Abstract
MiRNA molecules have been identified to play key roles in a broad range of physiologic and pathologic processes. Polymorphisms in microRNA target sites (PolymiRTSs) can disturb or obstruct miRNA binding and consequentially influence regulation of the target genes. A two-step study design was used in this study. A case-control study was designed to assess the relationship between miRNA-1 target site rs9548934C→T polymorphism in target gene (Component of Oligomeric Golgi Complex 6, COG6) and risk of coronary artery disease (CAD) in 1013 patients and 610 normal controls. This genetic variant was also evaluated for the association with major adverse cardiovascular events (MACE) of CAD in a follow-up study, including 785 (785/1013) patients followed up for 42 months. The phenotypes of circulating miRNA-1 levels in 34 cases were slightly lower than that of 40 controls but not significantly different (P = 0.090). The CT and CT/TT genotypes were associated with a 34% and 26% decreased risk of CAD, and the TT and CT/TT genotypes were associated with a 76% and 49% decreased risk of MACE of CAD. Cox regression analysis showed that rs9548937 C/T variant was associated with a decreased risk of MACE, while age, diabetes mellitus, higher levels of CRP (≥ 3.80 mg/L) and three pathological changes in the coronary artery were associated with an increased risk of MACE. Our findings implicate miRNA-1 target site rs9548934C→T genotypes, circulating miRNA-1 phenotype and CRP levels may modulate the occurrence and MACE of CAD.
Keywords: miRNAs, target gene, polymorphism, coronary artery disease, MACE
Introduction
Despite advances in the prevention, detection, and treatment in the last few decades, coronary artery disease (CAD) is the leading cause of morbidity and mortality in the world [1]. In China, more than 700,000 people die from CAD each year [2]. CAD is a complex disease caused by environmental and genetic factors [3]. Epidemiological studies show that smoking, diabetes, hypertension, obesity, physical inactivity, alcohol intake, high-fat diet and family history are associated with an increased risk of CAD [4,5]. Genome-wide association studies (GWAS) conducted both in European ancestry and Chinese populations also reveal some important genetic variants related to occurrence risks of CAD, such as 9p21 [2]. Nonetheless, only a few of these studies have detected both the genotype and phenotype associated with the occurrence of major adverse cardiovascular events (MACE) of CAD.
MicroRNAs (miRNAs, miRs) are a class of small noncoding RNA with a length of ~22 nucleotides. MiRNAs regulate gene expression through binding the 3’untranslated region (3’UTR) of mRNAs of target genes, causing inhibition of translation or even promoting degradation of the mRNA. It is estimated that about 30% of human genes are regulated by miRNAs [6]. Increasing evidence demonstrates that PolymiRTSs) can disturb or obstruct miRNAs binding [7,8], through regulating their mRNA expression [9] and consequentially influence regulation of the target genes, which might be associated with disease susceptibility, including risk of cancer [10], myocardial infarction [11], Alzheimer’s disease [12] and hypertension [13].
MiR-1 is a highly conserved miRNA and its expression is enriched specifically in cardiac and skeletal muscle. It is required for postnatal cardiac function and reinforces the striated muscle phenotype by regulating the smooth muscle gene expression network [14]. In mice, miR-1 regulates the transition from prenatal to neonatal stages by repressing the cardiac fetal gene program in the heart [15], and it also protects the heart from myocardial ischemia-reperfusion (IR) injury through regulating apoptosis-related genes [16]. However, very little is known about how genetic variation in miR-1 target sites might affect miR-1 biogenesis and function.
Recent studies have shown that some miRNAs are not only presented in tissues but also in systemic circulation, including serum and plasma [17]. Altered levels of circulating miRNAs have been found in patients with acute coronary syndrome (ACS) [18], acute myocardial infarction (AMI) [19], stable coronary artery disease [20], essential hypertension [21] and heart failure [22]. Ai, et al, found that miR-1 is over-expressed in ischemic myocardium of a rat model of AMI and of individuals with CAD as well, which indicate that circulating miR-1 is a molecule with multiple roles in the process of the cardiovascular disease [23].
In this large-scale case-control and follow-up study, we obtained the miR-1 target site SNP of gene COG6 (rs9548934C→T) from PolymiRTS Database 2.0 [24], and analyzed the association between the target site SNP and the occurrence and MACE risks of CAD together with the circulating miRNA-1 phenotype.
Materials and methods
Study population
This study included 1013 consecutive inpatients with newly-diagnosed CAD and 610 CAD-free controls. All patients and controls were recruited between July 2005 and December 2010 in ZhongDa Hospital Affiliated to Southeast University, Nanjing, China. All CAD patients received quantitative coronary angiography (QCA) with a cardiovascular measurement system (QCA-CMS: Angiostar plus Siemens German) shortly after being admitted to the hospital, and coronary angiograms were analyzed by two experienced interventional cardiologists. CAD patients were defined as having angiographic coronary stenosis of at least 50% lumen reduction in at least one major epicardial coronary artery.
610 CAD-free controls were randomly selected from individuals with no history of cardiovascular events or signs of dysfunction of the cardiovascular system as evidenced by physical examination. Electrocardiogram (ECG) and ultrasonic echocardiogram (UCG) examination were conducted to exclude those with myocardial ischemia, segmental ventricular wall motion dysfunction, or cardiac dysfunction from controls.
After informed consent was obtained, each subject was interviewed using a structured questionnaire administered by interviewers at enrollment to collect information on demographic data, risk factors related to CAD, and personal history of vascular events. The complete clinical history acquisition of each subject was collected, including sex, age, body mass index (BMI), hypertension, hyperglycemia, diabetes mellitus (DM), smoking, and family history of coronary disease. Individuals who smoked once a day for over one year were categorized as smokers. The study was approved by the Ethical Committee of Clinic Medical College, Southeast University, and Nanjing, China.
Follow-up and outcomes measure
A total of 785 (785/1013) patients had completed follow-ups for 3.5 years (42 months). MACE was defined as unstable angina pectoris, myocardial infarction, cardiac insufficiency, cerebral infarction, and death due to CAD. Outcomes were determined from patient interviews, hospital chart reviews, and telephone interviews with the patient, a close relative, or a referring physician.
Among the remaining 228 (22.51%) patients with incomplete follow-up, 61 (6.02%) had incorrect telephone numbers, 87 (8.59%) refused to participate, and 80 (7.89%) moved or were unavailable for unknown reasons. However, there was no significant difference in the distribution of demographic characters (e.g., age and sex), or smoking status between the CAD cases with and without follow-up/clinical information.
Biochemical variables and genotype analyses
Five milliliter venous blood samples were collected from subjects after fasting for at least 12 h. Plasma total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) levels were determined by kit procedures supplied by Merck Diagnostica (Merck KGaA, Darmstadt, Germany).
Genomic DNA was extracted from the leukocyte pellet obtained from the buffy coat of each blood sample. The miR-1 target gene COG6 rs9548934 C→T polymorphisms were detected by employing the polymerase chain reaction (PCR) using mismatch primer design method. Briefly, the primers were: 5’-TGTCACTACGTTCAATTCTTTGCCTT-3’ (forward), 5’-AGTCAGGGTGGAGCAGGTGACT-3’ (reverse) for hsa-miR-1 target gene COG6 rs9548934 C→T. The PCR products were digested with FspBI (New England BioLabs, Beverly, MA, USA). Genotyping was performed without knowing the subjects’ status and the same number of cases and controls were assayed in each 96-well PCR plate with a positive control of a DNA sample with known heterozygous genotype. Two research assistants independently read the gel pictures and performed the repeated assays in the cases when they did not reach a consensus on the tested genotype.
RNA preparation and circulating (plasma) miRNA-1 level detection
Blood samples for circulating (plasma) miRNA detection were collected from the 34 CAD patients and 40 controls and were processed within 1 h of collection by two-step centrifugation. The supernatant was transferred to RNase/DNase-free tubes and stored at -80°C. Total RNA in plasma were isolated by using mirVanaTM miRNA (Ambion, Austin, TX, USA) following the instructions from the manufacturer with appropriate modification. RNA concentration was determined by the NanoDrop® ND-1000 spectrophotometer (NanoDrop, USA), and the concentration of RNA (100-200 ng/µl), purity (ultraviolet 260 OD and 280 OD ratio range of 1.6-2.0) and gross (≥ 400 ng) all met the research requirement.
cDNA synthesis was performed according to the manufacturer’s instructions (Reverse Transcription System, Cat. #A3500, Promega) as described previously [25]. The miScript SYBR Green PCR kit (Qiagen) was used in real-time PCR for relative quantification of miRNAs in our study with U6 used as an internal control. qRT - PCR was performed on 7500 FAST Real-Time PCR System (Applied Biosystems). The data were analyzed with automatic setting for assigning baseline; the threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence exceeds the given threshold. The plasma levels of miRNA were detected and analyzed by two investigators who were blinded to the clinical data of patients. The relative circulating level for miR-1 was computed using the comparative Ct method.
ΔCt = Ct (miRNA)-Ct (RNU6B)
Statistical analyses
Differences in demographic characteristics, selected variables, frequencies and serum lipids levels between the cases and controls were evaluated using the χ2 or student’s t test. The associations between the rs9548934 C→T genotypes and risk of CAD were estimated by estimating adjusted ORs and 95% CIs from logistic regression analyses, with adjustment for age, sex, BMI and cigarette smoking. The Hardy-Weinberg equilibrium was tested by a goodness-of-fit χ2 test to compare the observed genotype frequencies with the expected ones among control subjects.
Follow-up time was calculated from the date of CAD diagnosis to the date of the last follow-up. The associations between overall cardiovascular events and demographic characteristics, serum lipids levels and miR-1 target site SNP were estimated using the Kaplan-Meier method and log-rank test. Univariate or multivariate Cox proportional hazards regression models were performed to estimate the crude HRs or adjusted HRs and their 95% confidence intervals (CI). The Cox stepwise regression model was also used to determine factors predictive of cardiovascular events, with a significance level of P < 0.05 for entering and P > 0.10 for removal of the respective explanatory variables.
All of the statistical analyses were performed with Statistical Analysis System software (v.9.1.3e; SAS Institute, Cary, NC).
Results
Characteristics of the study population
The distribution of demographic characteristics and clinical items of the 1013 CAD cases and 610 controls are shown in Table 1. There were no significant differences in the distribution of the age, sex, and hypertension between the cases and controls (P > 0.05), suggesting that our frequency-matching on age and sex was satisfactory. The average body mass index (BMI) of CAD cases was significantly higher than that of the controls (P < 0.001). In addition, the serum TG, TC and LDL-C levels were significantly higher in cases, and levels of HDL-C were significantly lower in cases as compared with the controls (P < 0.001). With regards to disease history, 277 (27.4%) patients had a history of DM, significantly more than amongst controls (11.9%) (P < 0.001), but no significant difference in hypertension was shown between cases and controls (P = 0.340). There were more smokers in the case group (50.4%) than in controls (28.0%) (P < 0.001). Among the cases, 936 (92.4%) had information regarding pathological changes in coronary arteries: 551 (58.9%) cases had one lesion, and 385 (41.1%) had more than one coronary artery lesion.
Table 1.
Distributions of selected variables in CAD cases and normal controls
| Variants | Control group (N = 610) | CAD (N = 1013) | P value |
|---|---|---|---|
| Age (M ± SD) | 65.55 ± 11.47 | 65.72 ± 9.95 | 0.759a |
| BMI (kg/m2) | 23.97 ± 3.08 | 24.89 ± 3.06 | < 0.001a |
| TG (mmol/L)c (M ± SD) | 1.38 ± 0.72 | 1.60 ± 1.13 | < 0.001a |
| TC (mmol/L)c (M ± SD) | 4.67 ± 1.03 | 4.30 ± 1.04 | < 0.001a |
| HDL-C (mmol/L)c (M ± SD) | 1.35 ± 0.54 | 1.11 ± 0.41 | 0.014a |
| LDL-C (mmol/L)c (M ± SD) | 2.53 ± 0.90 | 2.67 ± 1.27 | < 0.001a |
| CRP (mmol/L) (Median [range]) | 1.20 [0.72-48.80] | 3.80 [0.42-91.50] | < 0.001a |
| Sex (%) | |||
| Male | 407 (66.7) | 719 (71.0) | 0.075b |
| Female | 203 (33.3) | 294 (29.0) | |
| Diabetes Mellitusd (%) | |||
| no | 274 (88.1) | 734 (72.6) | < 0.001b |
| yes | 37 (11.9) | 277 (27.4) | |
| Hypertensione (%) | |||
| no | 89 (28.8) | 262 (26.0) | 0.340b |
| yes | 220 (71.2) | 744 (74.0) | |
| Smoking statusf (%) | |||
| no | 213 (72.0) | 486 (49.6) | < 0.001b |
| yes | 83 (28.0) | 493 (50.4) | |
| Numbers of pathological changes in coronary arteries (n = 936) (%) | |||
| 1 | 402 (42.9) | ||
| 2 | 233 (24.9) | ||
| 3 | 301 (32.2) | ||
| Pathological changes in coronary arteries (n = 936) (%) | |||
| Left bole branch | 109 (11.6) | ||
| anterior descending branch | 231 (24.7) | ||
| Convolution branch | 62 (6.6) | ||
| Right corona branch | 149 (15.9) | ||
| More than one branches | 385 (41.2) |
Two-sided t test.
Two-sided chi-square test.
Serum TC and TG levels were measured in 599 normal controls and 972 CAD cases.
Serum HDL-C levels were measured in 421 normal controls and 945 CAD cases. Serum LDL-C levels were measured in 598 normal controls and 953 CAD cases.
Information on DM status was obtained for 311 controls and 1011 CAD cases.
Information on hypertension status was obtained for 309 controls and 1006 CAD case.
Information on smoking status was obtained for 296 controls and 979 CAD case.
Circulating levels of miR-1 in cases and controls
Circulating levels of miR-1 in plasma from 34 CAD patients and 40 normal controls are shown in Figure 1. The mean ΔCt of circulating miR-1 in cases was 4.948 (± 1.315), and that of controls was 5.618 (± 1.918). The median of miR-1 in plasma of CAD patients was slightly lower than that of normal controls; however, there were no statistical significant differences in the circulating levels of miR-1 in plasma between CAD patients and normal controls (P = 0.090).
Figure 1.
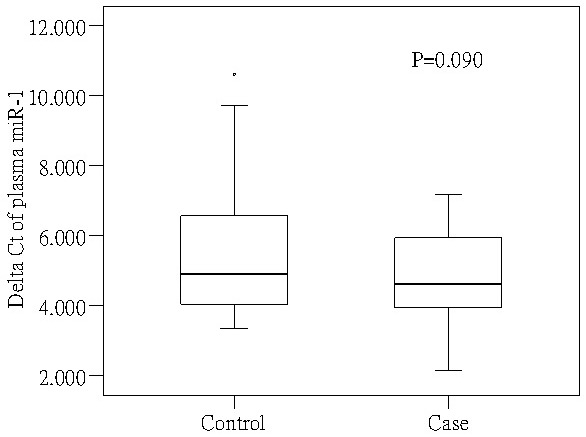
Circulating levels of miR-1 in plasma from CAD patients and normal controls.
Associations between miR-1 target gene COG6 rs9548934 C→T polymorphism and risk of CAD
The distribution of MiR-1 target gene COG6 binding site rs9548934 C→T genotypes in the CAD and controls are shown in Table 2. The observed genotype frequency among the control subjects was in agreement with the Hardy-Weinberg equilibrium (χ2 = 1.377, P = 0.240). After adjusting for age, sex, BMI and smoking status, we found that heterozygote CT and CT/TT genotypes were associated with a significantly reduced risk of CAD, compared with the wide genotype CC (adjusted OR = 0.66, 95% CI = 0.49-0.89 for CT, adjusted OR = 0.74, 95% CI = 0.56-0.97 for CT/TT, respectively), but homozygote TT in dominant model did not show a significant association with reduced CAD risk (adjusted OR = 0.98, 95% CI = 0.65-1.49).
Table 2.
Logistic regression analysis on the association between the rs9548934 C>T polymorphism in hsa-miR-1 binding site in the 3’UTR of COG6 and risk of CAD
| Genotypes | Controlsa (n = 610) | CAD (n = 1013) | Adjusted ORb (95% CI) | ||
|---|---|---|---|---|---|
|
|
|
||||
| N | % | N | % | ||
| hsa-miR-1 binding site in COG6 rs9548934 C→T | |||||
| CC | 200 | 32.8 | 451 | 44.5 | 1.00 |
| CT | 286 | 46.9 | 391 | 38.6 | 0.66 (0.49-0.89) |
| TT | 124 | 20.3 | 171 | 16.9 | 0.98 (0.65-1.49) |
| CT/TT | 410 | 67.2 | 562 | 55.5 | 0.74 (0.56-0.97) |
| C allele | 43.8 | 36.2 | |||
The observed genotype frequency among the control subjects was in agreement with the Hardy-Weinberg equilibrium (x2 = 1.377, P = 0.240).
Adjusted for age, sex, BMI and smoking status.
MiR-1 target gene C→T polymorphism and the hazard of MACE on the patients with CAD
We obtained genotypes and follow-up information for 664 CAD patients. Survival analyses showed that the differences of MACE hazards between carriers with rs9548934 TT, CT/TT and CC genotypes were statistically different (P < 0.001), but not for CT and CC genotypes (P = 0.085). Multivariate Cox proportional hazard regression analysis also showed that rs9548934 TT and CT/TT were significantly associated with MACE risks compared with CC wide genotype (adjusted HR = 0.24, 95% CI = 0.16-0.36 for TT; adjusted HR = 0.51, 95% CI = 0.39-0.68 for CT/TT) (Table 3).
Table 3.
Genotypes of hsa-miR-1 binding site rs9548934 C>T polymorphisms and the HR of the cardiovascular events on the patients with CAD
| Genotypes | Cases N | Events N | Log-rank P | Adjusted HRe (95% CI) | P |
|---|---|---|---|---|---|
| hsa-miR-1 binding site in COG6 rs9548934 C→Ta | |||||
| CC | 288 | 116 | < 0.001 | 1.00 | |
| CT | 242 | 103 | 0.78 (0.58-1.04) | 0.085 | |
| TT | 134 | 33 | 0.24 (0.16-0.36) | < 0.001 | |
| CT/TT | 376 | 136 | < 0.001 | 0.51 (0.39-0.68) | < 0.001 |
| CRP levels cut off by the median of the follow-up patients (mg/L)b | |||||
| < 3.80 | 321 | 65 | < 0.001 | 1.00 | |
| ≥ 3.80 | 322 | 181 | 3.44 (2.55-4.64) | < 0.001 | |
| Delta Ct of plasma miRNA-1 cut off by the median of the follow-up patientsc | |||||
| < 4.63 | 18 | 7 | 0.582 | 1.00 | 0.826 |
| ≥ 4.63 | 18 | 5 | 0.82 (0.14-4.77) | ||
| Numbers of pathological changes in coronary arteriesd,f | |||||
| 1 | 291 | 85 | < 0.001 | 1.00 | |
| 2 | 147 | 57 | 1.24 (0.87-1.75) | 0.235 | |
| 3 | 177 | 91 | 1.84 (1.36-2.50) | < 0.001 |
664 CAD patients were followed up with the detected genotype of rs9548934 and 252 patients obtained the CAD events.
643 CAD patients were followed up with the detected CRP levels and 246 patients obtained the CAD events.
36 CAD Patients were followed up with the detected plasma miRNA-1 levels and 12 patients obtained the CAD events.
615 CAD Patients were followed up with the detected plasma miRNA-1 levels and 233 patients obtained the CAD events.
Adjusted for age, sex, smoking status, TD, hypertension and numbers of pathological changes in coronary arteries.
Adjusted for age, sex, smoking status, TD, hypertension.
When the median of CRP levels and Delta Ct of plasma miRNA-1 in the follow-up patients were considered as the cut-off value, there was a significant increased risk of MACE in the higher CRP levels (≥ 3.8 mg/L) (adjusted HR = 3.44, 95% CI = 2.55-4.64), but there was no significantly decreased risk of serious prognosis of CAD in patients with higher Delta Ct of plasma miRNA-1 (≥ 4.63) (adjusted HR = 0.82, 95% CI = 0.14-4.77). Patients with three pathological changes in coronary arteries had an 84% increased risk of MACE, compared with those with one pathological change (adjusted HR = 1.84, 95% CI = 1.36-2.50).
As shown in Figure 2, cumulative hazard curves also showed that CAD patients carrying the COG6 rs9548934 CC, higher CRP level, and three pathological changes in coronary arteries had higher HR of MACE than others (Figure 2A, 2B, 2D). However, a higher delta Ct of plasma miR-1 variant was not associated with a significantly decreased hazard of MACE in CAD patients (Figure 2C).
Figure 2.
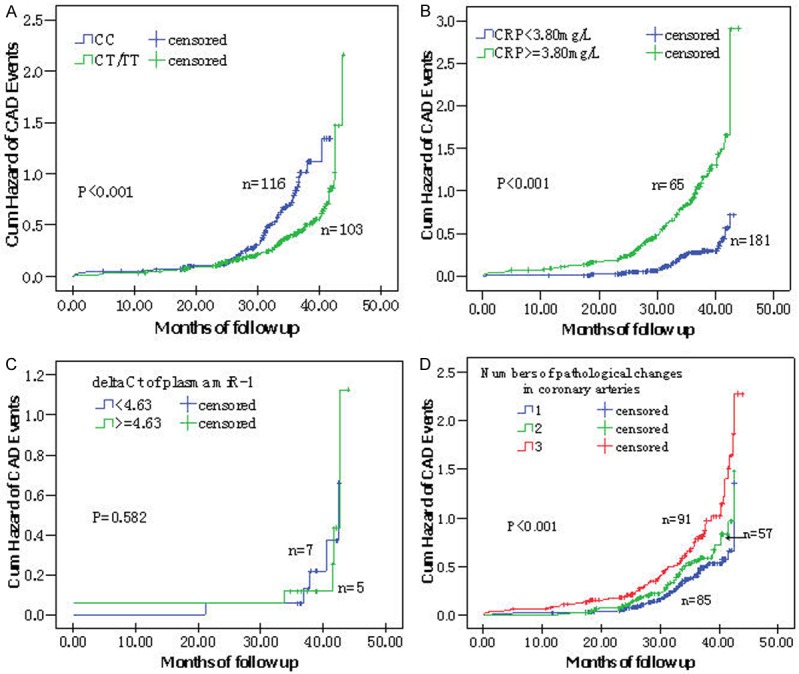
Kaplan-Meier survival curves of cardiovascular events in CAD patients with different genotypes, CRP levels, circulating miR-1 levels and numbers of pathological changes in coronary arteries. A: rs9548934 C→T, CC vs. CT/TT; B: CRP < 3.80 mg/L vs. ≥ 3.80 mg/L; C: Delta Ct of plasma miR-1 < 4.63 vs ≥ 4.63; D: Numbers of pathological changes in coronary arteries: 1 vs. 2 vs. 3.
Cox proportional hazard regression analysis on the cardiovascular events hazard of CAD patients
Both the univariate and multivariate Cox proportional hazard regression models were used to evaluate the correlation between variables including selected demographic characteristics, clinical features, and the miRNA SNPs and HR of MACE. Five variables (age, diabetes mellitus, COG6 rs9548934 C→T, CRP and numbers of pathological changes in coronary arteries,) were included in the both univariate and multivariate models, with a significance level of P < 0.05 for entering and P = 0.10 for removal from the model (Table 4). CAD patients with elder age, diabetes mellitus, rs9548934 CC genotype, higher CRP level and three pathological changes in coronary arteries had a higher HR of MACE.
Table 4.
Cox regression analysis on the risk factors of the prognosis of cardiovascular events on the patients with CAD
| Variables | β | SE | HR | 95% CI | P |
|---|---|---|---|---|---|
| Sex [1 = male, 2 = female] | 0.129 | 0.198 | 1.14 | 0.77-1.68 | 0.517 |
| Age | 0.017 | 0.008 | 1.02 | 1.00-1.03 | 0.034 |
| BMI (kg/m2) | 0.017 | 0.022 | 1.02 | 0.98-1.06 | 0.421 |
| TG (mmol/L) | -0.053 | 0.085 | 0.95 | 0.80-1.12 | 0.534 |
| TC (mmol/L) | -0.008 | 0.117 | 0.982 | 0.79-1.25 | 0.944 |
| HDL-C (mmol/L) | 0.225 | 0.222 | 1.25 | 0.81-1.93 | 0.310 |
| LDL-C (mmol/L) | -0.027 | 0.119 | 0.97 | 0.77-1.23 | 0.823 |
| Smoking status [0 = no, 1 = yes] | 0.146 | 0.171 | 1.16 | 0.83-1.62 | 0.396 |
| Diabetes Mellitus [0 = no, 1 = yes] | 0.446 | 0.143 | 1.56 | 1.18-2.07 | 0.002 |
| Hypertension [0 = no, 1 = yes] | -0.205 | 0.163 | 0.82 | 0.59-1.12 | 0.211 |
| rs9548934 CT/TT vs CC | -0.671 | 0.145 | 0.51 | 0.38-0.68 | <0.001 |
| CRP [lower than 3.80 mg/L = 0, more than 3.80 mg/L = 1] | 1.008 | 0.179 | 2.74 | 1.93-3.90 | <0.001 |
| Numbers of pathological changes in coronary arteries (1) [n = 2] | 0.234 | 0.181 | 1.26 | 0.89-1.80 | 0.197 |
| Numbers of pathological changes in coronary arteries (2) [n = 3] | 0.535 | 0.161 | 1.71 | 1.25-2.34 | 0.001 |
β: regression coefficient.
Discussion
Herein, we found that the genetic variant of miRNA-1 target gene COG6 binding site rs9548934C→T was associated with a significantly decreased risk and MACE of CAD, while the circulating miRNA-1 phenotype in CAD cases was slightly lower than that of controls but not statistically significant. In addition, age, diabetes mellitus, higher level of CRP and three pathological changes in coronary arteries were associated with the MACE of CAD.
MiRNA regulates gene expression by binding and modulating the translation of specific miRNAs [26], and it has been increasingly implicated in the control of various biological processes, including cell differentiation, cell proliferation, development and apoptosis, and many pathological processes such as cancer, diabetes, and cardiovascular disease [27-29]. Recently, miRNAs have been detected in serum and plasma, and circulating miRNA profiles have now been associated with a range of different tumor types [30,31], stroke and heart disease [32]. Several studies suggest that plasma/serum concentrations of miR-1 may be a useful indicator for AMI [33,34]. In this study, we detected the circulating expressions of miR-1 in CAD patients and controls, that level of miR-1 was lower in patients than in controls. Although it is not a significant finding (P = 0.090), the trend of miR-1 declines CAD patients was consisted with AMI [34]. Meanwhile, miR-1 expression level was significantly reduced in tumor tissues [35].
The presence of SNP within the 3’untranslated regions of target DNA gene could alter the binding with specific miRNAs, modulating gene expression, and ultimately could affect disease susceptibility. Evidence suggests that polymorphisms in miRNA-binding sites of genes are associated with increased risks of cancer, but few have demonstrated this effect on heart diseases. Lin et al, demonstrated that T allele genotypes (rs12537 CT and TT) of MTMR3, which was in miR-181a binding sites, were associated with significantly increased gastric risk and poor overall survival [36]. Naccarati et al, found that variants of miRNA-binding sites of nucleotide excision repair (NER) genes RPA2 rs7356 and GTF2H1 rs4596 were associated with colorectal cancer risk [10]. In this present study, we found that in the miR-1 target gene COG6 rs9548934 C→T variant, CT and CT/TT were associated with 34% and 26% significantly decreased risks of CAD, compared with the wide genotype CC, which suggests that polymorphism in microRNA target sites (PolymiRTSs) may be modulated in miR-1 targeting, whereas affected the mediate functions of miR-1.
C-reactive protein (CRP) is an acute phase protein which reflects a measure of the acute phase response. CRP is used as one of the markers of choice in monitoring the acute phase response because it increases to a relatively high concentration compared to basal concentration. Elevated plasma CRP has been suggested as a risk factor for ischemic stroke (IS) and coronary ischemic disease [37]. Huang et al, revealed that elevated plasma high sensitive CRP (> 3.0 mg/L) independently predicted risk of all-cause death within three months after acute IS in Chinese patients [38], while in this study, there was a significantly increased risk of cardiovascular events hazards in the Chinese patients with the higher CRP level (≥ 3.8 mg/L).
MiR-1 is being used in the oncology field to characterize tumors and predict the survival of cancer patients [39], while few studies were conducted on the prognosis of CAD. In this study, age, DM, higher level of CRP and three pathological changes in coronary arteries were associated with the increased risk of MACE, while the miR-1 target gene COG6 rs9548934 C→T variant was associated with decreased risk of MACE. This variant might be modulated the expression level of miR-1, which was associated with the decreased HR of MACE among CAD patients, but the biological mechanism of this regulation should be further studied.
Because small variation in the miRNA-binding sites of genes may have an effect on thousands of target mRNAs and result in diverse functional consequences, the most common genetic variation, SNPs, in miRNA sequences may also be functional and therefore may represent ideal candidate biomarkers for diseases prognosis. This is the first study to our knowledge to describe miR-1-binding site of gene COG6 rs9548934 C→T and CAD risk and prognosis outcome with a relatively large study population size and a high statistical power (despite the number of controls was less than that of cases--if we set α=0.05, based on our data set for COG6 rs9548934 TT, we have an 80% power to detect an OR of 0.72). Nevertheless, some limitations should be taken into consideration. First, the follow-up period was relatively short for some patients. Second, the mechanisms by which the miR-1 and its target mRNAs regulate CAD progression are unknown. This subject warrants further in vitro and in vivo studies, and further characterization of SNPs of miRNAs would improve our understanding of miRNA biogenesis and the potential contribution of these SNPs to disease development and prognosis, which may also favor therapeutic interventions.
Acknowledgements
This study was partly supported by the National Natural Science Foundation of China (30901230, 81273143).
Disclosure of conflict of interest
None.
References
- 1.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Xu CQ, He Q, Cai JP, Li XC, Wang D, Xiong X, Liao YH, Zeng QT, Yang YZ, Cheng X, Li C, Yang R, Wang CC, Wu G, Lu QL, Bai Y, Huang YF, Yin D, Yang Q, Wang XJ, Dai DP, Zhang RF, Wan J, Ren JH, Li SS, Zhao YY, Fu FF, Huang Y, Li QX, Shi SW, Lin N, Pan ZW, Li Y, Yu B, Wu YX, Ke YH, Lei J, Wang N, Luo CY, Ji LY, Gao LJ, Li L, Liu H, Huang EW, Cui J, Jia N, Ren X, Li H, Ke T, Zhang XQ, Liu JY, Liu MG, Xia H, Yang B, Shi LS, Xia YL, Tu X, Wang QK. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- 3.Bampali K, Mouzarou A, Lamnisou K, Babalis D. Genetics and coronary artery disease: present and future. Hellenic J Cardiol. 2014;55:156–163. [PubMed] [Google Scholar]
- 4.Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet. 2002;30:210–214. doi: 10.1038/ng827. [DOI] [PubMed] [Google Scholar]
- 5.Link N, Tanner M. Coronary artery disease: Part 1. Epidemiology and diagnosis. West J Med. 2001;174:257–261. doi: 10.1136/ewjm.174.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winter J, Diederichs S. MicroRNA biogenesis and cancer. Methods Mol Biol. 2011;676:3–22. doi: 10.1007/978-1-60761-863-8_1. [DOI] [PubMed] [Google Scholar]
- 7.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya A, Ziebarth JD, Cui Y. PolymiRTS Database 3.0: linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014;42:D86–91. doi: 10.1093/nar/gkt1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, Chen F, Gu J, Zhang W, Luo J, Guan X. Genetic variant rs1058240 at the microRNA-binding site in the GATA3 gene may regulate its mRNA expression. Biomed Rep. 2014;2:404–407. doi: 10.3892/br.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, Levy M, Polakova V, Lipska L, Vodicka P. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346–1351. doi: 10.1093/carcin/bgs172. [DOI] [PubMed] [Google Scholar]
- 11.Nossent AY, Hansen JL, Doggen C, Quax PH, Sheikh SP, Rosendaal FR. SNPs in microRNA binding sites in 3’-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am J Hypertens. 2011;24:999–1006. doi: 10.1038/ajh.2011.92. [DOI] [PubMed] [Google Scholar]
- 12.Mallick B, Ghosh Z. A complex crosstalk between polymorphic microRNA target sites and AD prognosis. RNA Biol. 2011;8:665–673. doi: 10.4161/rna.8.4.15584. [DOI] [PubMed] [Google Scholar]
- 13.Sethupathy P, Borel C, Gagnebin M, Grant GR, Deutsch S, Elton TS, Hatzigeorgiou AG, Antonarakis SE. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3’ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am J Hum Genet. 2007;81:405–413. doi: 10.1086/519979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidersbach A, Saxby C, Carver-Moore K, Huang Y, Ang YS, de Jong PJ, Ivey KN, Srivastava D. microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife. 2013;2:e01323. doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei Y, Peng S, Wu M, Sachidanandam R, Tu Z, Zhang S, Falce C, Sobie EA, Lebeche D, Zhao Y. Multifaceted roles of miR-1s in repressing the fetal gene program in the heart. Cell Res. 2014;24:278–292. doi: 10.1038/cr.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He B, Xiao J, Ren AJ, Zhang YF, Zhang H, Chen M, Xie B, Gao XG, Wang YW. Role of miR-1 and miR-133a in myocardial ischemic postconditioning. J Biomed Sci. 2011;18:22. doi: 10.1186/1423-0127-18-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 18.Widera C, Gupta SK, Lorenzen JM, Bang C, Bauersachs J, Bethmann K, Kempf T, Wollert KC, Thum T. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51:872–875. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 19.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, Rubino M, Carena MC, Spazzafumo L, De Simone M, Micheli B, Biglioli P, Achilli F, Martelli F, Maggiolini S, Marenzi G, Pompilio G, Capogrossi MC. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C, Weber M, Hamm CW, Roxe T, Muller-Ardogan M, Bonauer A, Zeiher AM, Dimmeler S. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 22.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 23.Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J, Li K, Yu B, Li Z, Wang R, Wang L, Li Q, Wang N, Shan H, Li Z, Yang B. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391:73–77. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Ziebarth JD, Bhattacharya A, Chen A, Cui Y. PolymiRTS Database 2.0: linking polymorphisms in microRNA target sites with human diseases and complex traits. Nucleic Acids Res. 2012;40:D216–221. doi: 10.1093/nar/gkr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- 26.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107:1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alshalalfa M, Bader GD, Bismar TA, Alhajj R. Coordinate microRNA-mediated regulation of protein complexes in prostate cancer. PLoS One. 2013;8:e84261. doi: 10.1371/journal.pone.0084261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng B, Cao Y, Chen S, Ruiz M, Chakrabarti S. miRNA-1 regulates endothelin-1 in diabetes. Life Sci. 2014;98:18–23. doi: 10.1016/j.lfs.2013.12.199. [DOI] [PubMed] [Google Scholar]
- 29.Catalucci D, Gallo P, Condorelli G. MicroRNAs in cardiovascular biology and heart disease. Circ Cardiovasc Genet. 2009;2:402–408. doi: 10.1161/CIRCGENETICS.109.857425. [DOI] [PubMed] [Google Scholar]
- 30.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7:e44398. doi: 10.1371/journal.pone.0044398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan CS, Wang CW, Tan KS. Circulatory microRNA-145 expression is increased in cerebral ischemia. Genet Mol Res. 2012;11:147–152. doi: 10.4238/2012.January.27.1. [DOI] [PubMed] [Google Scholar]
- 33.Long G, Wang F, Duan Q, Chen F, Yang S, Gong W, Wang Y, Chen C, Wang DW. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci. 2012;8:811–818. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ali R, Huang Y, Maher SE, Kim RW, Giordano FJ, Tellides G, Geirsson A. miR-1 mediated suppression of Sorcin regulates myocardial contractility through modulation of Ca2+ signaling. J Mol Cell Cardiol. 2012;52:1027–1037. doi: 10.1016/j.yjmcc.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Sasaki K, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N. Identification of novel molecular targets regulated by tumor suppressive miR-1/miR-133a in maxillary sinus squamous cell carcinoma. Int J Oncol. 2011;39:1099–1107. doi: 10.3892/ijo.2011.1096. [DOI] [PubMed] [Google Scholar]
- 36.Lin Y, Nie Y, Zhao J, Chen X, Ye M, Li Y, Du Y, Cao J, Shen B, Li Y. Genetic polymorphism at miR-181a binding site contributes to gastric cancer susceptibility. Carcinogenesis. 2012;33:2377–2383. doi: 10.1093/carcin/bgs292. [DOI] [PubMed] [Google Scholar]
- 37.Ramamoorthy RD, Nallasamy V, Reddy R, Esther N, Maruthappan Y. A review of C-reactive protein: A diagnostic indicator in periodontal medicine. J Pharm Bioallied Sci. 2012;4:S422–426. doi: 10.4103/0975-7406.100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Jing J, Zhao XQ, Wang CX, Wang YL, Liu GF, Wang CJ, Liu LP, Yang XM, Jiao Y, Jiao Y, Wang LS, Wang YJ, Gu WK. High-sensitivity C-reactive protein is a strong risk factor for death after acute ischemic stroke among Chinese. CNS Neurosci Ther. 2012;18:261–266. doi: 10.1111/j.1755-5949.2012.00296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB, Dorsey TH, Stephens RM, Croce CM, Ambs S. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]


