Abstract
Oncoprotein DEK plays an important role in cancer tumorigenesis. To explore the clinical implication of DEK expression on prognostic evaluation in small cell lung cancer (SCLC), 130 cases of SCLC with strict follow-up were selected for immunohistochemical (IHC) staining of DEK protein. The correlation between DEK expression and clinicopathological features of SCLC was evaluated using the Chi-square and Fisher’s exact tests, survival rates were calculated using the Kaplan-Meier method and univariate and multivariate analyses were performed using the Cox proportional hazards regression model. IHC analysis demonstrated that DEK protein staining was strongly positive and significantly higher (44.62%) in SCLC compared with either adjacent non-tumor or normal lung tissues (P < 0.001 for both). DEK expression correlated with large tumor size (P = 0.025) and late pathologic stage (P = 0.005). Moreover, it correlated with low disease-free (P = 0.004) and 5-year (P = 0.005) survival rates. In the late-stage group, disease-free and 5-year survival rates of patients with high level DEK expression were significantly lower than those with low level DEK expression (P = 0.006 and P = 0.001, respectively). Furthermore, Cox analysis revealed that DEK expression emerged as a significant independent hazard factor for the overall survival rate of patients with SCLC (HR: 1.594, 95% CI: 1.087-2.336, P = 0.017). In conclusion, DEK plays an important role in the progression of SCLC. DEK may potentially be used as an independent biomarker for the prognostic evaluation of SCLC.
Keywords: Small cell lung cancer, DEK, immunohistochemistry, survival analysis
Introduction
Small cell lung cancer (SCLC) is one of the leading causes of cancer-related deaths worldwide, and despite progress in the multimodal treatment of the disease, its prognosis is still poor with only about 5% of patients having a 5-year survival rate. Although it has been known that numerous signaling pathways, such as PI3K/Akt/mTOR [1], c-Kit/SCF [2] and c-Met/HGF [3] contribute to growth, survival status and chemotherapy resistance in SCLC, reliable biomarkers for recurrence and for identifying tumor type are still lacking. Therefore, the evaluation of predetermined candidate genes or loci attracts widespread research interest.
DEK is a highly evolutionarily conserved 43 kDa (375 amino acids) chromosomal factor with no known enzymatic activity and NO AND that has not been classified into any known protein family [4]. Its expression was first discovered in acute myeloid leukemia (AML) patients as a fusion gene with the CAN nuclear pore complex protein NUP214, which is involved in various cellular functions, such as chromatin remodeling, regulating transcription and mRNA processing [4,5].
Accumulating reports have suggested that DEK may promote tumorigenesis, at least in part, by its ability to interfere with cell division and DNA repair, by inhibiting cell differentiation, senescence and apoptosis, and by cooperating with transforming oncogenes. It has been shown that overexpression of the DEK gene was clearly connected with human malignancies, such as breast cancer [5,6], Merkel and small cell carcinoma [7], head and neck cancer [8], as well as others. Our previous studies also showed that overexpression of the DEK protein was closely related to poor prognosis of ovarian, breast and colorectal cancers [9-11].
However, to date, the expression status of DEK in SCLC and its relationship with clinicopathological features/prognosis is unknown. The critical role of DEK in numerous cancers compelled us to study the function of DEK in SCLC. Therefore, we performed immunohistochemical (IHC) staining of DEK in SCLC and normal lung tissues, and revealed that DEK might be an independent biomarker for prediction of SCLC prognosis.
Materials and methods
Clinical samples
One hundred and thirty cases of routinely processed and paraffin-embedded SCLC tissue sections stored in the Department of Pathology and Tumor Tissue Bank, with strict follow-up for survival status, were selected randomly from patients who underwent surgery between 2003-2008 at Yanbian University Medical College. The pathological parameters, including age, gender, smoking status, tumor size, location, subtypes, clinical stage, lymph node metastasis, and disease-free and 5-year survival data were carefully reviewed in all cases. Patient age ranged from 26-76 years with a mean age of 52.3 years. The male to female ratio was 71:59. Tumor staging was done according to the 7th edition of the American Joint Committee on Cancer. The 130 SCLCs encompassed 76 early-stage (I-II) and 54 late-stage (III) cases. Of the 130 cases, 69 were located in the central bronchus and 61 in the peripheral bronchus. None of the patients had any distant metastasis prior to surgery. By March 2013, 112 patients had died while 18 patients remained alive. The median survival time was 23.2 months.
IHC analysis
IHC analysis was performed using the DAKO LSAB kit (DAKO A/S, Glostrup, Denmark). Briefly, to eliminate endogenous peroxidase activity, 4 μm thick tissue sections were deparaffinized, rehydrated and incubated with 3% H2O2 in methanol for 15 min at room temperature (RT). The antigen was retrieved at 95°C for 20 min by placing the slides in 0.01 M sodium citrate buffer (pH 6.0). The slides were then incubated with the DEK antibody (1:50; BD Biosciences Pharmingen, San Diego, CA, USA) at 4°C overnight. After incubation with the biotinylated secondary antibody at RT for 30 min, the slides were incubated with a streptavidin-peroxidase complex at RT for 30 min. Immunostaining was developed using 3, 3’-diaminobenzidine, and Mayer’s hematoxylin was used for counterstaining. We used tonsil sections as the positive control and mouse IgG as an isotope control. In addition, tissue sections were processed omitting the primary antibody (mouse anti-DEK) as the negative control.
Two pathologists (Lin Z & Jin T) who did not possess knowledge of the clinical data examined and scored all tissue specimens. In case of discrepancies, a final score was established by reassessment by both pathologists on a double-headed microscope. Briefly, the IHC staining for DEK was semi-quantitatively scored as ‘-’ (negative) (no or less than 5% positive cells), ‘+’ (5-25% positive cells), ‘++’ (26-50% positive cells) and ‘+++’ (more than 50% positive cells). Only the nuclear expression pattern was considered as positive staining. Tissue sections scored as ‘++’ and ‘+++’ were considered as strong positives (high-level expression) of DEK.
Statistical analysis
Statistical analyses were performed using SPSS 17.0 software. Correlation between DEK expression and clinic pathological characteristics was evaluated using the χ2 test and Fisher’s exact test. Disease-free and 5-year survival rates after tumor removal were calculated using the Kaplan-Meier method, and differences in survival curves were analyzed using the log rank test. Multivariate analysis was performed using the Cox proportional hazards regression model on all significant characteristics measured for univariate analysis. A P-value of less than 0.05 was considered statistically significant.
Results
DEK protein expression in SCLC paraffin-embedded tissue sections
DEK protein expression showed a strictly nuclear staining pattern in SCLC using IHC. The positive and strongly positive rates of DEK protein expression were 69.23% and 44.62%, respectively, in 130 SCLC tissues, and significantly higher than in either adjacent non-tumor or normal lung tissues (P < 0.001) (Figure 1 and Table 1).
Figure 1.
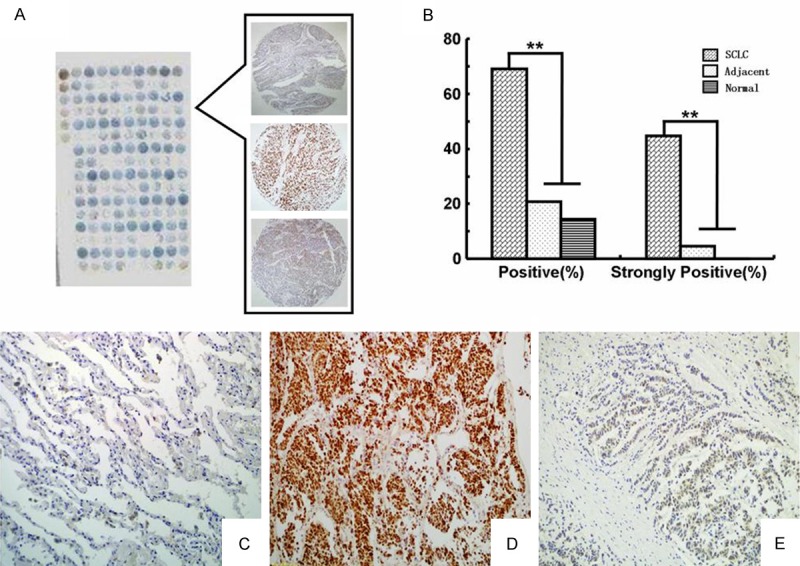
IHC staining of the DEK protein in SCLC and adjacent non-tumor tissue. DEK protein was detected in tissue microarray (A) by IHC staining. The histogram (B) showed that the positive rate and strongly positive rate of DEK protein expression were significantly higher in SCLC than that in either adjacent non-tumor or normal lung tissues. DEK was negative in normal lung tissue (C), but strongly positive in SCLC with lymph node metastasis (D) and weakly positive in SCLC with non-metastasis (E) (original magnification, (C-E) × 200).
Table 1.
DEK protein expression in SCLC
| Diagnosis | No. of cases | DEK protein expression | Positive rate (%) | Strongly positive rate (%) | |||
|---|---|---|---|---|---|---|---|
|
| |||||||
| - | + | ++ | +++ | ||||
| SCLC | 130 | 40 | 32 | 27 | 31 | 69.23%** | 44.62%** |
| Adjacent non tumor | 130 | 103 | 21 | 4 | 2 | 20.77% | 4.62% |
| Normal lung tissues | 14 | 12 | 2 | 0 | 0 | 14.29% | 0.00% |
P < 0.01 compared with either adjacent non-tumor or normal lung tissues.
Correlation between DEK expression and clinicopathological features of SCLC
To evaluate the role of the DEK protein in SCLC progression, we analyzed the correlation between DEK protein expression and major clinicopathological features of SCLC. The results showed that the DEK high-level expression rate was significantly related to tumor size and SCLC clinical stage (P = 0.025 and P = 0.005, respectively). However, DEK expression level was not related to age, gender, smoking status, location, subtype or lymph node metastasis of SCLC (P > 0.05) (Table 2).
Table 2.
Correlation between DEK expression and clinicopathological features of SCLC
| Variables | No. of cases | DEK positive cases (%) | x2 | P-value | |
|---|---|---|---|---|---|
|
| |||||
| +++/++ | +/- | ||||
| Age | |||||
| ≤ 52 | 65 | 31 (47.69%) | 34 (52.31%) | 0.498 | 0.482 |
| > 52 | 65 | 27 (41.54%) | 38 (58.46%) | ||
| Gender | |||||
| Male | 72 | 33 (45.83%) | 39 (54.17%) | 0.097 | 0.757 |
| Female | 58 | 25 (43.10%) | 33 (56.90%) | ||
| Tumor size | |||||
| ≤ 3 cm | 79 | 29 (36.71%) | 50 (63.29%) | 5.094 | 0.025* |
| > 3 cm | 51 | 29 (56.86%) | 22 (43.14%) | ||
| Location subtype | |||||
| Central | 69 | 31 (44.93%) | 38 (55.07%) | 0.012 | 0.912 |
| Peripheral | 61 | 27 (44.26%) | 34 (55.74%) | ||
| Clinical stage | |||||
| I-II | 76 | 26 (34.21%) | 50 (65.79%) | 8.016 | 0.005** |
| III | 54 | 32 (59.26%) | 22 (40.74%) | ||
| LN metastasis | |||||
| Positive | 63 | 32 (50.79%) | 31 (49.21%) | 1.888 | 0.171 |
| Negative | 67 | 26 (38.81%) | 41 (61.19%) | ||
| Smoking status | |||||
| Yes | 86 | 41 (47.67%) | 45 (52.33%) | 0.962 | 0.328 |
| No | 44 | 17 (38.64%) | 27 (61.36%) | ||
P < 0.05;
P < 0.01.
Correlation between survival rates and DEK expression status using the Kaplan-Meier method
To further confirm the role of DEK expression in SCLC progression, we analyzed disease-free and 5-year survival rates for the 130 SCLC cases using Kaplan-Meier survival curves and found that SCLC patients with DEK high-level expression had lower disease-free (Log-rank = 8.230, P = 0.004) and 5-year survival rates (Log-rank = 8.158, P = 0.004) than those with DEK low-level expression (Figure 2).
Figure 2.
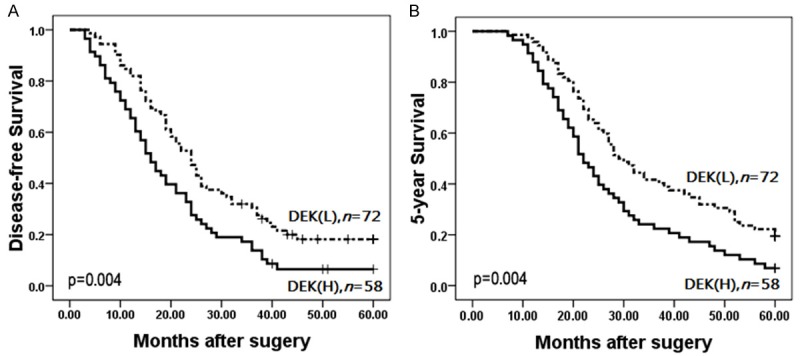
Kaplan-Meier survival curves illustrating the role of DEK expression in SCLC progression. SCLC patients with high DEK expression had lower disease-free (A) and 5-year survival (B) rates compared to those with low DEK expression (P < 0.001).
To further substantiate the importance of DEK expression in SCLC progression, we analyzed the correlation between the DEK high-level expression rate and SCLC clinical stage. In late-stage SCLC, patients with DEK high-level expression had lower disease-free and 5-year survival rates than those with DEK low-level expression (P = 0.002 and P = 0.001, respectively) (Figure 3A, 3B). However, disease-free and 5-year survival rates did not correlate with DEK expression status (P = 0.238 and P = 0.246) in early-stage SCLC (Figure 3C, 3D).
Figure 3.
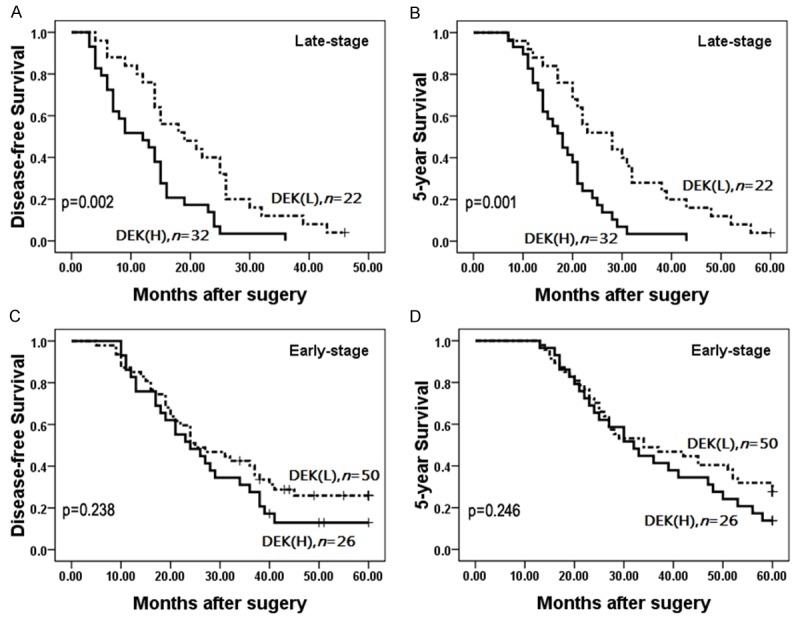
Kaplan-Meier survival curves illustrating the significance of DEK expression in SCLC in relation to clinical stage. In late-stage SCLC, patients with DEK high-level expression had lower disease-free (A) and 5-year survival (B) rates than those with DEK low-level expression (P = 0.002 and P = 0.001, respectively). In early-stage SCLC, disease-free (C) and 5-year survival (D) rates did not correlate with DEK expression status (P = 0.238 and P = 0.246).
DEK is an independent prognostic factor in SCLC using the Cox proportional hazards regression model
On univariate analysis, SCLC patients who had DEK high-level expression tumors had significantly lower 5-year survival (P = 0.007) than those with DEK low-level expression tumors. Additionally, patient’s age (P = 0.036), tumor size (P = 0.032), pathologic stage (P = 0.000), and lymph node metastasis (P = 0.012) were also significantly associated with the 5-year survival rate. Therefore, multivariate survival analysis was performed using the Cox proportional hazards model for all of NO OF the significant variables in the univariate survival analysis. We found that clinical stage (HR: 2.454, 95% CI: 1.665-3.616, P < 0.001) proved to be an independent prognostic factor of 5-year survival rates in SCLC. More importantly, DEK expression also emerged as a significant independent prognostic factor in the prognosis of SCLC (HR: 1.593, 95% CI: 1.088-2.334, P = 0.017) (Table 3).
Table 3.
Univariate and multivariate analyses of clinicopathological factors used to determine overall survival rates for 130 patients with SCLC
| Characteristics | B | SE | Wald | HR | 95% CI | P value | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Lower | Upper | ||||||
| Univariate analysis | |||||||
| Age | 0.386 | 0.184 | 4.404 | 1.472 | 1.026 | 2.111 | 0.036* |
| Gender | 0.294 | 0.182 | 2.611 | 1.342 | 0.939 | 1.917 | 0.106 |
| Tumor size | 0.392 | 0.182 | 4.624 | 1.480 | 1.035 | 2.115 | 0.032* |
| Location | 0.253 | 0.177 | 2.049 | 1.288 | 0.911 | 1.821 | 0.152 |
| Stage | 0.930 | 0.190 | 23.939 | 2.535 | 1.747 | 3.680 | 0.000** |
| LN metastasis | 0.453 | 0.180 | 6.360 | 1.573 | 1.106 | 2.238 | 0.012* |
| Smoking status | 0.028 | 0.185 | 0.023 | 1.028 | 0.715 | 1.479 | 0.880 |
| DEK | 0.486 | 0.179 | 7.373 | 1.626 | 1.145 | 2.311 | 0.007** |
| Multivariate analysis | |||||||
| Age | 0.141 | 0.197 | 0.511 | 1.151 | 0.783 | 1.692 | 0.475 |
| Tumor size | 0.314 | 0.186 | 2.860 | 1.369 | 0.951 | 1.969 | 0.091 |
| Stage | 0.898 | 0.198 | 20.607 | 2.454 | 1.665 | 3.616 | 0.000** |
| LN metastasis | 0.241 | 0.184 | 1.723 | 1.273 | 0.888 | 1.825 | 0.189 |
| DEK | 0.466 | 0.195 | 5.716 | 1.593 | 1.088 | 2.334 | 0.017* |
P < 0.05;
P < 0.01.
Discussion
The DEK protein is an abundant and ubiquitous chromatin protein in multicellular organisms (although not found in unicellular organisms such as yeast). It is expressed at more than a million copies/nucleus in rapidly proliferating mammalian cells [12]. DEK can promote the formation of various tumor cell types, by affecting cell division, DNA repair, aging and apoptosis, by inhibiting cell differentiation, as well as by oncogenic transformation [13-15]. It has been proved that DEK introduces constrained positive supercoils into DNA, can facilitate the ligation of linear DNA molecules in vitro [16,17], and plays a critical role in chromatin organization and maintenance of genome stability in vivo. It also inhibits p53-dependent and independent apoptosis and has been found to mediate the apoptotic response to clastogenic chemotherapeutic agents such as doxorubicin and cisplatin [18,19]. Moreover, DEK is a potential target of chromosome 6p22 gains in retinoblastoma, as well as in bladder cancer, suggesting that DEK gain of function may provide a selective advantage for tumor development [20].
DEK upregulation can interfere with cellular differentiation, apoptosis, senescence, and response to chemotherapy, justifying the classification of DEK as a bona fide oncogene that plays a role in tumor growth and survival [18,21]. Recent reports have shown that DEK overexpression occurs in various malignancies, including bladder cancer [22], breast cancer [5,6], hepatocellular carcinoma [23], retinoblastoma [24], melanoma [21], glioblastoma [25] as well as other cancers. Liu et al. [26] demonstrated that DEK inhibition resulted in significant growth repression of CaSki cells by inducing cell apoptosis and senescence. Privette Vinnedge et al. [19] identified DEK as being a novel estrogen receptor α (ERα) target gene, using chromatin immunoprecipitation experiments, whose expression promotes estrogen-induced proliferation. Datta et al. [22] examined the expression of DEK protein using western blotting on samples from 38 paired transitional cell carcinoma (TCC) bladder tumor and adjacent normal tissues, and identified DEK as a potential urinary and tissue biomarker of TCC of the bladder for both low and high-grade tumors, as well as for stage and progression. In addition, DEK was involved in the pathogenesis of Merkel cell carcinoma and may have therapeutic implications for this carcinoma [7].
However, few studies to date have reported an association between DEK expression and clinicopathological parameters, or DEK as a prognostic marker in lung cancer. Wang et al. [27] analyzed DEK using IHC on 112 NSCLC cases and reported that DEK-positive tumors correlated with poor differentiation, advanced p-TNM stage and nodal metastasis, and DEK expression in lung adenocarcinoma was significantly higher compared with DEK expression in squamous cell carcinoma. Shibata et al. [28] showed that DEK overexpression, partly through an increase in its gene dose, mediated the activity of global transcriptional regulators and was associated with tumor initiation activity and poor prognosis in high-grade neuroendocrine carcinoma of the lung.
The overall goal of this study was to explore the role of DEK upregulation in SCLC progression. Here we performed IHC staining of the DEK protein and survival data analysis using 14 cases of normal lung tissue, 130 cases of SCLC and their adjacent normal tissue counterparts, and found that positive and strongly positive rates of DEK oncoprotein expression were significantly higher in SCLC than in normal lung or adjacent normal tissues. These findings indicated that DEK potentially plays an important role in the progression of SCLC.
Despite the strong association between DEK expression and cancer, reports of DEK expression-based outcomes in tumor patients are limited. Kappes et al. [21] indicated that DEK overexpression was a frequent event in invasive melanomas, and WAS associated with the acquisition of ominous features such as deep dermal invasion and metastasis. However, the correlation between DEK expression and clinical outcomes of invasion and metastasis in patients with SCLC cannot be concluded from this study, nor from the works of Wang et al. [27] or Shibata et al. [28]. Here we found that DEK overexpression was significantly correlated with large tumor size and late stage of SCLC, suggesting that DEK might be useful for predicting the prognosis of patients with SCLC.
Moreover, our previous study [11] demonstrated that DEK protein expression was closely related to disease-free and overall survival rates of patients with colorectal cancers, and its overexpression was an independent risk factor for mortality in colorectal cancer. Similarly, with detailed clinical follow-up data, this study showed that SCLC patients with DEK high-level expression had lower disease-free (P = 0.004) and 5-year survival rates (P = 0.004) than those with DEK low-level expression. In late-stage SCLC, patients with DEK high-level expression had lower survival rates than those with DEK low-level expression (P = 0.002 and P = 0.001, respectively). In addition, multivariate analysis using Cox proportional hazards regression model confirmed that DEK was an independent prognostic factor in SCLC along with clinical stage.
In conclusion, DEK plays an important role in SCLC progression; it might be a new attractive biomarker for prognostic evaluation and a molecular therapeutic target in patients with SCLC.
Acknowledgements
This study was supported by grants from The National Natural Science Funds of China (61371067) and The International Cooperative Project from Jilin Provincial Science & Technology Department in China (20140414050GH).
Disclosure of conflict of interest
None.
References
- 1.Fischer B, Marinov M, Arcaro A. Targeting receptor tyrosine kinase signaling in small cell lung cancer (SCLC): what have we learned so far? Cancer Treat Rev. 2007;33:391–406. doi: 10.1016/j.ctrv.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Murray N, Salgia R, Fossella FV. Targeted molecules in small cell lung cancer. Semin Oncol. 2004;31(Suppl 1):106–111. doi: 10.1053/j.seminoncol.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, Johnson BE, Salgia R. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 4.D’Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer. Cancer Biol Ther. 2010;10:1–10. doi: 10.4161/cbt.10.1.12045. [DOI] [PubMed] [Google Scholar]
- 5.Privette Vinnedge LM, Ho SM, Wikenheiser-Brokamp KA, Wells SI. The DEK oncogene is a target of steroid hormone receptor signaling in breast cancer. PLoS One. 2012;7:e46985. doi: 10.1371/journal.pone.0046985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Privette Vinnedge LM, Benight NM, Wagh OK, Pease NA, Nashu MA, Serrano-Lopez J, Adams AK, Cancelas JA, Waltz SE, Wells SI. The DEK oncogene promotes cellular proliferation through paracrine Wnt signaling in Ron receptor-positive breast cancers. Oncogene. 2014;173:1–12. doi: 10.1038/onc.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel RM, Walters LL, Kappes F, Mehra R, Fullen DR, Markovitz DM, Ma L. DEK expression in Merkel cell carcinoma and small cell carcinoma. J Cutan Pathol. 2012;39:753–757. doi: 10.1111/j.1600-0560.2012.01941.x. [DOI] [PubMed] [Google Scholar]
- 8.Wise-Draper TM, Draper DJ, Gutkind JS, Molinolo AA, Wikenheiser-Brokamp KA, Wells SI. Future directions and treatment strategies for head and neck squamous cell carcinomas. Transl Res. 2012;160:167–177. doi: 10.1016/j.trsl.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han S, Xuan Y, Liu S, Zhang M, Jin D, Jin R, Lin Z. Clinicopathological significance of DEK overexpression in serous ovarian tumors. Pathol Int. 2009;59:443–447. doi: 10.1111/j.1440-1827.2009.02392.x. [DOI] [PubMed] [Google Scholar]
- 10.Liu S, Wang X, Sun F, Kong J, Li Z, Lin Z. DEK overexpression is correlated with the clinical features of breast cancer. Pathol Int. 2012;62:176–181. doi: 10.1111/j.1440-1827.2011.02775.x. [DOI] [PubMed] [Google Scholar]
- 11.Lin L, Piao J, Gao W, Piao Y, Jin G, Ma Y, Li J, Lin Z. DEK over expression as an independent biomarker for poor prognosis in colorectal cancer. BMC Cancer. 2013;13:366. doi: 10.1186/1471-2407-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein-an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF, Wells JM, Drissi R, Bissler JJ, Stambrook PJ, Andreassen PR, Wiesmüller L, Wells SI. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39:7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brázda V, Laister RC, Jagelská EB, Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrer J, Popp O, Malanga M, Beneke S, Markovitz DM, Ferrando-May E, Bürkle A, Kappes F. High-affinity interaction of poly (ADP-ribose) and the human DEK oncoprotein depends upon chain length. Biochemistry. 2010;49:7119–7130. doi: 10.1021/bi1004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 18.Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM, Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams AK, Hallenbeck GE, Casper KA, Patil YJ, Wilson KM, Kimple RJ, Lambert PF, Witte DP, Xiao W, Gillison ML, Wikenheiser-Brokamp KA, Wise-Draper TM, Wells SI. DEK promotes HPV-positive and -negative head and neck cancer cell proliferation. Oncogene. 2014;0:1. doi: 10.1038/onc.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kappes F, Khodadoust MS, Yu L, Kim DS, Fullen DR, Markovitz DM, Ma L. DEK expression in melanocytic lesions. Hum Pathol. 2011;42:932–938. doi: 10.1016/j.humpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta A, Adelson ME, Mogilevkin Y, Mordechai E, Sidi AA, Trama JP. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer. 2011;11:234. doi: 10.1186/1471-2407-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin LJ, Chen LT. The role of DEK protein in hepatocellular carcinoma for progression and prognosis. Pak J Med Sci. 2013;29:778–782. doi: 10.12669/pjms.293.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 25.Kroes RA, Jastrow A, McLone MG, Yamamoto H, Colley P, Kersey DS, Yong VW, Mkrdichian E, Cerullo L, Leestma J, Moskal JR. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156:191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 26.Liu K, Feng T, Liu J, Zhong M, Zhang S. Silencing of the DEK gene induces apoptosis and senescence in CaSki cervical carcinoma cells via the up-regulation of NF-κB p65. Biosci Rep. 2012;32:323–332. doi: 10.1042/BSR20100141. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Sun L, Yang M, Luo W, Gao Y, Liu Z, Qiu X, Wang E. DEK depletion negatively regulates Rho/ROCK/MLC pathway in non-small cell lung cancer. J Histochem Cytochem. 2013;61:510–521. doi: 10.1369/0022155413488120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibata T, Kokubu A, Miyamoto M, Hosoda F, Gotoh M, Tsuta K, Asamura H, Matsuno Y, Kondo T, Imoto I, Inazawa J, Hirohashi S. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 2010;29:4671–4681. doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]


