Abstract
Zinc is an essential trace element for human health and is a critical component of many proteins and transcription factors involved in DNA damage response and repair. The prostate is known to accumulate high levels of zinc, but levels are markedly decreased with cancer development. We hypothesized that zinc plays a critical role in maintaining DNA integrity in the prostate and zinc deficiency would lead to increased DNA damage and altered DNA damage response mechanisms. To test this hypothesis, the goal of this study was to determine the effects of zinc deficiency on DNA damage and DNA repair mechanisms by examining changes in global gene expression and transcription factor binding abilities in normal prostate epithelial cells (PrEC). Increased single-strand DNA breaks (Comet assay) were observed in PrEC grown in zinc-deficient media compared with cells grown in zinc-adequate media for 7 d. Using Affymetrix HG-U133A gene chips, differential expression of genes involved in cell cycle, apoptosis, transcription, and DNA damage response and repair were identified with low cellular zinc. Among genes involved in DNA damage response and repair were identified with low cellular zinc. Among genes involved in DNA damage response and repair, tumor protein p73,,tumor protein p73,MRE11 meiotic recombination 11 homolog A,X-ray repair complementing defective repair in Chinese hamster cells 4, and breast cancer 2, early onset were down-regulated and TP53 was up-regulated. Additionally, western blotting showed increased nuclear p53 protein expression with zinc deficiency. Despite increased p53 gene and nuclear protein expression, there was no significant change in p53 binding activity. Zinc deficiency also induced an increase in binding activity of transcription factors involved in regulating cell proliferation and apoptosis. Thus, zinc deficiency may compromise DNA integrity in the prostate by impairing the function of zinc-containing proteins.
Introduction
Zinc is an essential mineral that is integral to many proteins and transcription factors that regulate key cellular functions such as the response to oxidative stress, DNA replication, DNA damage repair, cell cycle progression, and apoptosis. In particular, several proteins involved in DNA damage signaling and repair, replicative enzymes, such as DNA and RNA polymerases, and transcription factors, such as tumor protein p53 (p53),4 require zinc for proper function (1–3). Consequently, zinc deficiency could disrupt the function of both signaling molecules and proteins directly involved in DNA replication and repair. Limited availability of cellular zinc due to zinc deficiency could result in loss of activity of these zinc-dependent proteins involved in the maintenance of DNA integrity and may contribute to the development of cancer. In vitro and in vivo studies have revealed that zinc deficiency leads to increased oxidative stress and DNA damage (4–6). Zinc deficiency has also been shown to upregulate expression of the tumor suppressor protein, p53, but impair the DNA binding abilities of p53, nuclear factor κ B (NFκB), and AP-1 transcription factors in rat glioma C6 cells (7). These studies suggest that a decrease in cellular zinc alone causes DNA damage and impairs DNA damage response mechanisms, resulting in a loss of DNA integrity and potential for increased cancer risk.
The prostate contains the highest concentrations of zinc compared with other soft tissues in the body (8). Interestingly, as prostate cells develop cancer, zinc concentrations decrease by 60–70% (9).Moreover, the main region of zinc accumulation in the prostate, the peripheral zone, is also the main region of prostate cancer development (9). Although the connections between prostate zinc concentrations and prostate cancer have been well established, the precise function of zinc in the prostate remains unknown. We hypothesized that zinc deficiency would disrupt the function of critical zinc dependent proteins that maintain DNA integrity in prostate cells, ultimately resulting in increased DNA damage.
To further understand the role of zinc in maintaining DNA integrity in the prostate, we used genomic approaches to examine the molecular changes that occur with zinc deficiency. The goal of this study was to investigate the impact of zinc deficiency on DNA damage and subsequent alterations in gene expression and protein function by examining global gene expression changes and alterations in transcription factor binding activities using microarray and transcription factor array technologies in normal prostate epithelial cells (PrEC). These studies will offer insight into the function of zinc in the maintenance of DNA integrity as well as help identify zinc-regulated genes and zinc-dependent transcription factors in the prostate.
Methods
Cell culture
Clonetics normal human PrEC were purchased from Cambrex and maintained in Prostate Epithelial Cell medium (PrEGM) (Cambrex). Custom made zinc-deficient media (ZnDF) was purchased from Cambrex. Zinc-adequate media (ZnAD) were prepared by adding 0.864 g/L (3 µmol/L) zinc sulfate heptahydrate to ZnDF, which was equivalent to zinc concentration in normal PrEGM media (3 µmol/L zinc). Zinc concentrations in normal, ZnDF, and zinc-free plus 3 µmol/L zinc (ZnAD) media were 0.027, 0.006, and 0.026 mg/L, respectively. PrEC were seeded at 3000 cells/cm2 in T75 flasks and allowed to attach overnight. The cells were maintained in ZnDF or ZnAD media for 7 d, with fresh media replaced every third day. Following the 7 d treatment period, cells were harvested by trypsinization and cell pellets were stored at −80°C for further analysis.
Inductively coupled plasma-optical emission spectroscopy
Zinc levels in ZnDF and ZnAD cells and media were determined using inductively coupled plasma-optical emission spectroscopy (ICP-OES). Cell pellets containing 5 million cells, or 1 mL media, were vortexed with 1 mL 70% ultrapure nitric acid (EMD Omnitrace) overnight. Following incubation, samples were diluted with chelex-treated nanopure water to a 7% acid solution, centrifuged, and analyzed by the Prodigy High Dispersion ICP-OES instrument (Teledyne Leeman Labs) against known standards.
Comet assay
Detection of single-strand DNA breaks were determined by alkali single-cell gel electrophoresis as described by Singh (10). Cells were suspended in 0.5% agarose and applied to microscope slides. Cells were subsequently lysed in Comet lysis buffer (Trevigen) for 1 h, subjected to alkali buffer for 20 min, and underwent electrophoresis. Nuclear material was stained with Sybr-green (Molecular Probes). Fifty cells from 4 independent samples were scored without knowledge of the treatments on a scale of 0–4 for tail migration intensity.
RNA isolation and microarray
Total RNA was isolated from ZnDF and ZnAD PrEC cells using the Qiagen RNeasy Mini kit. RNA integrity was determined using OD260/280 ratios and agarose gel electrophoresis with ethidium bromide staining. Global alterations in RNA transcripts from ZnDF and ZnAD cells were determined using the Affymetrix human genome HG-U133A GeneChip (Affymetrix). Three replicates per treatment group were performed. For microarray analysis, RNA samples were sent to the Center for Genome Research and Biocomputing Core Laboratories at Oregon State University, Corvallis, OR for RNA integrity screening, probe synthesis, hybridization, and scanning according to the GeneChip Expression Analysis technical manual (701021 Rev. 5).
Array data analysis
GeneSifter software (VizX Labs) was used for all analysis of microarray data. Array data were normalized to the all median signal intensity value for each experiment (each gene chip) and signal values were log base 2 transformed. Genes that did not have present calls for all 3 replicates in at least 1 treatment group were omitted. Fold-change was calculated and genes with ≥2- or ≤2-fold change in the ZnDF group compared with ZnAD group and a P-value <0.05 after the t test was applied were identified as differentially expressed by zinc deficiency. Genesifter grouped differentially expressed genes according to gene function using Gene Ontology and KEGG databases.
Real-time quantitative PCR
P53, Calpain 6, tyrosine 3-monooxygenase, nuclear factor of κ light polypeptide gene enhancer in B-cells 2 (NFκB2), ribosomal P2, myeloid cell leukemia sequence 1 (BCL-2 related), metallothionein 1 (MT-1), cyclin-dependent kinase inhibitor 1A (p21), BCL2-associated X protein (BAX), transformed 3T3 cell double minute 2, p53 binding protein (mouse) (Mdm2), growth arrest and DNA-damage-inducible, α (GADD45), and insulin-like growth factor binding protein 3 (IGF-BP3) were analyzed by quantitative PCR (qPCR). Five micrograms total RNA was reverse transcribed to cDNA using SuperScript First Strand Synthesis system for RT-PCR (Invitrogen). Primer sequences and annealing temperatures are listed in Supplemental Table 1. Presence of double-stranded PCR product was monitored using DyNAmo SYBR Green qPCR kit (New England Biolabs) using the Chromo4 Real Time PCR detections system (MJ Research). Melting curve analysis and agarose gel electrophoresis with ethidium bromide staining was conducted to ensure single PCR product of correct amplicon length. Each sample was run in triplicate. Normalized intensity was determined using the standard curve method. Fold-change for each gene was assessed after normalization of intensity value to β-actin (ACTB).
Western blot
Nuclear extracts were isolated from ZnDF and ZnAD PrEC cells using NE-PER Nuclear Cytoplasmic Extraction Reagents (Pierce). Standard Western blot procedure was preformed using 20 µg nuclear extract loaded and separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred onto nitrocellulose membrane (BioRad Laboratories). Antibody dilutions were as follows: p53, 1:1000 [p53 (DO-1), Santa Cruz Biotechnologies]; ACTB, 1:4000 (A5441, Sigma-Aldrich); and secondary antibody concentrations were 1:20,000 (goat anti-mouse IgG-horseradish peroxidase, Santa Cruz). Detection was by SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) with image analysis on an AlphaInnotech photodocumentation system. Quantification of signal intensity was determined using Image J 1.37v (NIH) software. Triplicate samples for each treatment group were analyzed.
Transcription factor array
Transcription factor binding activity was analyzed using Panomics Protein/DNA Array I. Nuclear extracts were prepared as described previously and hybridized to the Protein/DNA Array I membrane as directed by the manufacturer. The membrane was spotted with 54 different consensus-binding sequences, each corresponding to a different transcription factor or family of transcription factors. Chemiluminescence detection of consensus sequence binding was obtained with Hyperfilm ECL. Quantification of signal intensity was determined using Image J software. Triplicate Arrays were conducted per treatment group. Fold-change was calculated as the ratio of the signal intensity as ZnDF:ZnAD for each spot on the membrane.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) for p53 was analyzed with the Odyssey Infrared Imaging System (LI-COR Biosciences) using the p53 IRDye 700 Infrared Dye Labeled Oligonucleotides as directed by the manufacturer. Briefly, 5 µg nuclear extract, 50 fmol p53 oligo IRDye 700 Infrared Dye, 1 µg poly [poly(2′-deoxyinosinic-2′-deoxycytidylic acid)], 2 µL 10× binding buffer (1 mol/L Tris, 1 mol/L KCl, 1 mol/L dithiothreitol, and 500 mmol/L EDTA), 2 µL 25mmol/L dithiothreitol plus 2.5% Tween-20, and 2 µL loading dye was incubated at room temperature in a total reaction volume of 20 µL for 30 min. For specific competitor reaction, the sample was incubated for 10 min with 5 pmol of unlabeled p53 oligo before addition of 50 fmol of labeled probe. Reaction mixture was separated on a 6% acrylamide gel at 150 V for 2 h and then imaged and quantified using the Odyssey Infrared Imaging System.
Statistics
Statistical analysis of microarray data were performed using GeneSifter software as described above. Western blot and EMSA were analyzed by unpaired t test using GraphPad Prism Version 4.01. For the transcription factor array, T-ratio was calculated using the mean fold-change and was compared with 1, representing no change in transcription factor binding ability. The critical 2-tailed P-value was determined using T-distribution table with α = 0.05. Values in the text are means ± SEM.
Results
Zinc concentrations and DNA damage
PrEC grown in ZnDF media for 7 d had significantly lower zinc levels compared with PrEC grown in ZnAD media. Zinc concentrations in ZnAD and ZnDF cells were 0.1969 ± 0.0211 and 0.0479 ± 0.0083 nmol/million cells in ZnDF cells. This represented a 75% decrease, confirming the loss of cellular zinc (P < 0.001). No effects on cell growth and confluency were observed in ZnDF cells (data not shown). Comet scores in ZnDF cells (1.38 ± 0.065) were more than twice that in ZnAD cells (0.640 ± 0.028; P < 0.05), indicating an increase in single-strand DNA breaks with loss of cellular zinc.
Microarray and qPCR
A total of 286 of ~22,000 genes represented on the Affymetrix HG-U133A gene chip were significantly differentially expressed at least 2-fold with zinc deficiency. Of these genes, 146 genes were down-regulated (Supplemental Table 2) by zinc deficiency and 140 genes were up-regulated by zinc deficiency (Supplemental Table 3). Further analysis using Genesifter showed that differentially expressed genes were involved in a variety of biological processes, such as cellular metabolism, regulation of physiological processes, transport, cell organization and biogenesis, cell growth, cell homeostasis, and response to extracellular stimuli (Supplemental Fig. 1). Microarray data revealed none of the well-characterized genes involved in regulating zinc homeostasis were differentially expressed. Transcription of metallothionein, which is known to be largely controlled by zinc levels (11), was not differentially expressed. qPCR analysis showed that MT-1 expression tended to be lower in ZnDF than ZnAD cells (P = 0.06; Table 1).
TABLE 1.
qPCR analysis of PrEC treated with ZnAD or ZnDF media for 7 d
| Gene | qPCR fold-change1 |
Genesifter fold-change |
Gene | qPCR fold-change2 |
|---|---|---|---|---|
| p53 | 1.91 ± 0.03 | 4.83 | p21 | 1.45 ± 0.23 |
| Calpain 6 | 2.24 ± 0.12 | 3.49 | BAX | 1.13 ± 0.25 |
| Tyrosine 3 monooxygenase | 2.18 ± 0.38 | 2.48 | GADD45 | 1.11 ± 0.11 |
| Cyclin T1 | 1.43 ± 0.32 | 3.84 | MDM2 | 1.50 ± 0.45 |
| NFKB2 | −1.54 ± 0.09 | −2.6 | IGF-BP3 | 0.992 ± 0.05 |
| Ribosomal P2 | 1.74 ± 0.44 | −2.24 | MT-1 | 0.5767 ± 0.07 |
| Myeloid cell leukemia sequence 1 (BCL2-related) | −1.23 ± 0.09 | −2.41 |
Mean fold-change ± SEM, calculated from qPCR analysis, representing 3 biological replicates. Intensity values were normalized to ACTB and fold-change was calculated as ZnDF/ZnAD (using normalized intensity values).
Downstream targets of p53 analyzed by qPCR.
To gain a better understanding of the relationship between zinc deficiency and the increased DNA damage observed in these cells, we focused on down-regulated (Table 2) and up-regulated (Table 3) genes associated with DNA damage response and repair, cell cycle regulation, and apoptosis that were identified using gene ontology and KEGG pathway databases. There were 18 genes involved in cell cycle and proliferation, 9 involved in apoptosis and cell death, and 6 involved in DNA damage response and DNA repair. Genes involved in cell cycle included p53, cyclin T1, and cell division cycle 25A, which were upregulated, and adenomatosis polyposis coli, breast cancer 2, early onset (BRCA2), growth factor independent 1B, meiotic recombination 11 homolog A (MRE11A), and tumor protein p73 (p73), which were downregulated. Genes involved in apoptosis and cell death included p53, adrenergic, α-1-A, receptor, BCL2-associated athanogene 5, and nucleolar protein 3, which were upregulated; and complement component 7, FOS-like antigen 2, Harakiri, BCL2-interacting protein, p73, and myeloid cell leukemia sequence 1, which were down-regulated. Genes such as p53, p73, BRCA2, and MRE11A have overlapping functions as they are also involved in DNA damage response and repair. Additionally, the gene, X-ray repair complementing defective repair in Chinese hamster cells 4 (XRCC4) was also downregulated. XRCC4 is also involved in DNA repair.
TABLE 2.
A list of selected genes that were downregulated at least 2-fold in PrEC treated with ZnDF compared with ZnAD media for 7 d
| Fold-change | Gene identifier |
Gene name (downregulated genes) |
|---|---|---|
| Cell cycle | ||
| −2.44 | S67788 | Adenomatosis polyposis coli |
| −4.47 | NM_000059 | BRCA2 |
| −2.77 | NM_004188 | Growth factor independent 1B |
| −4.94 | AK026910 | Ciliary rootlet coiled-coil, rootletin |
| −2.05 | BC005241 | Meiotic recombination (S. cerevisiae) 11 homolog A |
| −2.36 | NM_005427 | p73 |
| Cell proliferation | ||
| −4.76 | U01134 | Fms-related tyrosine kinase 1 |
| −2.77 | NM_004188 | Growth factor independent 1B |
| −2.04 | NM_017409 | Homeo box C10 |
| −4.78 | NM_000590 | Interleukin 9 |
| −4.38 | NM_002309 | Leukemia inhibitory factor (cholinergic differentiation factor) |
| Apoptosis/cell death | ||
| −3.68 | NM_000587 | Complement component 7 |
| −2.65 | NM_005253 | FOS-like antigen 2 |
| −5.9 | U76376 | Harakiri, BCL2-interacting protein (contains only BH3 domain) |
| −2.36 | NM_005427 | p73 |
| −2.41 | H71805 | Myeloid cell leukemia sequence 1 (BCL2-related) |
| DNA damage response and DNA repair | ||
| −2.36 | NM_005427 | p73 |
| −2.05 | BC005241 | MRE11A (S. cerevisiae) |
| −2.51 | AB017445 | XRCC4 |
| −4.47 | NM_000059 | BRCA2 |
| Transcription | ||
| −2.04 | NM_017409 | Homeo box C10 |
| −2.18 | AL136823 | Thyroid hormone receptor coactivating protein |
| −2.18 | NM_006914 | RAR-related orphan receptor B |
| −2.21 | AF061192 | Ectodermal dysplasia 1, anhidrotic |
| −2.29 | NM_000280 | Paired box gene 6 (aniridia, keratitis) |
| −2.36 | NM_005427 | p73 |
| −2.39 | BC001161 | Zinc finger protein 174 |
| −2.6 | U09609 | Nuclear factor of κ light polypeptide gene enhancer in B-cells 2 |
| −2.65 | NM_005253 | FOS-like antigen 2 |
| −2.77 | NM_004188 | Growth factor independent 1B |
| −2.78 | AL117663 | Transcription factor 3 |
| −4.47 | NM_000059 | BRCA2 |
| −4.92 | U11701 | LIM homeobox protein 2 |
| −5.47 | NM_002196 | Insulinoma-associated 1 |
TABLE 3.
A list of selected genes that were upregulated at least 2-fold in PrEC treated with ZnDF compared with ZnAD media for 7 d
| Fold-change | Gene identifier | Gene name (upregulated genes) |
|---|---|---|
| Cell cycle | ||
| 4.83 | K03199 | p53 (Li-Fraumeni syndrome) |
| 3.84 | NM_001240 | Cyclin T1 |
| 2.48 | AI343459 | Cell division cycle 25A |
| Cell proliferation | ||
| 4.83 | K03199 | p53 (Li-Fraumeni syndrome) |
| 3.74 | NM_000618 | Insulin-like growth factor 1 (somatomedin C) |
| 3.68 | U02569 | Adrenergic, alpha-1A-, receptor |
| 2.48 | AI343459 | Cell division cycle 25A |
| Apoptosis/cell death | ||
| 4.83 | K03199 | p53 (Li-Fraumeni syndrome) |
| 3.68 | U02569 | Adrenergic, alpha-1A-, receptor |
| 3.62 | AA457021 | BCL2-associated athanogene 5 |
| 2.26 | AF043244 | Nucleolar protein 3 (apoptosis repressor with CARD domain) |
| DNA damage response and DNA repair | ||
| 4.83 | K03199 | p53 (Li-Fraumeni syndrome) |
| 4.59 | NM_005692 | ATP-binding cassette, sub-family F(GCN20), member 2 |
| Transcription | ||
| 7.35 | X79990 | Core-binding factor, runt domain, alpha subunit 2; translocated to, 1; cyclin D-related |
| 5.23 | AW589975 | Sin3A-associated protein, 30kDa |
| 4.83 | K03199 | p53 (Li-Fraumeni syndrome) |
| 4.59 | NM_005692 | ATP-binding cassette, sub-family F(GCN20), member 2 |
| 3.84 | NM_001240 | Cyclin T1 |
| 3.09 | NM_003153 | Signal transducer and activator of transcription 6, interleukin-4 induced |
| 2.74 | AF072814 | Metal response element binding transcription factor 2 |
| 2.7 | NM_002919 | Regulatory factor X, 3 (influences HLA class II expression) |
| 2.63 | BE675435 | Kruppel-like factor 6 |
| 2.37 | NM_002135 | Nuclear receptor subfamily 4, group A, member 1 |
| 2.32 | NM_003441 | Zinc finger protein 141 (clone pHZ-44) |
| 2.18 | AF231056 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily f, member 1 |
| 2 | NM_000937 | Polymerase (RNA) II (DNA directed) polypeptide A (220 kDa) |
Additionally, several genes that encode for transcription factors were also differentially expressed due to zinc deficiency. NFκB2, Fos-like antigen 2, and TP73 were downregulated, whereas p53, metal responsive element binding factor 2, and Kruppel-like factor 6 were upregulated with zinc deficiency. Furthermore, genes directly involved in transcription such as the AT-rich interactive domain 1A (SWI-like), which encodes a protein that is part of the chromatin remodeling complex (SNF/SWI), and polymerase (RNA) II (DNA) directed polypeptide A, which encodes for the protein that makes up the largest subunit of RNA polymerase II, were also upregulated with zinc deficiency. Overall, several key genes that regulate cell cycle progression, apoptosis, DNA damage response and repair, as well as genes that function in transcription and as transcriptional regulators were influenced by zinc deficiency.
Gene expression changes of p53, Calpain 6, tyrosine 3-monooxygenase, NFκB2, and myeloid cell leukemia sequence 1 (BCL-2 related) were confirmed by qPCR. ACTB was chosen to normalize copy numbers before assessment of fold-change, because microarray and qPCR analysis of ACTB showed no change in mRNA expression with zinc deficiency in PrEC cells. The direction and magnitude of the fold-changes found with qPCR for these genes were similar to microarray results, with the exception of ribosomal P2 [p53 and Calpain 6, P < 0.05; tyrosine 3-monooxygenase, P = 0.08; NFκB2, P = 0.06; myeloid cell leukemia sequence 1 (BCL-2 related), P = 0.16; and ribosomal P2, P = 0.23] (Table 1).
Nuclear p53 protein expression
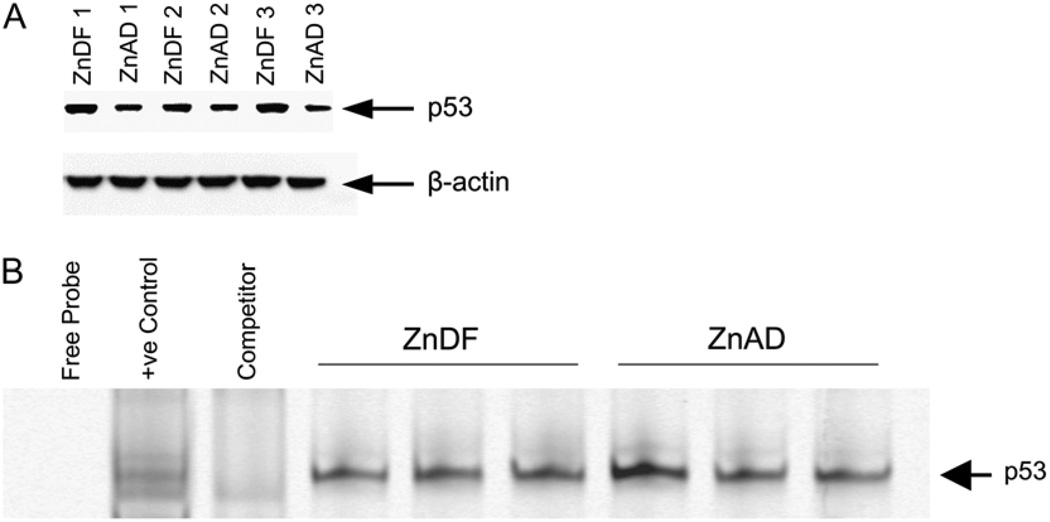
Western blot analysis revealed a significant increase in nuclear p53 expression in ZnDF PrEC compared with ZnAD PrEC. After normalization to ACTB, the OD for ZnDF cells (0.8606 ± 0.0932) was higher than that for ZnAD cells (0.4386 ± 0.0406; P < 0.05) (Fig. 1A). Importantly, the expression of targets downstream of p53, including p21, BAX, GADD45, Mdm2, and IGF-BP3, showed no significant change in expression with zinc deficiency by qPCR analysis (Table 3).
Figure 1.
Western blot and EMSA for p53 from nuclear proteins obtained from PrEC cells treated with ZnAD or ZnDF media for 7 d. A, Western blot for nuclear p53 protein. Triplicate samples, representing 3 independent experiments, for each treatment were analyzed. B, EMSA of p53 using nuclear extracts from ZnAD and ZnDF PrEC cells. Positive control represents HeLa extracts and specific competitor reaction (competitor) represents HeLa extracts preincubated with unlabeled p53 oligonucleotides. Three samples per treatment group were analyzed.
Transcription factor binding activity
The DNA binding ability of 54 different transcription factors was simultaneously assessed using Transcription Factor Array technology. The consensus sequences on the array were spotted in duplicate as well in 2 different dilutions. Table 4 summarizes transcription factors that showed at least 2-fold significant changes in binding activity and representative membranes hybridized with nuclear extracts from ZnDF and ZnAD cells are depicted in Supplemental Figure 2. The transcription factor with the highest change in binding activity was retinoid X receptor (RXR), which increased 4.5-fold in binding activity due to zinc deficiency. Other transcription factors with increased activity due to zinc deficiency included Sp1, vitamin D receptor, POU domain, class 4, transcription factor 1 (Brn-3), signal transducer and activator of transcription 4 (Stat4), cAMP responsive element-binding protein (CREB), forkhead box H1 (FAST-1), MAD, mothers against decapentaplegic homolog 3/4 (Smad3/4), and thyroid hormone receptor. Additionally, binding to the serum-inducible element responsive factor, heat shock transcription factor, and GATA binding protein (globin transcription factor) (GATA) consensus sequences on the array were also increased with zinc deficiency. Importantly, Transcription Factor Array data did not indicate a change in p53 binding activity with zinc deficiency. Further analysis of p53 binding activity by EMSA confirmed p53 binding activity in ZnDF (integrated intensity 34.27 ± 1.18) did not increase compared with ZnAD cells (41.07 ± 7.96) (Fig. 1B) despite increased gene and nuclear protein expression, indicating possible p53 dysfunction with zinc deficiency.
TABLE 4.
Transcription factors that showed >2-fold increase in binding activity that differed significantly between ZnDF and ZnAD PrEC cells
| Mean ratio1 | T-ratio2 | Transcription factor | Description |
|---|---|---|---|
| 2.015 | 9.47 | SIERF | Serum inducible element responsive factor |
| 2.027 | 5.12 | VDR | VDR: vitamin D receptor |
| 2.158 | 3.45 | Brn-3 | POU4F1: POU domain, class 4, transcription factor 1 |
| 2.243 | 6.87 | Sp1 | SP1: Sp1 transcription factor |
| 2.265 | 4.29 | TR(2) | Thyroid hormone receptor |
| 2.270 | 3.72 | Stat4 | Signal transducer and activator of transcription 4 |
| 2.362 | 2.61 | c-Myb | MYB: v-myb myeloblastosis viral oncogene homolog |
| 2.364 | 4.46 | CREB | CREB1 |
| 2.808 | 3.38 | FAST-1 | FOXH1 |
| 2.887 | 2.73 | Ets-1/PEA3 | ETS-domain transcription factor pea3 |
| 2.906 | 4.58 | TR(1) | Thyroid hormone receptor |
| 3.246 | 2.59 | CDP | CCAAT displacement protein |
| 3.486 | 2.85 | Smad3/4 | MADH3/4: MAD, mothers against decapentaplegic homolog 3/4 |
| 4.086 | 3.93 | HSF | Heat shock transcription factor |
| 4.187 | 4.96 | GATA | GATA: GATA binding protein (globin transcription factor) |
| 4.451 | 3.52 | RXR | RXR |
Ratio calculated as intensity of ZnDF:ZnAD.
A T-ratio value of ≥2.571 indicates the mean fold-change was different from 1, P<0.05.
Discussion
The precise function of zinc in the prostate and the effects of zinc deficiency on the molecular and cellular processes in the prostate are relatively unknown. This study demonstrated that decreased cellular zinc in PrEC resulted in increased single-strand DNA breaks and differential expression of genes involved in cell cycle progression, apoptosis, transcription, and DNA damage response and repair. RNA transcript and nuclear protein expression of p53 was upregulated with decreased cellular zinc, but the DNA binding ability of p53 was compromised, resulting in impaired signaling of downstream p53 targets responsible for mediating the DNA damage response. These data were consistent with our hypothesis that zinc plays an essential function in maintaining DNA integrity in the prostate. To our knowledge, this is the first report to examine DNA integrity and global gene expression changes due to low cellular zinc in PrEC. Overall, these data suggest that an important function of zinc is to protect cellular DNA, adding to the body of literature that suggests that loss of zinc may play an important role in the development of prostate cancer.
Prostate epithelial cells have a unique ability to accumulate high levels of zinc and have the highest concentration of zinc compared with other soft tissues in the body. It has been hypothesized that zinc accumulation is required for the inhibition of m-aconitase so citrate can accumulate for secretion in prostatic fluid (12,13). Moreover, during malignancy, the ability of the prostate cells to accumulate zinc is lost (14). The mechanisms leading to loss of zinc during prostate cancer progression is unclear but have been linked to aberrant expression of the zinc transporters Zip1,2 and 3 in cancer cells (15,16). A loss of cellular zinc through dietary zinc deficiency may exacerbate or accelerate this process leading to malignancy. Likewise, alterations in a variety of genes, mostly involved in signal transduction, stress response, and metabolism, have also been observed other cell and tissue types (17–19). Ho et al. (4) reported that zinc deficiency in IMR 90 cells, a primary human lung fibroblast cell line, resulted in altered expression of genes involved in stress response, cell signaling, protein degradation, and DNA damage/repair. Studies of zinc deficiency in rats have also shown differential expression of genes involved in growth and metabolism, stress response, and transcription/translation (20,21). Together, these data support the diverse biological role of zinc and indicate regulation of genes involved in DNA damage/repair and transcription by zinc.
Unexpectedly, none of the well-described genes involved in regulating zinc homeostasis, such as metallothionein and zinc transporters, were differentially expressed during zinc deficiency in prostate cells. Metallothionein functions to regulate zinc levels, acts as an antioxidant (due to multiple thiol groups on the molecule), and serves to protect against heavy metal toxicity. Zinc, other heavy metals, and oxidative stress can induce metallothionein expression. Transcription of metallothionein is partly controlled through interactions between the metal-responsive transcription factor 1 and metal response elements (MRE) located in the pro-moter region (11,22,23). However, the presence of other pro-moter elements also controls metallothionein expression due to cadmium overload or oxidative stress (24). Increased expression of metallothionein and metal-responsive transcription factor 1 with zinc treatment has been reported in several different prostate cancer cell lines (25,26). Decreased MT-1 expression due to zinc deficiency has been observed in other cell and tissue types (4,20). In most cell types, zinc is often sequestered through binding to metallothionein, keeping free zinc concentrations fairly low. In contrast, in the prostate, there is considerable free zinc available in the cell, much of it bound to citrate. Thus, the control, regulation, and synthesis of metallothionein may be different in prostate cells compared with other cell types. Indeed, variable and low metallothionein expression intensity and localization patterns in normal human prostate samples and normal RWPE prostate epithelial cells have been observed (27). Thus, it is possible there are unique mechanisms to control zinc homeostasis in the prostate that could account for the lack of change in typical zinc-sensitive genes. In this study, zinc deficiency in normal PrEC resulted in a nonsignificant decrease in MT-1 gene expression, a 1.6-fold increased binding of metal response factors to MRE, and no change in zinc transporter expression. These data highlight the unique metabolic responses to zinc depletion that are specific to the prostate and are an important area of future research.
In addition to the direct increases in DNA damage, our data suggest that zinc deficiency in the prostate impaired p53 binding activity, leading to an inability to signal downstream targets responsible for carrying out essential mechanisms involved in DNA damage signaling and repair. Increased gene and/or nuclear p53 protein expression due to zinc deficiency has also been reported in other cell types both in vitro and in vivo (4,7,28–30). Studies have shown that removal of zinc from the DNA binding region of p53 results in a nonfunctional protein that has lost its site-specific DNA-binding activity (3,31). Increased p53 expression without a concomitant increase in downstream targets has also been seen in other cell types such as aortic endothelial cells (30,31). In vivo studies by Fong et al. (32–34) have shown dietary zinc deficiency results in increased esophageal cell proliferation, increased expression of p53, mutations in p53, and the Ha-ras oncogene as well as increased tumor development in rats. However, these effects were modified with zinc replenishment (35). These data further support the essential role of zinc in DNA damage response/repair mechanisms and protection against cancer development.
Many other transcription factors, in addition to p53, contain zinc finger DNA-binding motifs (36). Several studies have indicated that loss of zinc from zinc-dependent enzymes, or mutations in the zinc-finger domain, can result in loss of protein function (2,3,37,38). In this study, 4 of the 16 transcription factors that showed significant increases in DNA binding activity with zinc deficiency were zinc ion binding. In addition to p53, Sp1, RXR, and GATA also contain zinc finger motifs in their DNA binding region (39,40). A hierarchy may exist of proteins/transcription factors in which proteins acquire or lose zinc depending on the tissue type and extent of zinc deficiency. Determining this hierarchy could reveal novel biomarkers for zinc deficiency and identify specific mechanisms by which zinc deficiency may increase prostate cancer risk.
Several other genes involved in DNA damage response were also affected with zinc deficiency, including downregulation of p73, XRCC4, MRE11A, and BRCA2. Recent studies suggest that p73 may also play a role in regulating p53 transcription (41). The BRCA2 gene, which is involved in repair of double-strand DNA breaks through homologous recombination (42), and mutations in BRCA2 have been associated with increased risk of developing breast and prostate cancer (43–46). Finally, protein phosphatase 1A was also upregulated. Protein phosphatase 1A has been shown to be involved in activation of p53, possibly via its dephosphorylation activity (47).
In conclusion, low cellular zinc levels in PrEC cells resulted in DNA damage and altered expression of genes involved in cell cycle, apoptosis, DNA damage, and repair and transcription. Interestingly, zinc deficiency increased both the transcript and nuclear p53 protein expression but did not significantly upregulate the DNA binding activity of p53. In addition, there was no change in gene expression of the p53 downstream targets, p21, BAX, and Mdm2. Although epidemiological studies are often inconsistent (48–52), there is experimental evidence that supports the protective role of zinc against prostate cancer (9). This study confirms the role of zinc in protecting DNA integrity and demonstrates that zinc deficiency may compromise DNA integrity by impairing the function of zinc-dependent proteins involved in the DNA damage response. These data suggest that zinc deficiency may impair cellular mechanisms that respond to and repair DNA damage that could result in an accumulation of DNA mutations and increased cancer risk.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dr. Qinjie Li, Dr. Chantal Courte-manche, and Dr. Mindy Myzak for their assistance in conducting these studies.
Footnotes
Supported by USDA (2005-35200-15439 to E. Ho), by Oregon Agriculture Experiment Station (OR00735), and by the Environmental Health Science Center at Oregon State University (National Institute of Environmental Health Sciences P30 ES00210).
Author disclosures: M. Yan, Y. Song, C. P. Wong, K. Hardin, and E. Ho, no conflicts of interest.
Supplemental Tables 1–3 and Supplemental Figures 1 and 2 are available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: ACTB, β-actin; BAX, BCL2-associated X protein; BRCA2, breast cancer 2, early onset;Brn-3,POU domain, class 4, transcription; CREB, cAMP responsive element-binding protein; EMSA, electrophoretic mobility shift assay; FAST-1, forkhead box H1; GADD45, growth arrest and DNA-damage-inducible, α; GATA, GATAbinding protein; IGF-BP3, insulin-like growth factor binding protein 3; Mdm2, transformed 3T3 cell double minute, p53 binding protein (mouse); MRE, metal response elements; MRE11A, MRE11 meiotic recombination 11 homolog A; MT-1, metallothionein 1; NFκB, nuclear factor κ B; NFκB2, nuclear factor of κ light polypeptide gene enhancer in B-cells 2; p21, cyclin-dependent kinase inhibitor A1; PrEC, prostate epithelial cells; PrEGM, Prostate Epithelial Cell medium; qPCR, quantitative PCR; RXR, retinoid X receptor; Smad 3/4, MAD, mothers against decapentaplegic homolog 3/4; Stat4, signal transducer and activator of transcription 4;TP53 (p53), tumor protein p53;TP73 (p73), tumor protein p73;XRCC4, X-ray repair complementing defective repair in Chinese hamster cells 4; ZnAD, zinc adequate; ZnDF, zinc deficient.
Literature Cited
- 1.Witkiewicz-Kucharczyk A, Bal W. Damage of zinc fingers in DNA repair proteins, a novel molecular mechanism in carcinogenesis. Toxicol Lett. 2006;162:29–42. doi: 10.1016/j.toxlet.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Dong J, Park JS, Lee SH. In vitro analysis of the zinc-finger motif in human replication protein A. Biochem J. 1999;337:311–317. [PMC free article] [PubMed] [Google Scholar]
- 3.Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 4.Ho E, Courtemanche C, Ames BN. Zinc deficiency induces oxidative DNA damage and increases p53 expression in human lung fibroblasts. J Nutr. 2003;133:2543–2548. doi: 10.1093/jn/133.8.2543. [DOI] [PubMed] [Google Scholar]
- 5.Oteiza PI, Clegg MS, Zago MP, Keen CL. Zinc deficiency induces oxidative stress and AP-1 activation in 3T3 cells. Free Radic Biol Med. 2000;28:1091–1099. doi: 10.1016/s0891-5849(00)00200-8. [DOI] [PubMed] [Google Scholar]
- 6.Olin KL, Shigenaga MK, Ames BN, Golub MS, Gershwin ME, Hendrickx AG, Keen CL. Maternal dietary zinc influences DNA strand break and 8-hydroxy-2′-deoxyguanosine levels in infant rhesus monkey liver. Proc Soc Exp Biol Med. 1993;203:461–466. doi: 10.3181/00379727-203-43623. [DOI] [PubMed] [Google Scholar]
- 7.Ho E, Ames BN. Low intracellular zinc induces oxidative DNA damage, disrupts p53, NFkappa B, and AP1 DNA binding, and affects DNA repair in a rat glioma cell line. Proc Natl Acad Sci USA. 2002;99:16770–16775. doi: 10.1073/pnas.222679399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello LC, Franklin RB. Novel role of zinc in the regulation of prostate citrate metabolism and its implications in prostate cancer. Prostate. 1998;35:285–296. doi: 10.1002/(sici)1097-0045(19980601)35:4<285::aid-pros8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 9.Costello LC, Franklin RB, Feng P, Tan M, Bagasra O. Zinc and prostate cancer: a critical scientific, medical, and public interest issue (United States) Cancer Causes Control. 2005;16:901–915. doi: 10.1007/s10552-005-2367-y. [DOI] [PubMed] [Google Scholar]
- 10.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 11.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 12.Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Cancer. 2006;5:17. doi: 10.1186/1476-4598-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello LC, Franklin RB, Feng P. Mitochondrial function, zinc, and intermediary metabolism relationships in normal prostate and prostate cancer. Mitochondrion. 2005;5:143–153. doi: 10.1016/j.mito.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franklin RB, Costello LC. Zinc as an anti-tumor agent in prostate cancer and in other cancers. Arch Biochem Biophys. 2007;463:211–217. doi: 10.1016/j.abb.2007.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franklin RB, Feng P, Milon B, Desouki MM, Singh KK, Kajdacsy-Balla A, Bagasra O, Costello LC. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol Cancer. 2005;4:32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desouki MM, Geradts J, Milon B, Franklin RB, Costello LC. hZip2 and hZip3 zinc transporters are down regulated in human prostate adenocarcinomatous glands. Mol Cancer. 2007;6:37. doi: 10.1186/1476-4598-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kindermann B, Doring F, Pfaffl M, Daniel H. Identification of genes responsive to intracellular zinc depletion in the human colon adenocarcinoma cell line HT-29. J Nutr. 2004;134:57–62. doi: 10.1093/jn/134.1.57. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard RK, Moore JB, Green CL, Cousins RJ. Modulation of intestinal gene expression by dietary zinc status: effectiveness of cDNA arrays for expression profiling of a single nutrient deficiency. Proc Natl Acad Sci USA. 2001;98:13507–13513. doi: 10.1073/pnas.251532498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cousins RJ, Blanchard RK, Moore JB, Cui L, Green CL, Liuzzi JP, Cao J, Bobo JA. Regulation of zinc metabolism and genomic outcomes. J Nutr. 2003;133:S1521–S1526. doi: 10.1093/jn/133.5.1521S. [DOI] [PubMed] [Google Scholar]
- 20.tom Dieck H, Doring F, Roth HP, Daniel H. Changes in rat hepatic gene expression in response to zinc deficiency as assessed by DNA arrays. J Nutr. 2003;133:1004–1010. doi: 10.1093/jn/133.4.1004. [DOI] [PubMed] [Google Scholar]
- 21.Sun JY, Wang JF, Zi NT, Jing MY, Weng XY. Gene expression profiles analysis of the growing rat liver in response to different zinc status by cDNA microarray analysis. Biol Trace Elem Res. 2007;115:169–185. doi: 10.1007/BF02686028. [DOI] [PubMed] [Google Scholar]
- 22.Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–237. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- 23.Saydam N, Adams TK, Steiner F, Schaffner W, Freedman JH. Regulation of metallothionein transcription by the metal-responsive transcription factor MTF-1: identification of signal transduction cascades that control metal-inducible transcription. J Biol Chem. 2002;277:20438–20445. doi: 10.1074/jbc.M110631200. [DOI] [PubMed] [Google Scholar]
- 24.Haq F, Mahoney M, Koropatnick J. Signaling events for metallothionein induction. Mutat Res. 2003;533:211–226. doi: 10.1016/j.mrfmmm.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Hasumi M, Suzuki K, Matsui H, Koike H, Ito K, Yamanaka H. Regulation of metallothionein and zinc transporter expression in human prostate cancer cells and tissues. Cancer Lett. 2003;200:187–195. doi: 10.1016/s0304-3835(03)00441-5. [DOI] [PubMed] [Google Scholar]
- 26.Albrecht AL, Singh RK, Somji S, Sens MA, Sens DA, Garrett SH. Basal and metal-induced expression of metallothionein isoform 1 and 2 genes in the RWPE-1 human prostate epithelial cell line. J Appl Toxicol. doi: 10.1002/jat.1277. Epub 2007 Jun 22. [DOI] [PubMed] [Google Scholar]
- 27.Garrett SH, Sens MA, Shukla D, Flores L, Somji S, Todd JH, Sens DA. Metallothionein isoform 1 and 2 gene expression in the human prostate: downregulation of MT-1× in advanced prostate cancer. Prostate. 2000;43:125–135. doi: 10.1002/(sici)1097-0045(20000501)43:2<125::aid-pros7>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 28.Reaves SK, Fanzo JC, Arima K, Wu JY, Wang YR, Lei KY. Expression of the p53 tumor suppressor gene is up-regulated by depletion of intracellular zinc in HepG2 cells. J Nutr. 2000;130:1688–1694. doi: 10.1093/jn/130.7.1688. [DOI] [PubMed] [Google Scholar]
- 29.Fanzo JC, Reaves SK, Cui L, Zhu L, Wu JY, Wang YR, Lei KY. Zinc status affects p53, gadd45, and c-fos expression and caspase-3 activity in human bronchial epithelial cells. Am J Physiol Cell Physiol. 2001;281:C751–C757. doi: 10.1152/ajpcell.2001.281.3.C751. [DOI] [PubMed] [Google Scholar]
- 30.Fanzo JC, Reaves SK, Cui L, Zhu L, Lei KY. p53 protein and p21 mRNA levels and caspase-3 activity are altered by zinc status in aortic endothelial cells. Am J Physiol Cell Physiol. 2002;283:C631–C638. doi: 10.1152/ajpcell.00248.2001. [DOI] [PubMed] [Google Scholar]
- 31.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42:2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 32.Fong LY, Lau KM, Huebner K, Magee PN. Induction of esophageal tumors in zinc-deficient rats by single low doses of N-nitrosomethylbenzylamine (NMBA): analysis of cell proliferation, and mutations in H-ras and p53 genes. Carcinogenesis. 1997;18:1477–1484. doi: 10.1093/carcin/18.8.1477. [DOI] [PubMed] [Google Scholar]
- 33.Fong LY, Sivak A, Newberne PM. Zinc deficiency and methylbenzylnitrosamine-induced esophageal cancer in rats. J Natl Cancer Inst. 1978;61:145–150. doi: 10.1093/jnci/61.1.145. [DOI] [PubMed] [Google Scholar]
- 34.Fong LY, Li JX, Farber JL, Magee PN. Cell proliferation and esophageal carcinogenesis in the zinc-deficient rat. Carcinogenesis. 1996;17:1841–1848. doi: 10.1093/carcin/17.9.1841. [DOI] [PubMed] [Google Scholar]
- 35.Fong LY, Nguyen VT, Farber JL. Esophageal cancer prevention in zinc-deficient rats: rapid induction of apoptosis by replenishing zinc. J Natl Cancer Inst. 2001;93:1525–1533. doi: 10.1093/jnci/93.20.1525. [DOI] [PubMed] [Google Scholar]
- 36.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 37.Gibbs PN, Gore MG, Jordan PM. Investigation of the effect of metal ions on the reactivity of thiol groups in human 5-aminolaevulinate dehydratase. Biochem J. 1985;225:573–580. doi: 10.1042/bj2250573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu HW, Moomaw JF, Moomaw CR, Casey PJ. Identification of a cysteine residue essential for activity of protein farnesyltransferase. Cys299 is exposed only upon removal of zinc from the enzyme. J Biol Chem. 1996;271:28541–28548. doi: 10.1074/jbc.271.45.28541. [DOI] [PubMed] [Google Scholar]
- 39.Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Szanto A, Narkar V, Shen Q, Uray IP, Davies PJ, Nagy L. Retinoid X receptors: X-ploring their (patho)physiological functions. Cell Death Differ. 2004;11(Suppl 2):S126–S143. doi: 10.1038/sj.cdd.4401533. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66:6982–6989. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- 42.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 43.Dong JT. Prevalent mutations in prostate cancer. J Cell Biochem. 2006;97:433–447. doi: 10.1002/jcb.20696. [DOI] [PubMed] [Google Scholar]
- 44.Horsburgh S, Matthew A, Bristow R, Trachtenberg J. Male BRCA1 and BRCA2 mutation carriers: a pilot study investigating medical characteristics of patients participating in a prostate cancer prevention clinic. Prostate. 2005;65:124–129. doi: 10.1002/pros.20278. [DOI] [PubMed] [Google Scholar]
- 45.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, Ausems MG, Menko FH, Gomez Garcia EB, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–719. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 47.Ofek P, Ben-Meir D, Kariv-Inbal Z, Oren M, Lavi S. Cell cycle regulation and p53 activation by protein phosphatase 2C alpha. J Biol Chem. 2003;278:14299–14305. doi: 10.1074/jbc.M211699200. [DOI] [PubMed] [Google Scholar]
- 48.Andersson SO, Wolk A, Bergstrom R, Giovannucci E, Lindgren C, Baron J, Adami HO. Energy, nutrient intake and prostate cancer risk: a population-based case-control study in Sweden. Int J Cancer. 1996;68:716–722. doi: 10.1002/(SICI)1097-0215(19961211)68:6<716::AID-IJC4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 49.Vlajinac HD, Marinkovic JM, Ilic MD, Kocev NI. Diet and prostate cancer: a case-control study. Eur J Cancer. 1997;33:101–107. doi: 10.1016/s0959-8049(96)00373-5. [DOI] [PubMed] [Google Scholar]
- 50.Key TJ, Silcocks PB, Davey GK, Appleby PN, Bishop DT. A case-control study of diet and prostate cancer. Br J Cancer. 1997;76:678–687. doi: 10.1038/bjc.1997.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leitzmann MF, Stampfer MJ, Wu K, Colditz GA, Willett WC, Giovannucci EL. Zinc supplement use and risk of prostate cancer. J Natl Cancer Inst. 2003;95:1004–1007. doi: 10.1093/jnci/95.13.1004. [DOI] [PubMed] [Google Scholar]
- 52.Kristal AR, Stanford JL, Cohen JH, Wicklund K, Patterson RE. Vitamin and mineral supplement use is associated with reduced risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:887–892. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.