Abstract
The definition and phylogenetic placement of the autogenous molestus form of Culex pipiens has puzzled entomologists for decades. We identified genetic differences between Cx. p. pipiens (L.) and Cx. pipiens f. molestus Forskål in the SH60 fragment described previously. Single-strand conformation polymorphism analysis, cloning, and sequencing of this fragment demonstrated high polymorphism within and among individual Cx. p. pipiens, with common SH60 variants shared among individuals from distant locations. In contrast, Cx. pipiens f. molestus from New York City each contained a single SH60 variant, which was not identified in any other Cx. p. pipiens specimens analyzed. Supporting microsatellite analysis demonstrated significant but reduced gene flow between Cx. p. pipiens and Cx. pipiens f. molestus in New York relative to Cx. p. pipiens populations in New York and California. Results are discussed in the context of two contrasting hypotheses regarding the origin of Cx. pipiens f. molestus populations.
Keywords: Culex pipiens pipiens, Culex pipiens f. molestus, genetics, microsatellites, West Nile virus
Culex pipiens f. molestus Forskål is a morphologically identical ecological biotype of Culex pipiens pipiens (L.) defined by a host of behavioral and physiological characteristics. In contrast to Cx. p. pipiens, Culex pipiens f. molestus has the ability to produce eggs without a vertebrate bloodmeal (autogeny); can mate in confined spaces (stenogamy); foregoes winter diapause; occupies subterranean environments with limited surface access; and feeds readily on mammals, including humans (Mattingly 1952, Vinogradova 2000). The potential public health importance of the mammal-feeding molestus biotype of Cx. p. pipiens as a human disease vector has led to a search for markers to readily identify these two forms.
Despite the numerous biological differences that distinguish Cx. pipiens f. molestus from Cx. p. pipiens, reliable identification of Cx. pipiens f. molestus by using morphology and/or molecular tools and phylogenetic placement of this form within the Cx. pipiens complex remains unresolved. Morphological and biochemical studies using larval chaetotaxy, variation in length of dorsal and ventral arms of the phallus in adult males (DV/D ratios), and chromatography have all failed to reliably separate these two forms (Jobling 1938, Mattingly 1952, Micks 1954, Harbach et al. 1984). Micks and Scrollini (1954) identified biochemical differences among Cx. p. pipiens, Cx. p. quinquefasciatus Say, and Cx. pipiens f. molestus by using infrared spectrometry. Recently Fonseca et al. (2004) reported unique microsatellite signatures among worldwide populations of Cx. p. pipiens and Cx. pipiens f. molestus. This report was followed by Bahnck and Fonseca (2006) who developed a rapid molecular assay to differentiate between Cx. p. pipiens, Cx. pipiens f. molestus, and putative hybrid populations based on sequence differences in the genomic regions flanking the CQ11 microsatellite locus. These advances provide evidence of a molecular and genetic basis for the observable phenotypes that distinguish Cx. p. pipiens and Cx. pipiens f. molestus, but they also demonstrate that progress in identifying genetic differences between them has been challenging despite decades of work. Identification of additional markers and a more thorough characterization of genetic differences between Cx. p. pipiens and Cx. pipiens f. molestus are needed as well as further evidence supporting the evolutionary origin of Cx. pipiens f. molestus.
Two hypotheses predominate regarding the origin of Cx. pipiens f. molestus populations. Byrne and Nichols (1999) concluded with allozyme analysis that underground, autogenous Cx. pipiens f. molestus in London were most likely founded from a single colonization event from local aboveground Cx. p. pipiens populations. This hypothesis was also proposed by Dobrotworsky (1967), who through comparative studies of DV/D ratios of Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. fatigans Weidemann in the South Pacific postulated that Cx. pipiens f. molestus originated from Cx. p. pipiens. Alternatively, microsatellite data generated by Fonseca et al. (2004) supports a hypothesis that Cx. p. pipiens and Cx. pipiens f. molestus are genetically distinct forms and that northern European underground populations were founded instead by a southern autogenous, mammal-feeding form that moved northward and colonized underground habitats. This conclusion was reached because the genetic signature of the underground, autogenous specimens from London and northern Europe was more similar to African and Middle Eastern autogenous populations than with sympatric, aboveground Cx. p. pipiens. It was further postulated that North American Cx. p. pipiens had a hybrid ancestry between Old World Cx. p. pipiens and Cx. pipiens f. molestus (Fonseca et al. 2004).
Our aims were therefore to identify genetic differences between Cx. p. pipiens and Cx. pipiens f. molestus in New York and to address the question of whether Cx. pipiens f. molestus populations in New York City were derived from sympatric, aboveground Cx. p. pipiens or existed as a separate or hybrid genetic entities. We applied polymerase chain reaction (PCR); single-strand conformation polymorphism (SSCP) analysis; restriction fragment length polymorphism analysis; and sequencing to mitochondrial, ribosomal, and genomic DNA markers in colony and field-collected Cx. p. pipiens and Cx. pipiens f. molestus from New York. We also analyzed 12 microsatellite loci in field-collected specimens of Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. quinquefasciatus to compare gene flow between Cx. pipiens complex mosquitoes from New York and California. The collective results presented provide a comprehensive picture of the genetic makeup of New York Cx. p. pipiens and Cx. pipiens f. molestus through the integration of novel and existing genetic markers. Results are discussed in the context of both existing hypotheses regarding the origin of Cx. pipiens f. molestus populations.
Materials and Methods
Sample Sources and DNA Isolation
Colony and field-collected Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. quinquefasciatus were obtained from sources listed in Table 1. Cx. pipiens f. molestus from New York City (NYC) were defined at the time of collection by subterranean habitat, autogenous reproduction, stenogamy, and mammalian blood-feeding behavior. Cx. p. pipiens from NYC and Syracuse were collected aboveground by resting collections and larval collections from storm drains and containers. Specimens from Baltimore, Co., MD, were aspirated in mid-March from a known overwintering site for this species. Specimens from Pennsylvania also were collected aboveground as part of mosquito surveillance and control programs. All Cx. p. pipiens and Cx. pipiens f. molestus samples listed in Table 1 were evaluated by SSCP, described below. Microsatellite analysis was performed only on field-collected samples of Cx. p. pipiens, and Cx. pipiens f. molestus from NYC and Syracuse as well as Shasta, San Bernadino County, and Coachella Valley populations from California. Due to geographic proximity, Cx. p. quinquefasciatus populations from San Bernadino and Coachella Valley were treated as one population for the microsatellite analysis. DNA was extracted from whole mosquitoes by a modified salt procedure as described previously (Norris et al. 2001, Kent and Norris 2005).
Table 1.
Sources of Cx. pipiens complex mosquitoes used in genetic analyses
| Strain | Source | n | |
|---|---|---|---|
| Cx. p. pipiens | Colony: Cornell | Cornell University, Tompkins Co., Ithaca, NY | 20 |
| Colony: Shasta F3 | UC Davis, Shasta County, Davis, CA | 8 | |
| Colony: LIN | Sierra Pacific log decks, Placer Co., Lincoln, CA | 10 | |
| Colony: Iowa State University | AIDL, Colorado State University, Larimer Co., Ft. Collins, CO | 15 | |
| Colony: San Francisco, CA | AIDL, Colorado State University, Larimer Co., Ft. Collins, CO | 18 | |
| Colony: SJCMVC | San Joaquin Co., Rancho Cordova, CA | 18 | |
| Field-collected | St. Albans, Prospect Park, North Central Park, Clove Lake Park Staten Island, Split Rock Golf Course, New York Co., NYC, NY | 20 | |
| Field-collected | Onondaga Co., Syracuse, NY | 30 | |
| Field-collected | Philadelphia Co, Philadelphia; Union Co., Buffalo, Lewis, White Deer, Mifflinburg Boro and Kelly Townships; Chester Co., Wallace Township; Bucks Co., Falls Township; Montgomery Co., Norristown, and Erie Co., PA | 19 | |
| Field-collected | Fort Howard bunkers, Baltimore Co., Ft. Howard, MD | 11 | |
| C.x pipiens f. molestus | Colony: Cornell | New York Co., NYC, NY | 12 |
| Field-collected | 91st St. sewer, New York Co., NYC, NY | 16 | |
| Cx. pipiens quinquefasciatus | Field-collected | San Mateo Co., West Valley, CA | 20 |
| Field-collected | Orange, Riverside, and San Diego Counties, Coachella Valley, CA | 20 |
AIDL, Arthropod-borne Infectious Disease Laboratory; SJCMVC, San Joaquin County Mosquito and Vector Control District.
Polymerase Chain Reaction
The relative quality of all DNA extractions was evaluated by PCR amplification of a fragment of the mitochondrial NADH dehy-drogenase subunit four (ND4) by using arthropod-specific primers (Simon et al. 1994, Kent and Norris 2005). Molecular identification for all samples was obtained using the Crabtree et al. (1995) PCR diagnostic to confirm placement of each specimen in the Cx. pipiens complex. The Crabtree et al. (1997), Aspen and Savage (2003), and Smith and Fonseca (2004) PCR diagnostics also were used to differentially identify Cx. p. pipiens and Cx. p. quinquefasciatus as well as potential Cx. p. pipiens/quinquefasciatus hybrids. The Bahnck and Fonseca (2006) diagnostic was used to characterize the genetic background of specimens originating from NYC or Syracuse.
To further analyze the SH60 fragment amplified by the Crabtree et al. (1997) PCR assay, alternative forward and reverse primers were designed manually from the original SH60 sequence (GenBank accession no. U90782). The F4 forward primer was shifted five bases to the right, and the R4 reverse primer was similarly shifted both to the right 20 bases and to the left five bases. The 5′ to 3′ sequence of the alternative, left-shifted reverse primer used successfully with the reported F4 forward primer (Crabtree et al. 1997) was ACTGCCCACCCACTCCATAG. DNA amplifications were completed on a MJ Research PTC-200 thermal cycler (Bio-Rad Laboratories, Hercules, CA) and visualized on ethidium bromide-stained 2% agarose gels. All gels were run with GeneRuler 100-bp molecular mass marker (MBI Fermentas, Hanover, MD).
SSCP Analysis
To identify nucleotide substitutions, insertions, and deletions within and between mosquito populations, Cx. p. pipiens and Cx. pipiens f. molestus SH60 PCR products derived from the Crabtree et al. (1997) PCR diagnostic were compared by SSCP following the methods of Hiss et al. (1994) with some modification. Four microliters of PCR product was mixed with 8 µl of denaturing loading mix (DLM stock solution: 0.005 g of xylene cyanol blue + 0.005 g of bromophenol blue + 9.0 ml of deionized formamide + 0.2 ml of 1 M NaOH + 0.8 ml of double distilled H2O), heated at 95°C for 5 min, and then immediately plunged into ice. Samples were electrophoresed at 4°C at constant power on medium-format 5% native polyacrylamide gels and stained with SYBR Green (Cambrex Bio-Science Rockland Inc., Rockland, ME) for band visualization. Variation in banding patterns was assessed manually by comparing the relative mobility of both upper and lower bands among samples run on the same gel (Black and DuTeau 1997). To ensure accurate scoring, samples producing similar patterns on different plates were rerun on the same gel. After samples were condensed in this manner and unique banding patterns recognized, representative samples of each unique banding pattern were then selected and polymorphisms confirmed by sequencing. When trying to identify identical SH60 variants between Cx. p. pipiens and Cx. pipiens f. molestus, the same Cx. pipiens f. molestus SH60 samples were electrophoresed with each plate of Cx. p. pipiens samples as an internal standard.
Cloning and Sequencing
Individual Cx. p. pipiens contained multiple variants at the SH60 locus. Therefore, SH60 PCR products from 13 NYC Cx. p. pipiens, 25 Syracuse Cx. p. pipiens, three NYC Cx. pipiens f. molestus, one Cornell colony Cx. pipiens f. molestus, and one Cornell colony Cx. p. pipiens were purified with the QIAquick PCR Purification or Gel Extraction kits (QIAGEN, Valencia, CA) and cloned using chemically competent TOP10 One Shot Escherichia coli (TOPO TA Cloning kit with pCR 2.1-TOPO vector catalog K4500-01, Invitrogen, Carlsbad, CA) to evaluate differences between SH60 variants. Transformed E. coli were grown at 37°C overnight on LB agar plates containing 50 µg/ml carbenicillin, and transformed colonies were identified by blue-white selection. The SH60 insert was recovered from individual colonies by the Crabtree et al. (1997) PCR, and amplicons from 10 to 15 clones from each mosquito in a series of individual mosquitoes representing each taxon were evaluated for inter- and intra-individual variability by SSCP. Clones representative of unique SH60 mobility classes on SSCP were selected for sequencing. Sequencing was performed using the universal M13 forward and reverse primers to ensure sequence information for the entire fragment was obtained. Multiple sequence alignments were performed using Multalin (Corpet 1988). All SH60 sequences from four New York City Cx. p. pipiens, three New York City Cx. pipiens f. molestus, three Syracuse Cx. p. pipiens, one colony Cx. p. pipiens from Cornell University, and one colony Cx. pipiens f. molestus from Cornell University are available from GenBank (accession nos. AY923229–AY923239, EF015564–EF015568, and DQ421381–DQ421386) (Table 2).
Table 2.
Nucleotide substitutions present among selected Cx. p. pipiens and Cx. pipiens f. molestus SH60 clones from New York
| Biotype | Source | Accession no. |
Clone name |
Nucleotide position |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 | 40 | 44 | 49 | 69 | 87 | 102 | 187 | 207 | 209–217 | 239 | 246 | 249 | ||||
| Cx. p. pipiens | Cornell Univ. colony | AY923231 | P16B | T | C | T | C | C | T | G | C | C | GGAAGGGCC | T | A | C |
| Cx. p. pipiens | Syracuse, NY | AY923233 | Cpp50J | . | . | . | . | . | . | . | . | . | . | . | . | T |
| Cx. p. pipiens | Syracuse, NY | AY923234 | Cpp50D | G | . | . | . | . | . | A | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | AY923232 | Cpp50C | G | . | . | . | . | C | A | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | AY923236 | Cpp24G | G | . | C | . | . | . | . | . | T | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | AY923235 | Cpp24F | G | . | C | . | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | EF015565 | Cpp32A | G | . | C | . | T | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | EF015566 | Cpp42B | G | . | C | . | T | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | EF015567 | Cpp45H | G | T | C | . | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Syracuse, NY | EF015568 | Cpp45Q | G | . | C | . | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | St. Albans, NYC | DQ421382 | Cpp01G | G | . | C | T | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | St. Albans, NYC | DQ421381 | Cpp01H | G | . | C | T | . | . | . | . | T | ——— | . | G | T |
| Cx. p. pipiens | St. Albans, NYC | DQ421383 | Cpp01I | G | . | C | T | . | . | . | . | T | ——— | . | G | T |
| Cx. p. pipiens | Staten Island, NYC | DQ421385 | Cpp10A | G | . | C | . | T | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Staten Island, NYC | DQ421384 | Cpp10B | G | . | C | T | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Staten Island, NYC | AY923239 | Cpp10P | G | . | C | . | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Central Park, NYC | AY923230 | Cpp17A | G | . | C | T | . | . | . | . | . | ——— | . | G | T |
| Cx. p. pipiens | Prospect Park, NYC | EF015564 | Cpp19F | G | . | C | . | . | . | . | . | T | ——— | . | G | T |
| Cx. p. pipiens | Crabtree et al. (1997) | U90782 | U90782 | G | . | C | . | T | . | . | . | . | ——— | . | G | T |
| Cx. pipiens f. molestus | 91st St. sewer, NYC | DQ421386 | Cpm4A | G | . | C | T | . | . | . | A | T | ——— | . | G | T |
| Cx. pipiens f. molestus | 91st St. sewer, NYC | AY923229 | Cpm7B | G | . | C | T | . | . | . | A | T | ——— | . | G | T |
| Cx. pipiens f. molestus | 91st St. sewer, NYC | AY923238 | Cpm3C | G | . | C | T | . | . | . | A | T | ——— | C | G | T |
| Cx. pipiens f. molestus | Cornell Univ. colony | AY923237 | M4 | G | . | C | T | . | . | . | A | T | ——— | . | G | T |
Dots indicate nucleotides shared with the reference sequence (AY923231, P16B), whereas dashes indicate gaps.
Microsatellites
To corroborate the SSCP results, 12 microsatellite loci were selected for analysis (Table 3). Primers designed by Fonseca et al. (1998) and Keyghobadi et al. (2004) had previously demonstrated to be polymorphic within the Cx. pipiens complex and among related species (Fonseca et al. 2000, 2004; Smith et al. 2005). An alternate reverse primer for locus CQ11 was used due to nulls and misamplification by the original primer set (Smith et al. 2005). The Cx. tarsalis Coquillett microsatellite loci used for this study represent a subset of loci that were experimentally determined to amplify from Cx. pipiens complex mosquitoes. All PCR reactions were performed in 20-µl reaction volumes containing 10 mM Tris, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin; 1.0 mM dNTPs; 1.0 U of Taq polymerase, and 25 pmol (0.25 µM) each of forward and reverse primer. An initial denaturation of 5 min at 96°C was followed by 30 cycles of 94°C for 40 s, 54°C for 45 s, and 72°C for 45 s. The final 72°C extension step was 45 min (Norris et al. 2001). The forward primer in each reaction was labeled with a fluorescent marker (FAM, TET, or HEX) compatible with ABI PRISM (PerkinElmer Life and Analytical Sciences, Boston, MA) electrophoresis. Single locus PCR products were mixed for multiplexed analysis (CxpGT4 + CxpGT46 + CQ26; CxpGT9 + CQ29 + CQ11; CUTA1+CUTD107+CUTD4; CUTD120+CUTB1+CUTD113). Multiplexed products were evaluated on an ABI Prism 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA), and data were analyzed using GeneScan and Geno-Typer Fragment Analysis software packages to derive microsatellite genotypes and allele sizes (Applied Biosystems). Arlequin version 1.1 (Schneider et al. 1997) was used to calculate allele frequencies, evaluate for compliance to Hardy-Weinberg equilibrium, and estimate FST and number of migrants per generation (Nm) values for NYC Cx. p. pipiens, NYC Cx. pipiens f. molestus, Syracuse Cx. p. pipiens, Shasta Cx. p. pipiens, and San Bernadino Cx. p. quinquefasciatus populations.
Table 3.
Microsatellite loci examined in North American populations of Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. quinquefasciatus
| Locus | Designed for | Repeat motif | Reference |
|---|---|---|---|
| CUTA1 | Cx. tarsalis | (AC)n | Rasgon et al. (2006) |
| CUTD107 | Cx. tarsalis | (CAG)nCAA(CAG)n | Rasgon et al. (2006) |
| CUTD4 | Cx. tarsalis | (CAG)n | Rasgon et al. (2006) |
| CUTD120 | Cx. tarsalis | (CAG)n | Rasgon et al. (2006) |
| CUTB1 | Cx. tarsalis | (GA) | Rasgon et al. (2006) |
| CUTD113 | Cx. tarsalis | (CAG) | Rasgon et al. (2006) |
| CQ26 | Cx. p. quinquefasciatus | (GTGTGTAT)2 + (GT)10 + (GT)5 | Fonseca et al. (1998) |
| CQ29 | Cx. p. quinquefasciatus | (GT) GA(GT) | Fonseca et al. (1998) |
| CQ11 | Cx. p. quinquefasciatus | (GT)2ACTTC(GT)9 | Fonseca et al. (1998), Smith et al. (2005) |
| CxpGT4 | Cx. p. pipiens | (GT)5(GTTT)2GC(GT)2CT(GT)5 | Keyghobadi et al. (2004) |
| CxpGT9 | Cx. p. pipiens | (GT)13 | Keyghobadi et al. (2004) |
| CxpGT46 | Cx. p. pipiens | (TG)15 | Keyghobadi et al. (2004) |
Results
PCR and SSCP Analysis of the SH60 Fragment
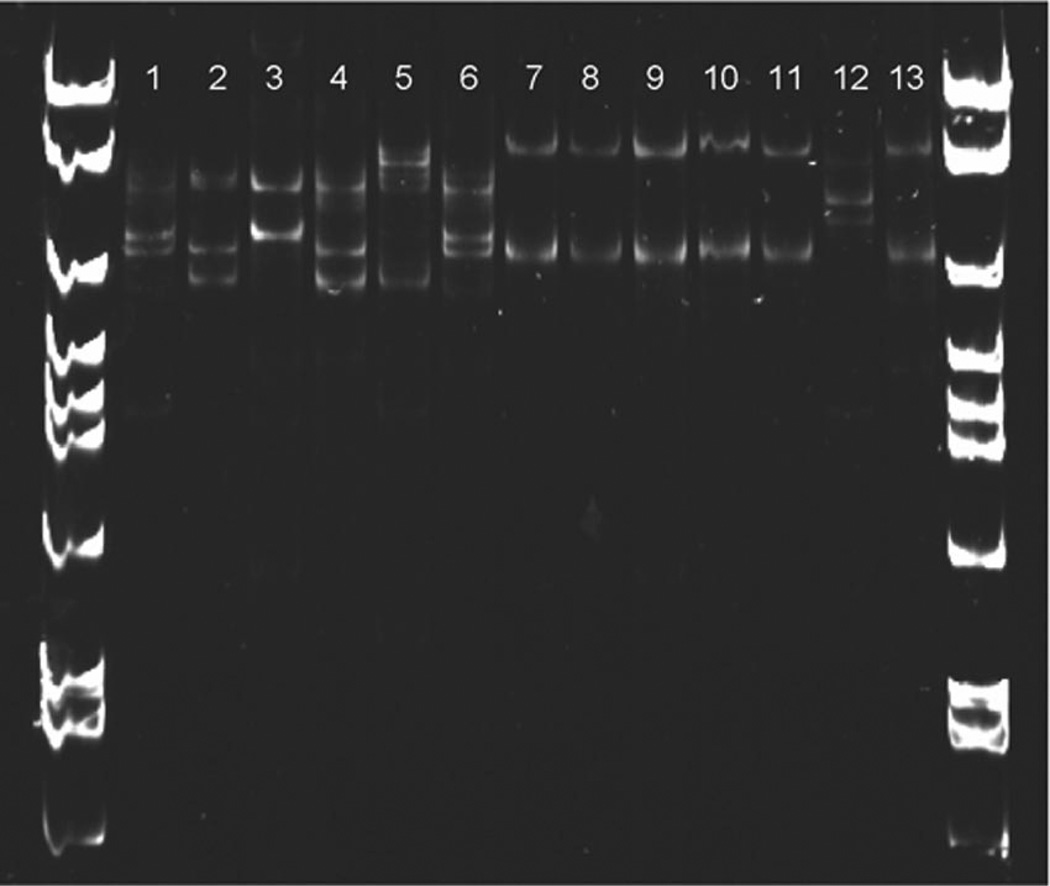
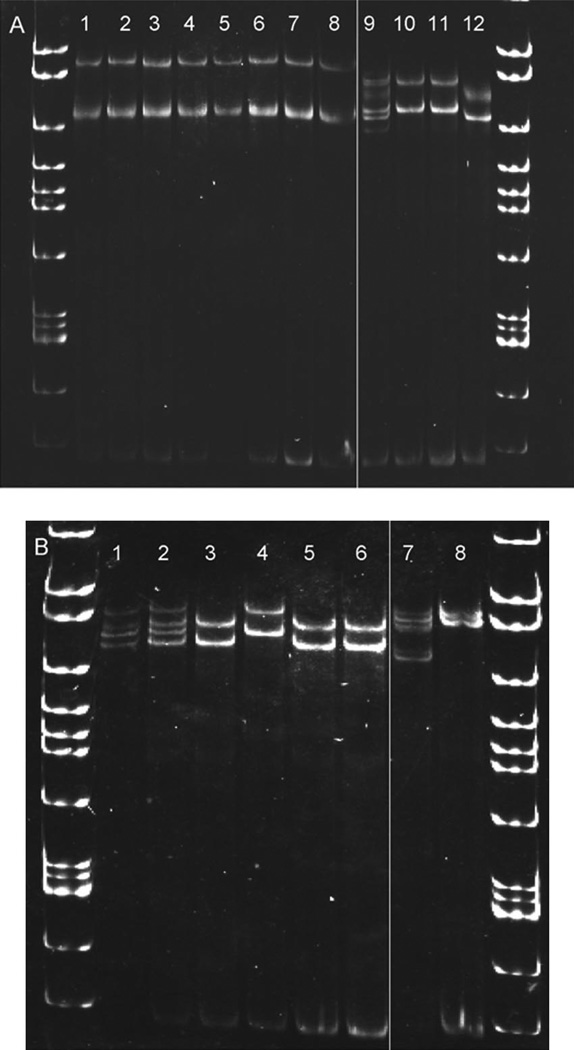
The identity of all specimens as Cx. p. pipiens, Cx. p. quinquefasciatus, or Cx. p. pipiens/quinquefasciatus hybrids was confirmed by PCR. Molecular results corroborated with original species designations listed in Table 1, except for the Cx. p. pipiens colony from San Joaquin County, CA. This colony seemed to be of hybrid origin between Cx. p. pipiens and Cx. p. quin-quefasciatus according to specific primer annealing of the ACE2 locus (Aspen and Savage 2003, Smith and Fonseca 2004). We identified consistent differences in genetic diversity in the Cx. p. pipiens-specific SH60 fragment described by Crabtree et al. (1997) between Cx. p. pipiens and Cx. pipiens f. molestus populations in New York. Because this fragment was Cx. p. pipiens-specific, no amplification could be obtained from Cx. p. quinquefasciatus for comparison, even when forward and primers were shifted in either direction. The identity and genomic location of the SH60 fragment remains unknown; however, the serendipitous observation of differential SSCP banding patterns between Cx. p. pipiens and Cx. pipiens f. molestus prompted further investigation into SH60 diversity between biotypes and across geographic locations. Both colony and field-collected Cx. p. pipiens are highly polymorphic at this locus, whereas Cx. pipiens f. molestus have greatly reduced diversity (Fig. 1). SSCP analysis of additional Cx. p. pipiens populations listed in Table 1 revealed similar patterns of inter- and intra-individual diversity as illustrated in Figs. 1 and 2 (data not shown). The complex banding patterns observed in individual Cx. p. pipiens specimens were the result of multiple SH60 variants amplified simultaneously during PCR (Fig. 2A, B). Variants were identified through SSCP analysis of PCR products from genomic DNA and clones from the same individual mosquito elec-trophoresed side by side. When evaluating all Cx. p. pipiens and Cx. pipiens f. molestus populations sampled, some SH60 variants were shared among Cx. p. pipiens individuals from widely separated locations, for example Cpp24F (Syracuse, NY) and Cpp10P (New York City, NY) (Table 2).
Fig. 1.
Genetic variability between Cx. p. pipiens and Cx. pipiens f. molestus in the SH60 fragment by SSCP analysis. Lanes 1–6, colony Cx. p. pipiens; lanes 7–11, colony Cx. pipiens f. molestus; lane 12, field-collected Cx. p. pipiens; and lane 13, field-collected Cx. pipiens f. molestus.
Fig. 2.
Genomic DNA and corresponding clones of the SH60 fragment illustrating that Cx. pipiens f. molestus contains a single SH60 variant, whereas Cx. p. pipiens specimens contain multiple variants that are additive to form the complex banding patterns seen in Fig. 1. (A) NYC Cx. p. pipiens and Cx. pipiens f. molestus. Lane 1, Cx. pipiens f. molestus genomic DNA; lanes 2–5, corresponding clones from the same specimen; lane 6, Cx. pipiens f. molestus genomic DNA; lanes 7–8, SH60 clones; lane 9, Cx. p. pipiens genomic DNA; and lanes 10–12, SH60 clones. (B) NYC and Syracuse Cx. p. pipiens. Lane 1, Syracuse Cx. p. pipiens genomic DNA; lanes 2–6, SH60 clones; lane 7, NYC Cx. p. pipiens genomic DNA; and lane 8, one corresponding clone.
To address the possibility that Cx. pipiens f. molestus contained additional SH60 variants that were not amplified due to mutations in the priming region, one additional forward and two reverse primers were manually selected by shifting the existing primers in either direction. Primer combinations consisting of a novel forward primer shifted inward five bases coupled with the either the original reverse or a novel reverse shifted to the right 20 bases, did not amplify the target sequence from either Cx. p. pipiens or Cx. pipiens f. molestus templates. The primer combination that used the existing F4 forward primer together with a novel reverse primer shifted to the left five bases did produce the expected product in both Cx. p. pipiens and Cx. pipiens f. molestus. Using the latter alternative primer set, still only one SH60 variant was amplified from Cx. pipiens f. molestus, whereas multiple variants were amplified from Cx. p. pipiens.
Finally, a multiple sequence alignment of SH60 clones from Cx. p. pipiens and Cx. pipiens f. molestus revealed that the observed diversity of SH60 banding patterns was generated by nucleotide polymorphism at a limited number of variable sites. Notably, all four Cx. pipiens f. molestus SH60 clones contained an adenine residue at position 187, whereas each of the 14 clones from Cx. p. pipiens contained cytosine (Table 2) at this position. Despite this finding, attempts to develop a reliable restriction fragment length polymorphism diagnostic to differentiate between Cx. p. pipiens and Cx. pipiens f. molestus based on this single mutation were unsuccessful. In addition, extreme sequence diversity was observed at the remaining variable positions among the Cx. p. pipiens SH60 variants (Table 2).
Microsatellites
Of the 12 microsatellite loci evaluated, only CUTB1, CUTD113, CxpGT4, CxpGT46, and CQ11 complied with Hardy-Weinberg equilibrium in at least three of five populations (Tables 3 and 4). These results could be due to small sample sizes in some populations or the existence of null alleles. Therefore, only these five loci were included in population analyses. Diversity indices for these loci are reported in Table 4 for New York Cx. p. pipiens and Cx. pipiens f. molestus. Cx. pipiens f. molestus in NYC were monomorphic at locus CQ11 for a 283 bp allele. Table 5 compares the FST values and corresponding estimated Nm among five populations of Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. quinquefasciatus. Microsatellite CQ11 allele sizes, Bahnck and Fonseca (2006) PCR results and SH60 variant data for field-collected New York Cx. p. pipiens and Cx. pipiens f. molestus are integrated in Table 6.
Table 4.
Genetic diversity of two populations of Cx. p. pipiens and one population of Cx. pipiens f. molestus from New York
| Locus | New York City Cx. p. pipiens Allele frequency |
New York City Cx. pipiens f. molestus allele frequency |
Syracuse Cx. p. pipiens allele frequency |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2N | A | MC | LC | Ho | He | P | Locus | 2N | A | MC | LC | Ho | He | P | Locus | 2N | A | MC | LC | Ho | He | P | |
| CUTBl | 32 | 5 | 0.41 | 0.09 | 0.625 | 0.786 | 0.632 | CUTBl | 32 | 3 | 0.59 | 0.19 | 0.438 | 0.605 | 0.17 | CUTBl | 20 | 5 | 0.35 | 0.05 | 1 | 0.77 | 0.619 |
| CUTD113 | 32 | 4 | 0.66 | 0.03 | 0.333 | 0.537 | 0.223 | CUTD113 | 32 | 2 | 0.56 | 0.44 | 0.625 | 0.508 | 0.609 | CUTD113 | 20 | 2 | 0.55 | 0.45 | 0.5 | 0.57 | 1 |
| CxpGT4 | 32 | 8 | 0.34 | 0.03 | 0.625 | 0.825 | 0.2 | CxpGT4 | 32 | 6 | 0.41 | 0.03 | 0.688 | 0.754 | 0.158 | CxpGT4 | 20 | 5 | 0.40 | 0.05 | 0.8 | 0.76 | 0.381 |
| CxpGT46 | 32 | 12 | 0.22 | 0.03 | 0.933 | 0.892 | 0.813 | CxpGT46 | 32 | 7 | 0.38 | 0.03 | 0.75 | 0.78 | 0.016* | CxpGT46 | 20 | 7 | 0.35 | 0.05 | 0.9 | 0.82 | 0.844 |
| CQ11 | 32 | 5 | 0.34 | 0.03 | 0.333 | 0.777 | 0.002* | CQ11 | 32 | 1 | 1.00 | 0.00 | Monomorphic | CQ11 | 20 | 2 | 0.80 | 0.20 | 0.2 | 0.41 | 0.301 | ||
2N is the number gene copies, A is the no. of alleles per locus, MC represents the frequency of the most common allele, LC is the frequency of the least common allele, Ho represents observed heterozygosity, and He represents expected heterozygosity.
Denotes significant deviation from Hardy-Weinberg equilibrium.
Table 5.
Relative gene flow among populations of Cx. p. pipiens, Cx. pipiens f. molestus, and Cx. p. quinquefasciatus in the United States
| NYC Cx. p. molestus | NYC Cx. p. pipiens | Syracuse Cx. p. pipiens | Shasta Cx. p. pipiens | |
|---|---|---|---|---|
| NYC Cx. p. molestus | ||||
| NYC Cx. p. pipiens | 61.5 (0.00806)* | |||
| Syracuse Cx. p. pipiens | 121.88 (0.004009) | Infinity (−0.01002) | ||
| Shasta Cx. p. pipiens | 65.47 (0.00758) | Infinity (−0.00205) | Infinity (0.00000) | |
| San Bernadino Cx. p. quinquefasciatus | 28.39 (0.01731)* | 49.01 (0.01010)* | 46.85 (0.01110)* | 44.53 (0.00000) |
Values presented are the estimated number of migrants per generation (Nm) followed by the FST in parentheses.
Denotes significant FST.
Table 6.
Combined CQ11 and SH60 data for field-collected New York Cx. p. pipiens and Cx. pipiens f. molestus. The no. of SH60 variants was determined by cloning and SSCP. “Cpp” denotes specimens identified as Cx. p. pipiens and “Cpm” denotes specimens identified behaviorally and physiologically as Cx. pipiens f. molestus. Column (P/M) refers to Bahnck and Fonseca (2006) PCR results, where P = “pipiens” and M = “molestus”. “+” indicates instances where not all SH60 variants present were isolated by cloning.
| Mosquito | Collection location | CQ11 alleles | P/M | # SH60 variants |
|---|---|---|---|---|
| Cpp01 | St. Albans, NYC | 251,279 | P/M | 3+ |
| Cpp02 | St. Albans, NYC | 283,283 | M | 2+ |
| Cpp06 | St. Albans, NYC | 251,251 | P | 4 |
| Cpp09 | St. Albans, NYC | 251,251 | P | 2 |
| Cpp10 | Clove Lake Park, Staten Is. | 283,283 | M | 2 |
| Cpp12 | North Central Park, NYC | 283,283 | M | 1 |
| Cpp14 | North Central Park, NYC | 251,251 | P | 4 |
| Cpp17 | North Central Park, NYC | 265,265 | P | 3 |
| Cpp18 | Silver Lake Golf Course, NYC | 259,283 | P/M | 5 |
| Cpp19 | Prospect Park, NYC | 259,283 | P/M | 4 |
| Cpp20 | Prospect Park, NYC | 259,265 | P | 1 |
| Cpp21 | Syracuse, NY | 259,259 | P | 2 |
| Cpp22 | Syracuse, NY | 283,283 | M | 3 |
| Cpp23 | Syracuse, NY | 259,283 | P | 2 |
| Cpp24 | Syracuse, NY | 259,283 | P/M | 2 |
| Cpp26 | Syracuse, NY | 259,283 | P | 2+ |
| Cpp27 | Syracuse, NY | 259,259 | P | 2+ |
| Cpp31 | Syracuse, NY | 259,259 | P | 2 |
| Cpp32 | Syracuse, NY | 251,251 | P | 2 |
| Cpp33 | Syracuse, NY | 259,259 | P | 4 |
| Cpp45 | Syracuse, NY | 259,283 | P | 2 |
| Cpp48 | Syracuse, NY | 259,259 | P | 4 |
| Cpp49 | Syracuse, NY | 283,283 | M | 3 |
| Cpp50 | Syracuse, NY | 259,259 | P | 3 |
| Cpm01 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm02 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm03 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm04 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm05 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm06 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm07 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm08 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm09 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm10 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm11 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm12 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm13 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm14 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm15 | 91st St. sewer, NYC | 283,283 | M | 1 |
| Cpm16 | 91st St. sewer, NYC | 283,283 | M | 1 |
indicates that not all SH60 variants present in that individual were identified by cloning, so the true no. is unknown.
Pure molestus are fixed at the CQ11 locus for allele 283, and contain one SH60 variant which in this case has the one point mutation unique from all pipiens analyzed.
Populations that have mixed gene flow between P and M have additional microsatellite alleles present and at the population level, show both P and M and P/M types on the rapid assay, and have more than one SH60 variant per individual. The rapid assay is only accurate for the F1 generation in showing true hybrids, after which recombination and backcrossing will skew results if viewed only on an individual basis. Therefore, hybrid populations may have individuals that are homozygous for either P or M CQ11 alleles or have one or more SH60 variant. In Cpp01, the 283 allele has been lost altogether.
There is or was at one point, gene flow between surface and subterranean populations.
Discussion
Numerous attempts were made to identify genetic differences between Cx. p. pipiens and Cx. pipiens f. molestus. There were no consistent differences between these forms in the small subunit ribosomal 12S (EF028702, EF028703), CO1 (DQ072277, DQ072278, DQ072279), or ND4 (EF028084, EF030092). Additionally, screening of Cx. p. pipiens and Cx. pipiens f. molestus from New York with 10 different random amplification of polymorphic DNA-PCR primers (Sigma-Genosys, GEN1-10, 50% G+C) did not reveal any consistently unique amplicons for either form. However, the serendipitous analysis of the Cx. p. pipiens-specific SH60 fragment identified by Crabtree et al. (1997) resulted in some interesting discoveries.
Our analysis of the SH60 fragment can be interpreted in two ways. First, taking into account the diversity of the SH60 amplicon in Cx. p. pipiens populations versus Cx. pipiens f. molestus, these data may suggest that this population of Cx. pipiens f. molestus in NYC was locally founded from aboveground populations of Cx. p. pipiens through a geographically independent colonization event associated with a dramatic genetic bottleneck. This colonization event resulted in a genetically similar sympatric population but with greatly reduced diversity. Alternatively, these PCR, SH60, and microsatellite data may support the idea that these observed differences distinguish Cx. pipiens f. molestus from Cx. p. pipiens as genetic entities. Consistent genetic differences were identified between Cx. p. pipiens and Cx. pipiens f. molestus at the SH60 locus, and these results are easily interpreted in the context of the rapid assay published by Bahnck and Fonseca (2006). Supporting data for these two hypotheses are presented below.
We have demonstrated that New York Cx. p. pipiens populations as well as additional Cx. p. pipiens populations from around the United States, are highly polymorphic at the SH60 locus, whereas NYC Cx. pipiens f. molestus are genetically consistent. Two SH60 variants differing by a single nucleotide substitution at position 239 were identified in Cx. pipiens f. molestus from NYC, with individual mosquitoes each containing only a single variant. In contrast, individual Cx. p. pipiens invariably contained multiple SH60 variants, with the number of variants existing in each collection of mosquitoes too large to be completely catalogued in this study. For example, 18 NYC Cx. p. pipiens comprised 11 unique SSCP banding patterns. From cloning, each banding pattern represented a composite of between two and five different SH60 variants that were amplified simultaneously during PCR. The 30 Cx. p. pipiens specimens from Syracuse, NY contributed an additional 20 SSCP banding patterns unique from those observed in NYC Cx. p. pipiens (data not shown). Some of the SH60 variants were shared between individuals in these two locations (Table 2). Cloning, SSCP, and sequencing analysis of amplicons generated by alternative primers flanking the SH60 fragment confirmed that Cx. pipiens f. molestus individuals contained only a single SH60 variant, whereas Cx. p. pipiens individuals contained multiple variants. Thus, diversity in Cx. pipiens f. molestus at the SH60 locus was not underestimated due to polymorphisms in the PCR priming region. Finally, because the SH60 fragment was Cx. p. pipiens-specific, it could only be amplified from Cx. p. pipiens and Cx. pipiens f. molestus, nullifying the use of a phylogenetic analysis due to absence of the outgroups necessary to provide context.
Contrary to what would be expected if the reduced diversity in Cx. pipiens f. molestus is indeed due to a founder’s effect, the diversity at this locus seems to be maintained in colony Cx. p. pipiens populations. Because the identity and genomic location of the SH60 fragment is unknown, it is difficult to interpret these results. One possible explanation is that the Cx. p. pipiens colonies evaluated did not experience a severe bottleneck due to recent colonization, founding from a larger number of individuals, reproduction of many individuals each generation, and/or subsequent introductions into the colony to maintain genetic diversity. A more plausible explanation is that the SH60 fragment is genetically linked to a Cx. pipiens f. molestus-associated trait, leading to its fixation in those populations and apparent absence of genetic selection or reduction in Cx. p. pipiens colonies. Linkage of phenotypes with genetic elements controlling autogeny has been investigated previously (Spielman 1957).
All SH60 clones from 38 Cx. p. pipiens, representing both NYC and Syracuse collections, were analyzed by SSCP simultaneously with SH60 amplicons from Cx. pipiens f. molestus. No shared SH60 variants were identified between either of these Cx. p. pipiens collections and Cx. pipiens f. molestus from NYC. However, due to the enormous variability in SH60 variants among Cx. p. pipiens, exhaustive sequencing of all SH60 variants was not completed, and the presence of shared variants between these two biotypes cannot be ruled out. Should shared SH60 variants be discovered, this finding would argue in favor of a founder’s effect and local origination of this Cx. pipiens f. molestus population from surface-dwelling Cx. p. pipiens. Because the analysis stands however, NYC Cx. pipiens f. molestus embodied a unique SSCP banding pattern characterized in part by an adenosine residue at position 187 that was not shared by any other sequenced Cx. p. pipiens clones.
These PCR and SSCP experiments were supplemented with a population analysis at five microsatel-lite loci, including Cx. p. pipiens and Cx. p. quinque-fasciatus collections from California as geographic outgroups. The Cx. pipiens f. molestus population had less microsatellite allelic diversity at each locus as compared with sympatric populations of Cx. p. pipiens from NYC (Table 4). Additionally, there was at least one shared allele between Cx. pipiens f. molestus and Cx. p. pipiens from both New York locations at all five loci. The shared alleles present in the Cx. pipiens f. molestus population were likely derived from contemporary or ancestral genetic exchange with surface-dwelling Cx. p. pipiens. The reduced allelic diversity across all microsatellite loci in general in Cx. pipiens f. molestus relative to Cx. p. pipiens mirrors the results of the SH60 fragment analysis.
At microsatellite locus CQ11, which is purportedly diagnostic in size for Cx. pipiens f. molestus (Bahnck and Fonseca 2006), all 16 NYC Cx. pipiens f. molestus were monomorphic for a 283-bp allele (Tables 4 and 6). This finding is similar with that reported by Fon-seca et al. (2004) in which the majority of Cx. pipiens f. molestus contained a 285-bp allele, and Bahnck and Fonseca (2006), in which CQ11 for Cx. pipiens f. molestus was 284 bp. Slight differences in allele size between this study and previous publications could be due to differences in the settings used to designate alleles.
Microsatellite analysis also revealed largely unrestricted gene flow among populations of Cx. p. pipiens collected from East and West coasts of the continental United States, indicating that this species exists as a panmictic population in much of North America. In contrast, genetic structuring was confirmed among the different members of the species complex. There was relatively more gene flow estimated between New York Cx. pipiens f. molestus and Cx. p. pipiens populations than between either Cx. pipiens f. molestus and Cx. p. quinquefasciatus or Cx. p. pipiens and Cx. p. quinquefasciatus (Table 5). Because this latter cross is known to occur in the United States throughout the hybrid zone between the 36th and 39th parallels (Barr 1957), the estimated gene flow between Cx. p. pipiens and Cx. pipiens f. molestus is also likely to be biologically significant. The finding of relatively greater gene flow between Cx. pipiens f. molestus and Cx. p. pipiens could be the result of a common ancestry between these two biotypes, the founding or derivation of Cx. pipiens f. molestus from Cx. p. pipiens, or the existence of some degree of ongoing but restricted gene flow between sympatric Cx. p. pipiens and Cx. pipiens f. molestus populations.
The key data from all aspects of this study are summarized in Table 6, from which we posit the following hypotheses. Our collection of Cx. pipiens f. molestus collected from the 91st St. sewer in NYC represents a population of true molestus and is characterized by one SH60 variant per individual mosquito and fixation of allele 283 at the CQ11 microsatellite locus. In contrast, Cx. p. pipiens populations from New York and elsewhere exhibit a much greater diversity of CQ11 alleles as well as SH60 variants. From these data, either of the following interpretations could be argued. (1) NYC Cx. pipiens f. molestus were locally founded from surface Cx. p. pipiens populations. Evidence supporting this theory is the greatly reduced genetic diversity in Cx. pipiens f. molestus relative to Cx. p. pipiens in the SH60 amplicon as well as the five microsatellite loci statistically analyzed. Microsatellite alleles, including CQ11 283, were shared between NYC Cx. p. pipiens and Cx. pipiens f. molestus. Although no common SH60 variant was identified between biotypes, it likely exists given the extreme polymorphism seen among Cx. p. pipiens at this locus. Furthermore, there was relatively more gene flow between Cx. p. pipiens and Cx. pipiens f. molestus than between any other combination of members of the species complex. (2) Alternatively, the patterns observed between Cx. p. pipiens and Cx. pipiens f. molestus in New York at the CQ11 microsatellite locus and SH60 locus could represent true genetic differences that characterize these biotypes, which also readily hybridize. For populations where hybridization is occurring between Cx. p. pipiens and Cx. pipiens f. molestus, CQ11 alleles and SH60 variants once present in Cx. p. pipiens or Cx. pipiens f. molestus are introduced and dispersed throughout the larger geographic population through introgression and recombination. As a result, we also observe “M” types that have multiple SH60 variants (Cpp10), and “P” types that have only one SH60 variant (Cpp20), and individuals such as Cpp01 which have the P/M molecular type and multiple SH60 variants but not the CQ11 283 allele (Table 6). Bahnck and Fonseca (2006) cautioned that their PCR results must be interpreted on the population level, because after the F1 generation the assay is not accurate in identifying individual mosquitoes of hybrid ancestry due to independent assortment and recombination among microsatellite alleles. This phenomenon is evident through the presence of “P,” “M,” and “P/M” types among individual mosquitoes collected from the same geographic location (i.e., St. Albans, NYC, and Prospect Park, NYC, Table 6). This latter hypothesis is also supported by the micro-satellite analysis which demonstrates the presence of reduced but biologically significant degree of gene flow between Cx. p. pipiens and Cx. pipiens f. molestus (Table 5). Despite an extensive genetic analysis of these populations, we are unable to indisputably side with one hypothesis over the other. Evidence of gene flow and “hybridization” between Cx. p. pipiens and Cx. pipiens f. molestus could be interpreted as two genetically distinct forms that are converging and hybridizing in ecological zones of sympatry. Alternatively, Cx. pipiens f. molestus represents an autogenous biotype derived from surface dwelling Cx. p. pipiens with the residual appearance of hybridization as the two forms are behaviorally and physiologically divergent but are not yet reproductively isolated. A more thorough evaluation of sympatric populations of Cx. P pipiens and Cx. pipiens f. molestus biotypes in the United States and globally is needed before these results can be validated and placed in a broader phylogenetic context.
Acknowledgments
Specimens were kindly provided by Anton Cornel and Rajeev Vaidyanathan (University of California, Davis, CA); Jason Rasgon (Johns Hopkins Bloomberg School of Public Health, Baltimore, MD); Mike Hutchinson (Pennsylvania Department of Environmental Protection); Scott Shone (New Jersey Department of Health and Senior Services, Trenton, NJ); Stacy Bearden (San Joaquin County Mosquito and Vector Control District, Stockton, CA); and Chris Bosio and Cynthia Meredith (Arthropod-borne Infectious Disease Laboratory, Colorado State University, Ft. Collins, CO). We appreciate the assistance of Varuni Kulasekera with the New York City collections. We also thank Jason Rasgon for providing Cx. tarsalis microsatellite primers and Katie Provost for optimizing them for use on Cx. pipiens complex, and Gregory Lanzaro for constructive and helpful comments. This research was supported in part by Centers for Disease Control Cooperative Agreement Awards U50/CCU320520 (to D.E.N.) and U50/CCU220512 (to L.C.H.); and National Institute of Environmental Health Sciences Training Award T32ES07141, Johns Hopkins Bloomberg School of Public Health Frederik B. Bang Award, and Johns Hopkins Malaria Research Institute Predoctoral Fellowship (to R.J.K.).
References Cited
- Aspen S, Savage HM. Polymerase chain reaction assay identifies North American members of the Culex pipiens complex based on nucleotide sequence differences in the acetylcholinesterase gene Ace. 2. J. Am. Mosq. Control Assoc. 2003;19:323–328. [PubMed] [Google Scholar]
- Bahnck CM, Fonseca DM. Rapid assay to identify the two genetic forms of Culex (Culex) pipiens L. (Diptera: Culicidae) and hybrid populations. Am. J. Trop. Med. Hyg. 2006;75:251–255. [PubMed] [Google Scholar]
- Barr AR. The distribution of Culex p. pipiens and C. p. quinquefasciatus in North America. Am. J. Trop. Med. Hyg. 1957;6:153–165. doi: 10.4269/ajtmh.1957.6.153. [DOI] [PubMed] [Google Scholar]
- Black WC, IV, DuTeau NM. RAPD-PCR and SSCP analysis for insect population genetic studies. In: Crampton JM, Beard CB, Louis C, editors. The molecular biology of insect disease vectors: a methods manual. London, United Kingdom: Chapman & Hall; 1997. pp. 361–373. [Google Scholar]
- Byrne K, Nichols RA. Culex pipiens in London Underground tunnels: differentiation between surface and subterranean populations. Heredity. 1999;82:7–15. doi: 10.1038/sj.hdy.6884120. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree MB, Savage HM, Miller BR. Development of a species-diagnostic polymerase chain reaction assay for the identification of Culex vectors of St. Louis encephalitis based on interspecies sequence variation in ribosomal DNA spacers. Am. J. Trop. Med. Hyg. 1995;53:105–109. [PubMed] [Google Scholar]
- Crabtree MB, Savage HM, Miller BR. Development of a polymerase chain reaction for differentiation between Culex pipiens pipiens, Cx. p. quinquefasciatus (Diptera: Culicidae) in North America based on genomic differences identified by subtractive hybridization. J. Med. Entomol. 1997;34:532–537. doi: 10.1093/jmedent/34.5.532. [DOI] [PubMed] [Google Scholar]
- Dobrotworsky NV. The problem of the Culex pipiens complex in the South Pacific (including Australia) Bull. World Health Organ. 1967;37:251–255. [PMC free article] [PubMed] [Google Scholar]
- Fonseca DM, Atkinson CT, Fleischer RC. Microsatellite primers for Culex pipiens quinquefasciatus, the vector of avian malaria in Hawaii. Mol. Ecol. 1998;7:613–621. [PubMed] [Google Scholar]
- Fonseca DM, LaPointe DA, Fleischer RC. Bottlenecks and multiple introductions: population genetics of the vector of avian malaria in Hawaii. Mol. Ecol. 2000;9:1803–1814. doi: 10.1046/j.1365-294x.2000.01070.x. [DOI] [PubMed] [Google Scholar]
- Fonseca DM, Keyghobadi N, Malcolm CA, Mehmet C, Schaffner F, Mogi M, Fleischer RC, Wilkerson RC. Emerging vectors in the Culex pipiens complex. Science (Wash., D.C.) 2004;303:1535–1538. doi: 10.1126/science.1094247. [DOI] [PubMed] [Google Scholar]
- Harbach RE, Harrison BA, Gad AM. Culex (Culex) molestus Forskål (Diptera: Culicidae): neotype designation, description, variation, and taxonomic status. Proc. Entomol. Soc. Wash. 1984;86:521–542. [Google Scholar]
- Hiss RH, Norris DE, Dietrich CH, Whitcomb RF, West DF, Bosio CF, Kambhampati S, Piesman J, Antolin MF, Black WC., IV Molecular taxonomy using single strand conformation polymorphism (SSCP) analysis of mitochondrial ribosomal DNA genes. Insect Mol. Biol. 1994;3:171–182. doi: 10.1111/j.1365-2583.1994.tb00164.x. [DOI] [PubMed] [Google Scholar]
- Jobling B. On two subspecies of Culex pipiens L. Trans. R. Entomol. Soc. Lond. 1938;87:193–216. [Google Scholar]
- Kent RJ, Norris DE. Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome b. Am. J. Trop. Med. Hyg. 2005;73:336–342. [PMC free article] [PubMed] [Google Scholar]
- Keyghobadi N, Matrone MA, Ebel GD, Kramer LD, Fonseca DM. Microsatellite loci from the northern house mosquito (Culex pipiens), a principal vector of West Nile virus in North America. Mol. Ecol. Notes. 2004;4:20–22. [Google Scholar]
- Mattingly PF. The problem of biological races in the Culex pipiens complex. Proc. Linn. Soc. Lond. 1952;163:53–55. [Google Scholar]
- Micks DW. Paper chromatography as a tool for mosquito taxonomy: the Culex pipiens complex. Nature (Lond.) 1954;174:217. doi: 10.1038/174217a0. [DOI] [PubMed] [Google Scholar]
- Micks DW, Scrollini F. An infrared spectro-metric study of the Culex pipiens complex. Riv. Parassitol. 1954;15:161–165. [Google Scholar]
- Norris DE, Shurtleff AC, Toure´ YT, Lanzaro GC. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae s.s. (Diptera: Culicidae) J. Med. Entomol. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Venkatesan M, Westbrook CJ, Hauer MC. Polymorphic microsatellite loci from the West Nile virus vector Culex tarsalis . Mol. Ecol. Notes. 2006 Published online May 23, 2006. [Google Scholar]
- Schneider S, Kueffer J-M, Roessli D, Excoffier L. Genetics and Biometry Laboratory. Switzerland: University of Geneva; 1997. Arlequin ver. 1.1: a software for population genetic data analysis. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994;87:651–701. [Google Scholar]
- Smith JL, Keyghobadi N, Matrone MA, Escher RL, Fonseca DM. Cross-species comparison of mic-rosatellite loci in the Culex pipiens complex and beyond. Mol. Ecol. Notes. 2005;5:697–700. [Google Scholar]
- Smith JL, Fonseca DM. Rapid assays for identification of members of the Culex (Culex) pipiens complex, their hybrids, and other sibling species (Diptera: Culicidae) Am. J. Trop. Med. Hyg. 2004;70:339–345. [PubMed] [Google Scholar]
- Spielman A. The inheritance of autogeny in the Culex pipiens complex of mosquitoes. Am. J. Hyg. 1957;65:404–425. doi: 10.1093/oxfordjournals.aje.a119878. [DOI] [PubMed] [Google Scholar]
- Vinogradova EB. Culex pipiens pipiens mosquitoes: taxonomy, distribution, ecology, physiology, genetics, applied importance and control. Sofia, Bulgaria: Pensoft; 2000. [Google Scholar]