = moderate reaction | 4-8 = moderate |
| 3 = 51-80% positive cells | 3 = intense reaction | 9-12 = strong positive |
| 4 ≥ 80% positive cells | ||
|
| ||
| Her2/neu Score | Reaction Format | Impression |
|
| ||
| 0 | No or less than 10% cells | Negative |
| 1+ | > 10% cells with minimal staining intensity | Negative |
| 2+ | > 10% cells with moderate staining intensity | Mildly positive |
| 3+ | > 10% cells with strong staining intensity | Strongly positive |
In order to minimize the inter-observer bias, the immunohistochemical analysis was performed by two independent investigators on each tissue section. All immunohistochemically stained slides were digitized using an VS120 slide scanner (Olympus® U.S.A). Then, Definiens software (Definiens Developer XD and Definiens Tissue Studio®; Munich, Germany) was used for the image analysis of the virtual tissue slides. In a first step a simple image analysis solution was developed and was applied on the virtual slides by using Definiens Developer XD. We used low magnification (5x) of the virtual slides. This first image analysis solution employs chessboard segmentation and threshold classification algorithms for the separation of tissue from non-tissue regions on the slide, assigns tissue regions to different semantic classes according to their stain intensity in “positive”, “strong positive” and “negative”. “Positive” are stained and “negative” are non-stained tissue regions. Areas of these regions and relations between them are calculated automatically. Such relations are then correlated to Her2- and IRS-score as calculated by the pathologist. In a next step we used Definiens Tissue Studio® (Munich, Germany) for the automated cell-by-cell image analysis and quantification of the virtual slides. Definiens Tissue Studio® identifies regions of interest (ROI) automatically or with learn-by-example approaches, operates through a simple, intuitive user interface and offers unlimited throughput with parallel batch processing. This image analysis solution, separates tumor from non-tumor regions, calculates SSTR (1, 2A, 4, 5) staining intensity in the tumor regions, calculates areas of tumor and non-tumor regions, calculates and quantifies morphological properties of single cells and single cell compartments like nuclei (area, symmetry, staining intensity) and determines relations between tumor and non-tumor regions. Using the image analysis results, the calculation of user defined features in analogy to the HER2/neu score and to the IRS was done.
Similar to the immunoreactive score of Remmele and Stegner, we developed a virtual score “Bad Berka Score 1 (BB1)” which is calculated by the percentage of cells with high marker intensity multiplied with the mean immunohistochemical marker intensity (BB1 = % cell high marker intensity x mean immunohistochemical marker intensity). In a next step, we used data mining methods in order to find correlations between all the previous data of the same and of different modalities for all patients. 124 immunohistochemical slides from 25 patients were digitized (31 each of the SSTR-subtypes 1, 2A, 4, 5). The SSTR3 slides were not digitized due to logistical problems. From these 124 slides, 23 SSTR1, 23 SSTR2A, 25 SSTR4 and 22 SSTR5 slides were used as virtual slides for the present study. 31 slides of SSTR5 (stained by a new monoclonal anti-SSTR5 antibody) were taken out. All slides were from the same patients who had received the PET/CT scan before (PET/CT data shown in Table 3). Most of the patients had more than one tissue sample, sometimes up to five specimens (primary tumor and metastases). We employed Definiens AG Image Analysis (Definiens Tissue Studio®) for the automated analysis of these virtual slides (Figure 2). SSTR-stained regions on the slides were assigned as positive and tissue regions without SSTR expression as negative. Regions of interest (ROIs), color intensities, numbers of objects like nuclei and cells and their morphological features among other properties were calculated. Data mining methods were applied in order to quantify correlations between image analysis results of SSTR-stained tissue and the corresponding IRS and HER2/neu scoring data, which have been manually evaluated.
Table 3.
Patients data for SSTR2A results and PET/CT data
| Patient no./lesion, slide | IRS SSTR2A | Her2/neu SSTR2A | BB1 SSTR2A | SUVmax | SUVmean |
|---|---|---|---|---|---|
| 1/1 | 6 | 3+ | 0.4291 | 5.7 | 3.4 |
| 2/1 | 3 | 1+ | 9.2348 | 8.2 | 2.0 |
| 3/1 | 3 | 1+ | N/D | N/D | N/D |
| 3/2 | 4 | 1+ | 0.0906 | 4.3 | 2.1 |
| 4/1 | 12 | 3+ | 1.0434 | 13.6 | 8.1 |
| 5/1 | 9 | 3+ | N/D | 6.8 | 3.9 |
| 6/1 | 12 | 3+ | N/D | 33.7 | 21.0 |
| 7/1 | 4.5 | 3+ | 8.9353 | 8.3 | 4.9 |
| 8/1 | 12 | 3+ | 90.511 | 25.9 | 15.8 |
| 9/1 | 4 | 1+ | 0.8317 | 5.7 | 2.9 |
| 10/1 | 6 | 3+ | 4.5260 | 10.6 | 6.9 |
| 11/1 | 12 | 3+ | 0.9260 | 5.6 | 4.6 |
| 12/1 | 4 | 1+ | N/D | N/D | N/D |
| 12/2 | 12 | 3+ | N/D | 4.2 | 2.7 |
| 12/3 | 12 | 3+ | N/D | N/D | N/D |
| 12/4 | 12 | 3+ | N/D | 4.3 | 2.8 |
| 13/1 | 6 | 3+ | 1.0818 | 6.4 | 4.0 |
| 14/1 | 6 | 3+ | 4.9974 | 6.2 | 4.9 |
| 15/1 | 6 | 3+ | 10.024 | 17.4 | 10.0 |
| 16/1 | 4.5 | 3+ | 0.1238 | 14.8 | 9.5 |
| 16/2 | 6 | 3+ | 119.52 | 6.5 | 3.6 |
| 17/1 | 12 | 3+ | N/D | 10.5 | 6.5 |
| 18/1 | 12 | 3+ | 0.8516 | 12.6 | N/D |
| 19/1 | 12 | 3+ | 15.176 | 9.4 | 5.5 |
| 20/1 | 9 | 3+ | 4.5053 | 13.5 | 9.6 |
| 20/2 | 12 | 3+ | 14.520 | 9.6 | 5.9 |
| 21/1 | 5 | 3+ | 7.2947 | 14.6 | 9.0 |
| 22/1 | 12 | 3+ | 1.9985 | 12.4 | 9.1 |
| 23/1 | 12 | 3+ | 1.8545 | 8.4 | 4.8 |
| 24/1 | 3 | 1+ | 0.2441 | 4.9 | 2.8 |
| 25/1 | 8 | 2+ | 2.7681 | 6.7 | 2.0 |
Abbreviations: N/D - no data available; IRS - Immunoreactive score; Her2/neu - Her2/neu-score; BB1 - digital immunohistochemical score “Bad Berka 1”; SUVmax,-mean standardized uptake value maximum, -mean.
Figure 2.
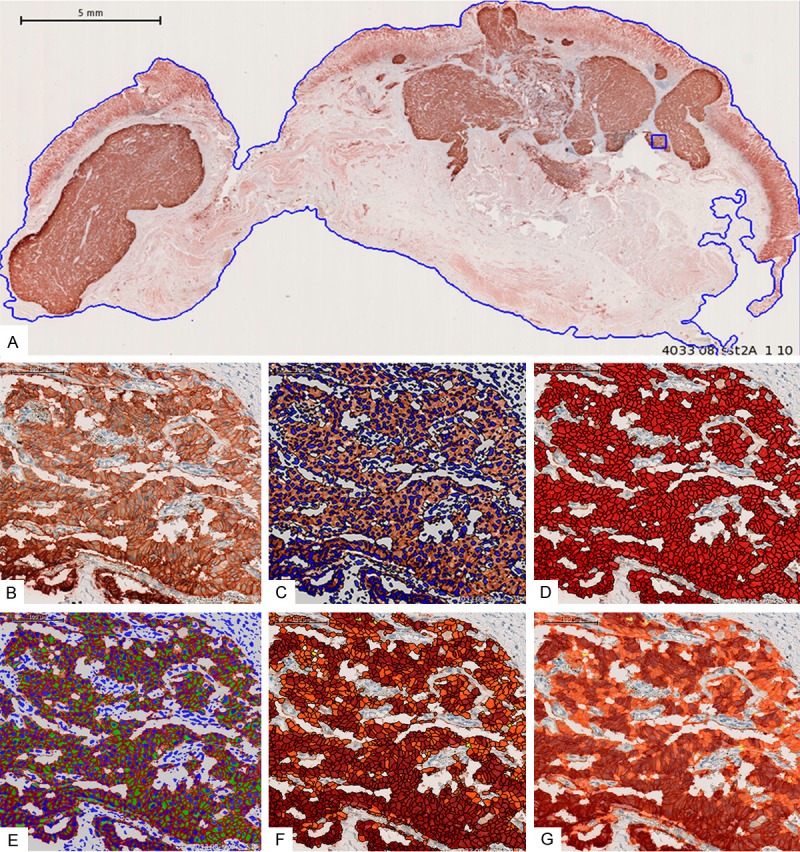
A: Digitized immunohistochemical slide from a gastric metastasis of a neuroendocrine pancreatic tumor showing the selected region of interest (ROI; blue line). The section was stained with a monoclonal antibody against the SSTR2A and displays a strong SSTR2A expression in the tumor cells (brown color; magnification 100 ×). B: Detail of A, showing the predominant localization of the SSTR2A at the plasma membrane of the tumor cells; Initialization of cellular analysis (magnification 600 ×). C: Automated nucleus segmentation (magnification 600 ×). D: Automated cell segmentation (magnification 600 ×). E: Automated cell membrane classification (SSTR2A is a membrane-bound receptor) (magnification 600 ×). F: Automated cell classification according to intensity of cell membrane staining (magnification 600 ×). G: Automated cell classification (transparent) according to plasma membrane staining (magnification 600 ×).
Definiens image analysis technology
The Definiens Software we used for image analysis is based on the Definiens Cognition Network Technology (CNT). The technology is object-oriented, multi-scale, context-driven and knowledge-based [21,22]. Images are interpreted on the properties of networked image objects, which results in numerous advantages. This approach enables users to bring in detailed expert knowledge and enables complex analyses to be performed with unprecedented accuracy, even on poor quality data or for structures exhibiting heterogeneous properties or variable phenotypes. Extracted structures are the basis for detailed morphometric, structural and relational measurements which can be exported for each individual structure. These data can be used for decision support or correlated against experimental or molecular data. Especially the Definiens Tissue Studio® software enables the user to apply ready to use image analysis solutions in a butch processing [23,24]. There are several of such solutions already. These solutions are developed in order to analyze single objects. Single objects are for instance single and their compartments. These image analysis solutions segment and classify individual cells and cell compartments like nuclei, cytoplasm and membranes. So it is possible to classify each individual cell based on the stain of the individual cell membrane, or the stain intensity of the cell cytoplasm, or of the cell nucleus. Visual inspection of the virtual slides showed strong expression of SSTR 1, 4 and 5 in the cytoplasm of each individual cell (Figure 3). In contrast to SSTR2A, which showed a strong membrane-bound receptor expression (Figure 2B). Therefore we used the Tissue Studio® Solutions for classifying each individual cell according to the SSTR expression in the cytoplasm for all these stains. For the SSTR2A we applied the Tissue Studio® Solution, which classifies cells according on the stain in the cell membrane. Both solutions allow the automatically calculation of stain intensity und of morphological properties of each individual object. The results are automatically arranged and are available for further statistical calculations.
Figure 3.
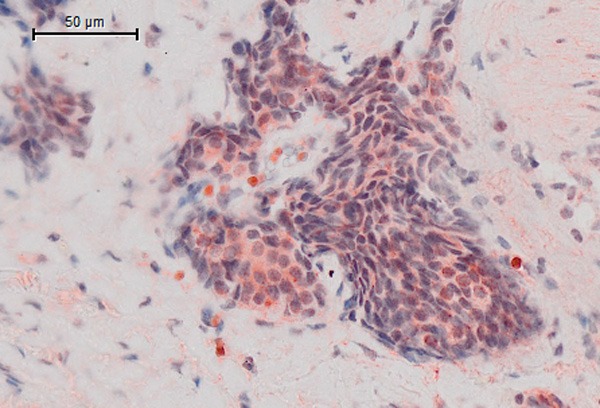
Cytoplasmatic staining of SSTR.
Statistical analysis
Statistical analysis was performed using SigmaPlot for Windows Version 11.0, Build 11.0.0.75, Systat Software 2008. Spearman’s rank order correlation (rs) was used to investigate correlations between automated and manually examined immunohistochemical image analysis results (IRS and Her2/neu scoring) and PET/CT uptake values. A P-value ≤ 0.05 was considered statistically significant.
Results
The SSTR-subtypes 2A and 5 were mostly confined to the plasma membrane, whereas SSTR1 and SSTR4 were predominantly located in the cytoplasm of the tumor cells. All tumor slides were characterized by a remarkable heterogeneity of staining, both within and between the different samples of the same patient.
Automated analysis of SSTR
The virtual BB1 score was significantly positively correlated (range rs: 0.43-0.57) to the corresponding manually evaluated IRS and Her2/neu score of the SSTR-subtypes 2A and 5 (Table 4). BB1 of SSTR4 only displayed a significant positive correlation with Her2/neu (rs: 0.44; P = 0.028), whereas no significant correlation was seen to IRS (rs: 0.25; P = 0.229). BB1 of SSTR2A exhibited a significant correlation with the SUVmax (rs: 0.41; P = 0.049) and the SUVmean (rs: 0.50; P = 0.019) of the PET/CT. With all other subtypes, SSTR1, SSTR4 and SSTR5, the correlation factor was below 0.1 and no significant correlation was observed (Table 4). Separated analysis of patients imaged by DOTA-NOC (n = 16) and DOTA-TATE (n = 8) peptides demonstrated a significant correlation of BB1 of SSTR2A to SUVmax (rs: 0.58; P = 0.018) and SUVmean (rs: 0.52; P = 0.039) whereas no significant correlation was detectable for imaging with DOTA-TATE (Supplementary Tables 1 and 2).
Table 4.
Digital analysis of BB1 in correlation to PET/CT- and manually evaluated immunohistochemical scores
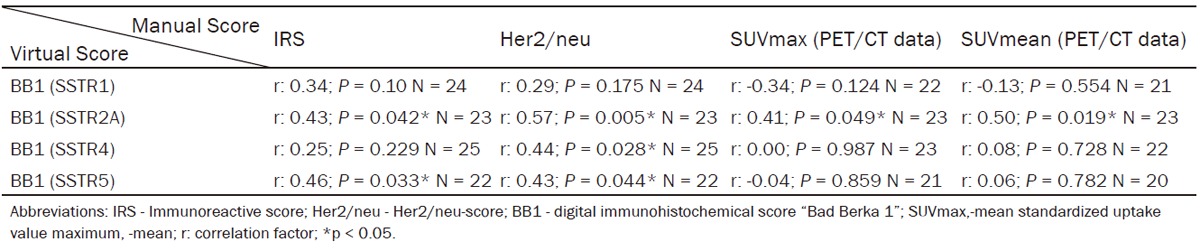
|
Manually evaluated SSTR data
The SSTR2A expression as evaluated manually by means of the Her2/neu-score was significantly positively correlated to the SUVmax (rs: 0.42; P = 0.028) and SUVmean (rs: 0.62; P < 0.001). In contrast, the SSTR2A expression as determined by the IRS score exhibited no significant correlation to SUVmax (rs: 0.24; P = 0.224) and SUVmean (rs: 0.34; P = 0.081). This was also the case for all other SSTR subtypes (SSTR1, 4, 5) which did not show any significant association to the PET/CT values.
PET/CT data
SUVmax data could be detected in 28/31 lesions with a range from 4.2 to 33.7 (median 8.35). SUVmean data were determined in 27/31 lesions (range 2.0 to 21.0, median 4.9). Some small lesions were not measured or even not detected by molecular imaging but were histologically proven as small metastases.
Discussion
Since few years automated cellular imaging systems are available for improvement of histopathological investigations [10]. In many studies, automatic measurements of cell proliferation and of immunohistochemical markers, automated vessel identification in immunohistochemical sections, automated in-situ hybridization (ISH) and semiautomated image analysis of tissue microarrays (TMAs), have been proven to be comparable to manual analysis [11-15]. Additionally, these automated procedures have been shown to be cost-effective and time saving [16]. Particularly in large clinical trials they have a proven precision, are less observer dependent and have shown a better reproducibility of data in comparison to manual ones [17,18].
However, although many automatic procedures are used during routine histopathology already, quantitative correlations between automated and manually evaluated modalities used for neuroendocrine tumor patients are not available so far.
Previous studies have demonstrated the significant positive association between the SUV of SSTR-based PET/CT and in-vitro immunohistochemical analysis [25,26]. In the present study, we were also able to detect a significant positive correlation between the SSTR2A expression as evaluated manually according to the Her2/neu score and the PET/CT SUVmax and -mean. However, SSTR2A expression as determined by IRS did not show any significant association to PET/CT data. This latter result is in contradiction to the studies of Miederer and Kaemmerer et al. who were able to demonstrate a strong correlation between SUV in PET/CT and SSTR2A expression [19,26]. Both of these studies reported data from patients who were injected and analyzed by only one SSTR-analogue (DOTA-TOC or DOTA-NOC), whereas in the present investigation PET/CT data were obtained with two different peptides (DOTA-TATE and DOTA-NOC), displaying quite different SSTR-affinities, were included. DOTA-TATE showed a 10 times higher affinity to SSTR2A than DOTA-NOC [7]. This could be one explanation for our results and for the discrepancy between the two scoring systems.
BB1 was created as a virtual immunohistochemical score. Our data show a significant positive correlation between the BB1 of the SSTR2A, the manually evaluated SSTR2A expression and the SUVmax and SUVmean of the PET/CT. The BB1s of the other SSTRs, in contrast, displayed no correlation to the PET/CT data. With regard to the somatostatin analogues used for the PET/CT measurements in the present investigation, these results seem to be reasonable, because the affinity of these peptides to the SSTR2A is much higher than to all other SSTR subtypes [27]. In the present investigation correlation factors ranging from 0.25 to 0.57 were observed between the BB1 and the corresponding manual data. These correlations were significant for the SSTR2A and the SSTR5 expression both with respect to the IRS and to the Her2/neu scoring system. Concerning the SSTR4 expression, a significant correlation between the BB1 and the respective manual data was seen for the HER2/neu scoring system only.
In clinical practice, in vivo molecular imaging by somatostatin-analogue-based PET/CT has become the golden standard for diagnostics of GEP-NET. To further optimize molecular imaging and to save financial and staff resources, it is useful to evaluate the SSTR subtype status of the tumor beforehand. Without a molecular sstr targets a molecular sstr based imaging procedure is useless. For this, the immunohistochemical evaluation of the SSTR status on formalin-fixed, paraffin-embedded resected tumor specimens has become the method of choice [28,29]. It is cheaper and faster than autoradiography, does not require radioactive material and can be done during routine histopathological examinations. Our data provide further clinical evidence for the feasibility of an automated evaluation of immunohistochemical stainings e.g. by means of the Definiens software tools [17,18]. Thus, in future, the immunohistochemical slides can be digitized and analyzed by an automated image analysis procedure to evaluate the SSTR subtype profile prior to further diagnostic and therapeutic recommendations.
Conclusion
In summary, it can be concluded that the evaluation of the main interesting SSTR subtypes (2A and 5) of a given tumor by an automatic SSTR analysis is a reliable method to further improve diagnostics and the assignment of the patients to a stratified therapy with one of the different somatostatin analogues available for PRRT or pharmacological treatment.
Disclosure of conflict of interest
Prof. G. Binnig is founder and Dr. M. Athelogou employee of the company Definiens AG, Munich, Germany. Prof. G. Binning developed Definiens Cognition Network Technology®. All other authors declare no conflicts of interest regarding this manuscript.
Supporting Information
References
- 1.Kloppel G, Anlauf M. Epidemiology, tumour biology and histopathological classification of neuroendocrine tumours of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2005;19:507–517. doi: 10.1016/j.bpg.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Kloppel G, Rindi G, Anlauf M, Perren A, Komminoth P. Site-specific biology and pathology of gastroenteropancreatic neuroendocrine tumors. Virchows Arch. 2007;451(Suppl 1):S9–27. doi: 10.1007/s00428-007-0461-0. [DOI] [PubMed] [Google Scholar]
- 3.Anlauf M. Neuroendocrine neoplasms of the gastroenteropancreatic system: pathology and classification. Horm Metab Res. 2011;43:825–831. doi: 10.1055/s-0031-1291307. [DOI] [PubMed] [Google Scholar]
- 4.Reubi JC. Somatostatin and other Peptide receptors as tools for tumor diagnosis and treatment. Neuroendocrinology. 2004;80(Suppl 1):51–56. doi: 10.1159/000080742. [DOI] [PubMed] [Google Scholar]
- 5.Baum RP, Kulkarni HR. THERANOSTICS: From Molecular Imaging Using Ga-68 Labeled Tracers and PET/CT to Personalized Radionuclide Therapy - The Bad Berka Experience. Theranostics. 2012;2:437–447. doi: 10.7150/thno.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baum RP, Kulkarni HR, Carreras C. Peptides and receptors in image-guided therapy: theranostics for neuroendocrine neoplasms. Semin Nucl Med. 2012;42:190–207. doi: 10.1053/j.semnuclmed.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Antunes P, Ginj M, Zhang H, Waser B, Baum RP, Reubi JC, Maecke H. Are radiogallium-labelled DOTA-conjugated somatostatin analogues superior to those labelled with other radiometals? Eur J Nucl Med Mol Imaging. 2007;34:982–993. doi: 10.1007/s00259-006-0317-x. [DOI] [PubMed] [Google Scholar]
- 8.Ocak M, Demirci E, Kabasakal L, Aygun A, Tutar RO, Araman A, Kanmaz B. Evaluation and comparison of Ga-68 DOTA-TATE and Ga-68 DOTA-NOC PET/CT imaging in well-differentiated thyroid cancer. Nucl Med Commun. 2013;34:1084–1089. doi: 10.1097/MNM.0b013e328364eaab. [DOI] [PubMed] [Google Scholar]
- 9.Kaemmerer D, Peter L, Lupp A, Schulz S, Sanger J, Baum RP, Prasad V, Hommann M. Comparing of IRS and Her2 as immunohistochemical scoring schemes in gastroenteropancreatic neuroendocrine tumors. Int J Clin Exp Pathol. 2012;5:187–194. [PMC free article] [PubMed] [Google Scholar]
- 10.Minot DM, Kipp BR, Root RM, Meyer RG, Reynolds CA, Nassar A, Henry MR, Clayton AC. Automated cellular imaging system III for assessing HER2 status in breast cancer specimens: development of a standardized scoring method that correlates with FISH. Am J Clin Pathol. 2009;132:133–138. doi: 10.1309/AJCPJV0SKAF2PCMY. [DOI] [PubMed] [Google Scholar]
- 11.Alvarenga AW, Coutinho-Camillo CM, Rodrigues BR, Rocha RM, Torres LF, Martins VR, da Cunha IW, Hajj GN. A Comparison between Manual and Automated Evaluations of Tissue Microarray Patterns of Protein Expression. J Histochem Cytochem. 2013;61:272–282. doi: 10.1369/0022155413477661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grin A, Brezden-Masley C, Bauer S, Streutker CJ. HER2 In Situ Hybridization in Gastric and Gastroesophageal Adenocarcinoma: Comparison of Automated Dual ISH to FISH. Appl Immunohistochem Mol Morphol. 2013;21:561–6. doi: 10.1097/PAI.0b013e3182849826. [DOI] [PubMed] [Google Scholar]
- 13.Fiore C, Bailey D, Conlon N, Wu X, Martin N, Fiorentino M, Finn S, Fall K, Andersson SO, Andren O, Loda M, Flavin R. Utility of multispectral imaging in automated quantitative scoring of immunohistochemistry. J Clin Pathol. 2012;65:496–502. doi: 10.1136/jclinpath-2012-200734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Laak JA, van Engelen N, Melissen M, Hebeda KM. Automated measurement of MIB-1 positive area as an alternative to counting in follicular lymphoma. Cytometry A. 2012;81:527–531. doi: 10.1002/cyto.a.22053. [DOI] [PubMed] [Google Scholar]
- 15.Mikalsen LT, Dhakal HP, Bruland OS, Nesland JM, Olsen DR. Quantification of angiogenesis in breast cancer by automated vessel identification in CD34 immunohistochemical sections. Anticancer Res. 2011;31:4053–4060. [PubMed] [Google Scholar]
- 16.Lopez C, Lejeune M, Salvado MT, Escriva P, Bosch R, Pons LE, Alvaro T, Roig J, Cugat X, Baucells J, Jaen J. Automated quantification of nuclear immunohistochemical markers with different complexity. Histochem Cell Biol. 2008;129:379–387. doi: 10.1007/s00418-007-0368-5. [DOI] [PubMed] [Google Scholar]
- 17.Rohner F, Zeder C, Zimmermann MB, Hurrell RF. Comparison of manual and automated ELISA methods for serum ferritin analysis. J Clin Lab Anal. 2005;19:196–198. doi: 10.1002/jcla.20077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanaian NK, Cohen C, Waldrop S, Wang J, Shehata BM. Epstein-Barr virus (EBV)-encoded RNA: automated in-situ hybridization (ISH) compared with manual ISH and immunohistochemistry for detection of EBV in pediatric lymphoproliferative disorders. Pediatr Dev Pathol. 2009;12:195–199. doi: 10.2350/07-07-0316.1. [DOI] [PubMed] [Google Scholar]
- 19.Kaemmerer D, Peter L, Lupp A, Schulz S, Sanger J, Prasad V, Kulkarni H, Haugvik SP, Hommann M, Baum RP. Molecular imaging with 68Ga-SSTR PET/CT and correlation to immunohistochemistry of somatostatin receptors in neuroendocrine tumours. Eur J Nucl Med Mol Imaging. 2011;38:1659–1668. doi: 10.1007/s00259-011-1846-5. [DOI] [PubMed] [Google Scholar]
- 20.Kaemmerer D, Lupp A, Peter L, Fischer E, Schulz S, Kloppel G, Hommann M. Correlation of monoclonal and polyclonal somatostatin receptor 5 antibodies in pancreatic neuroendocrine tumors. Int J Clin Exp Pathol. 2013;6:49–54. [PMC free article] [PubMed] [Google Scholar]
- 21.Baatz M, Zimmermann J, Blackmore CG. Automated analysis and detailed quantification of biomedical images using Definiens Cognition Network Technology. Comb Chem High Throughput Screen. 2009;12:908–916. doi: 10.2174/138620709789383196. [DOI] [PubMed] [Google Scholar]
- 22.Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- 23.Schutze S, Loleit T, Zeretzke M, Bunkowski S, Bruck W, Ribes S, Nau R. Additive microglia-mediated neuronal injury caused by amyloid-beta and bacterial TLR agonists in murine neuron-microglia co-cultures quantified by an automated image analysis using cognition network technology. J Alzheimers Dis. 2012;31:651–657. doi: 10.3233/JAD-2012-120856. [DOI] [PubMed] [Google Scholar]
- 24.Schonmeyer R, Athelogou M, Sittek H, Ellenberg P, Feehan O, Schmidt G, Binnig G. Cognition Network Technology prototype of a CAD system for mammography to assist radiologists by finding similar cases in a reference database. Int J Comput Assist Radiol Surg. 2011;6:127–134. doi: 10.1007/s11548-010-0486-8. [DOI] [PubMed] [Google Scholar]
- 25.Haug AR, Assmann G, Rist C, Tiling R, Schmidt GP, Bartenstein P, Hacker M. Quantification of immunohistochemical expression of somatostatin receptors in neuroendocrine tumors using 68Ga-DOTATATE PET/CT. Radiologe. 2010;50:349–354. doi: 10.1007/s00117-009-1972-2. [DOI] [PubMed] [Google Scholar]
- 26.Miederer M, Seidl S, Buck A, Scheidhauer K, Wester HJ, Schwaiger M, Perren A. Correlation of immunohistopathological expression of somatostatin receptor 2 with standardised uptake values in 68Ga-DOTATOC PET/CT. Eur J Nucl Med Mol Imaging. 2009;36:48–52. doi: 10.1007/s00259-008-0944-5. [DOI] [PubMed] [Google Scholar]
- 27.Reubi JC, Schar JC, Waser B, Wenger S, Heppeler A, Schmitt JS, Macke HR. Affinity profiles for human somatostatin receptor subtypes SST1-SST5 of somatostatin radiotracers selected for scintigraphic and radiotherapeutic use. Eur J Nucl Med. 2000;27:273–282. doi: 10.1007/s002590050034. [DOI] [PubMed] [Google Scholar]
- 28.Fischer T, Doll C, Jacobs S, Kolodziej A, Stumm R, Schulz S. Reassessment of sst2 somatostatin receptor expression in human normal and neoplastic tissues using the novel rabbit monoclonal antibody UMB-1. J Clin Endocrinol Metab. 2008;93:4519–4524. doi: 10.1210/jc.2008-1063. [DOI] [PubMed] [Google Scholar]
- 29.Kulaksiz H, Eissele R, Rossler D, Schulz S, Hollt V, Cetin Y, Arnold R. Identification of somatostatin receptor subtypes 1, 2A, 3, and 5 in neuroendocrine tumours with subtype specific antibodies. Gut. 2002;50:52–60. doi: 10.1136/gut.50.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


