Abstract
Developmental processes are highly dependent on transcriptional regulation by RNA polymerase II, which initiates transcription at the core promoter. The dorsal-ventral gene regulatory network (GRN) includes multiple genes that are activated by different nuclear concentrations of the Dorsal transcription factor along the dorsal-ventral axis. Downstream core promoter element (DPE)-containing genes are conserved and highly prevalent among Dorsal target genes. Moreover, the DPE motif is functional in multiple Dorsal target genes, as mutation of the DPE results in the loss of transcriptional activity. Furthermore, analysis of hybrid enhancer-promoter constructs reveals that the core promoter composition plays a pivotal role in the transcriptional output. Importantly, we provide in vivo evidence that expression driven by the homeotic Antennapedia P2 promoter during Drosophila embryogenesis is dependent on the DPE. Taken together, we propose that transcriptional regulation results from the interplay between enhancers and core promoter composition, thus establishing a novel dimension in developmental GRNs.
Keywords: transcriptional regulation, RNA polymerase II, core promoter, DPE, gene regulatory network, Drosophila, development, gene expression, Dorsal
Introduction
Developmental processes are highly dependent on transcriptional regulation by RNA polymerase II.1-4 Multiple sequence-specific DNA binding transcription factors and co-regulators control gene expression,5,6 but the ultimate target of the transcription machinery is the initiation of transcription at the core promoter, which could hence be referred to as the gateway to transcription.7-12 The RNA pol II core promoter is typically ~80 nucleotides in length (-40 to +40 relative to the +1 transcription start site) and encompasses the transcription start site.8,11-13 Core promoters may contain one or more functional DNA sequence elements, termed core promoter elements or motifs, such as the TATA box, TFIIB recognition elements (BREu and BREd), initiator (Inr), the TCT motif, motif 10 element (MTE), and downstream core promoter element (DPE), which confer specific properties to the core promoter.11,14,15 In addition, it is well documented that certain basal transcription factors can bind specific core promoter motifs, suggesting the regulation of transcription through specific core promoter motifs.16
Specific core promoter motifs have previously been implicated in the regulation of specific regulatory networks. The polypyrimidine initiator (TCT) was demonstrated to direct the transcription of genes encoding factors involved in protein synthesis, including ribosomal proteins and other protein synthesis factors.14 The downstream core promoter element (DPE) has previously been identified in multiple developmentally-regulated genes.17 The homeotic (Hox) genes determine the identity of the segments along the anterior-posterior axis in the developing embryo. Most of the Drosophila Hox gene promoters (all of which lack the TATA box element), contain functionally important DPE motifs.17 Moreover, Caudal, a master regulator of the Hox genes, exhibits preferential activation of DPE-containing promoters as opposed to TATA-containing promoters.17 It is likely that the distribution of specific core promoter elements in the genome is not random, but rather is clustered within functional groups.
Dorsal-ventral patterning of the Drosophila embryo is one of the most critical events during early embryogenesis. The regulation of dorsal-ventral axis formation is mediated through DNA elements, referred to as cis-regulatory modules, which can be bound by transcription factors. The overall interactions between the modules regulating the same process can be mapped to generate a gene regulatory network (GRN).5,18 The dorsal-ventral patterning network includes multiple genes that are activated by different nuclear concentrations of the Dorsal transcription factor along the dorsal-ventral axis (e.g., refs. 19–23). Activation of Dorsal target genes is achieved by the recruitment of Dorsal to the enhancers of these genes, which contain Dorsal-binding sites hundreds or even thousands of base pairs upstream of the transcription start site.
We have recently discovered that the dorsal-ventral developmental GRN is dependent on the presence of the DPE motif.24 We demonstrated that over two-thirds of Dorsal target genes contain DPE sequence motifs, which is significantly higher than the proportion of DPE-containing promoters in Drosophila genes. Furthermore, we showed that multiple Dorsal target genes are evolutionarily conserved and functionally dependent on the DPE. Here, we discuss our recent findings and provide additional evidence that supports the concept of specialized transcription systems, which are dependent on specific core promoter elements.
Activation of DPE-Containing Dorsal Target Genes Differs from Activation of TATA Box-Containing Target Genes
The TATA box is the first core promoter motif identified.25 It is conserved from archaebacteria to humans.26 The TATA box is bound by the TATA box-binding protein (TBP) subunit of TFIID, which was shown to be necessary for TATA-dependent transcription. The DPE was originally discovered as a TFIID recognition site that is located downstream of the initiator element (precisely from +28 to +33 relative to the A+1 of the Inr) and is conserved from Drosophila to humans.27,28 Interestingly, TBP downregulates DPE-dependent transcription.17 NC2 and MOT1, which have been shown to be positive regulators of DPE-dependent transcription, do so by counteracting TBP, thus relieving its inhibition of DPE transcription.29-31 While the overall frequency of a TATA box among Drosophila promoters is 18%, less than 8% of the Dorsal target genes appear to contain a TATA box without a DPE motif.24 In the mesoderm, where Dorsal nuclear concentration is the highest, there are no Dorsal target genes containing solely the TATA box motif. To examine Dorsal activation of TATA-containing promoters, we mapped the putative Dorsal DNA binding sites within the enhancers of Dorsal target genes Aspartyl β-hydroxylase (asph) and Actin 5C (act5C). Consensus Dorsal DNA-binding sites (GGG(W)nCCM, where (W)n is 4–5 repeats of T/A20), were identified using JASPAR (http://jaspar.genereg.net). The asph promoter contains a TATA box, an Inr and a DPE motif. We examined the activation of an asph genomic fragment encompassing from -1373 to +114 relative to the A+1 of the Inr. The genomic fragment, which contains 8 putative Dorsal binding sites, was cloned upstream of a firefly luciferase reporter gene to generate a wt asph reporter. In addition, we generated an asph reporter with a mutation in the TATA box (mTATA) and an asph reporter with a mutation in the DPE motif (mDPE). The activation of these constructs by co-transfected Dorsal (kindly provided by Prof Albert Courey, UCLA) was analyzed by dual luciferase reporter assays in Drosophila S2R+ cells. A mutation of the TATA box of asph causes a substantial reduction in basal activity (in the absence of transfected Dorsal), whereas a mutation of the DPE motif does not result in reduced basal activity (Fig. 1A). Mutation of either the TATA box or the DPE motif does not significantly reduce the activation of asph by Dorsal. Hence, Dorsal is able to activate the asph promoter either through the DPE or the TATA box, and each motif is sufficient to allow activation of the asph promoter by Dorsal.
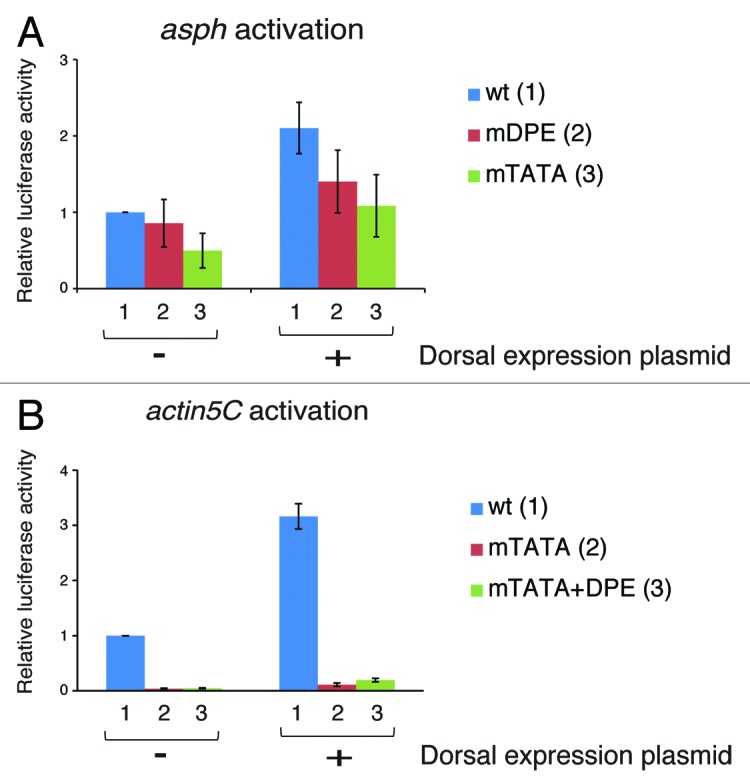
Figure 1. Activation of TATA box-containing promoters by Dorsal. (A) Activation of the asph gene by Dorsal. The asph promoter contains a TATA box, an Inr, and a DPE sequence motifs. Drosophila S2R+ cells were co-transfected with a firefly luciferase reporter construct driven by the natural enhancer-promoter of asph (which includes 8 putative Dorsal binding sites) containing wt (1), mDPE (2), or mTATA box promoter (3), as well as expression vectors of either Dorsal or an empty vector, as indicated. To control for transfection efficiency variations, cells were co-transfected with a control Renilla luciferase reporter driven by the Pol III promoter and assayed for dual luciferase. The activities are reported relative to the wild-type promoter in the absence of co-transfected Dorsal expression plasmid, which was defined to be 1. n = 3 and error bars represent S.E.M. (B) Activation of the actin5C gene by Dorsal. The actin5C promoter contains both TATA box and Inr sequence motifs. Drosophila S2R+ cells were co-transfected with a Renilla luciferase reporter construct driven by the natural enhancer-promoter of actin5C (which includes 7 putative Dorsal binding sites) containing wt (1), mTATA (2), or mTATA +DPE promoter (3), as well as expression vectors of either Dorsal or an empty vector, as indicated. To control for transfection efficiency variations, cells were co-transfected with a control firefly luciferase reporter driven by the Scr (Sex combs reduced) enhancer-promoter that is not responsive to Dorsal and assayed for dual luciferase as in (A).
It has previously been reported that Dorsal activates the actin5C gene,32 which contains both a TATA box and an initiator (Inr) motif. To analyze the activation of the actin5C, we used an actin5C genomic fragment encompassing from -2560 to +88 relative to the A+1 of the Inr that was cloned upstream of the Renilla luciferase gene (kindly provided by Prof Norbert Perrimon, Harvard Medical School). This genomic fragment contains 7 putative Dorsal binding sites. We generated two additional actin5C-Renilla reporter constructs; the first contained a mutation in the TATA box of the actin5C promoter, while the other contained a DPE motif in addition to the mutated TATA box (mTATA+DPE). The three constructs were analyzed for Dorsal activation by co-transfection into Drosophila S2R+ cells. We have performed dual luciferase reporter assays using a firefly luciferase reporter driven by the Scr (Sex combs reduced) enhancer-promoter that is not responsive to Dorsal, as a normalization control. We show that actin5C is dependent on the TATA box, while the addition of a DPE to a mutated TATA box-containing actin5C reporter could not restore its activity in the absence of transfected Dorsal. As can be seen by comparing between the activation of each reporter in the presence and absence of transfected Dorsal, Dorsal activates the wt, as well as the mTATA and the mTATA+DPE containing reporters (Fig. 1B). In general, despite the TATA box being a strong core promoter element, only a few of the Dorsal targets that are regulated by high or intermediate nuclear concentrations of Dorsal, contain a TATA box without a DPE motif. Interestingly, as shown in Figure 1, both asph and actin5C are only activated 2- to 3-fold by Dorsal, whereas the previously tested TATA-less DPE-containing Dorsal target promoters tinman, brinker, twist, and leak were activated 6-fold to over 100-fold24 under the same experimental conditions. Taken together, not only are the TATA-containing promoters underrepresented in genes that are regulated by high or intermediate nuclear concentrations of Dorsal, these findings further suggest that they are activated to lower extents as compared with DPE-containing genes.
The Core Promoter Composition Determines the “Signature” of Gene Expression
We have shown that both twist and tinman are DPE-dependent promoters.24 We suggested that the core promoter has a major impact on the transcriptional output, as hybrid enhancer-promoter constructs of tinman enhancer and twist core promoter resulted in an expression pattern similar to the native twist enhancer-promoter constructs.24 These results were obtained by in vitro transcription reactions with Drosophila embryo nuclear extracts followed by primer extension analysis.24 To further explore this effect of the core promoter, we have generated the complementary hybrid constructs, which contain the twist enhancer upstream of the tinman core promoter. Similarly to the already published experiments, we generated a series of constructs with three different core promoter variations—wt, mDPE, and mDPE, to which a TATA box was added (mDPE+TATA). The activity of the natural tinman enhancer-promoter construct is reduced to 10% following a mutation of the DPE (Fig. 2A). Interestingly, the addition of a TATA box to a mDPE-containing tinman promoter results in an alternatively initiating transcript (Fig. 2A). It is fascinating that the transcription activities of the hybrid reporter constructs using Drosophila embryo nuclear extracts are very similar to the activities of the natural enhancer-promoter constructs (Fig. 2B), suggesting that even though the enhancers are present in these constructs, the activity of the constructs in the embryo nuclear extracts is dictated by the core promoters. The alternatively initiating transcript that was detected for the tinman enhancer-promoter mDPE+TATA can also be detected in the hybrid twist enhancer-tinman core promoter mDPE+TATA construct (Fig. 2), implying that the TATA box directs alternative transcription initiation. Collectively, the analysis of the hybrid enhancer-promoter constructs in nuclear extracts derived from Drosophila embryos highlights the contribution of the core promoter to the transcriptional output both in terms of the transcription levels and start site selection.
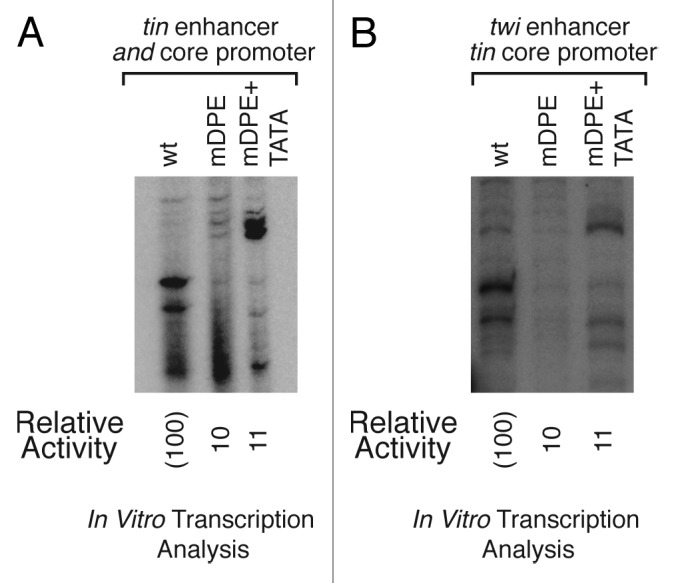
Figure 2. The core promoter is a major contributor to the transcriptional output. Transcription of both the natural tinman enhancer-promoter and the twist enhancer-tinman core promoter hybrid is dependent on the tinman DPE motif. The addition of a TATA box to a mDPE tinman-containing construct directs alternative transcription initiation. (A) The natural enhancer-promoter constructs of tinman containing either wild type (wt), mutated DPE (mDPE), or mutated DPE with a TATA box (mDPE+TATA) versions of the tinman core promoter, were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected by primer extension-reverse transcription analysis. (B) The hybrid twist enhancer-tinman promoter constructs containing either wt, mDPE, or mDPE+TATA motifs, were subjected to in vitro transcription analysis with a Drosophila embryo nuclear extract. The resulting transcripts were detected as in (A).
The DPE Motif Is Necessary for Transcriptional Regulation in Vivo
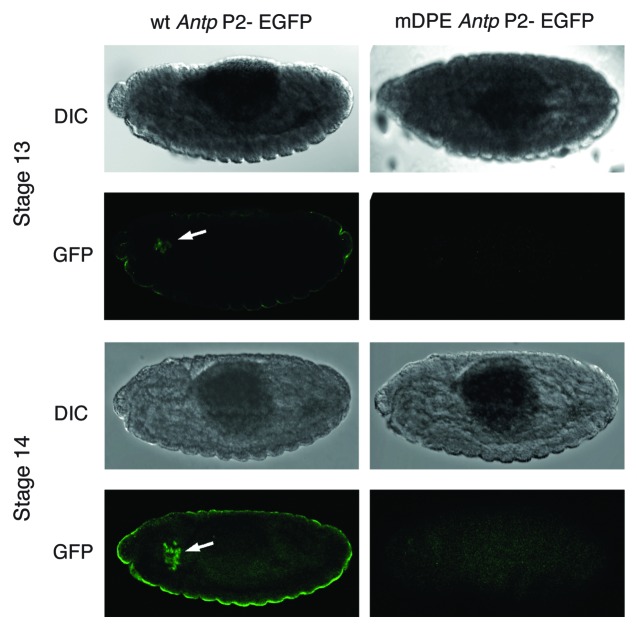
To examine the importance of the DPE motif in vivo, we have used the φC31 site-specific integration system33,34 to generate transgenic flies expressing the EGFP reporter gene driven by the natural enhancer and downstream promoter of one of the Hox genes, Antennapedia (Antp P2). The Antp P2, as well as the majority of the promoters of the Hox genes, was previously analyzed using Drosophila embryo nuclear extracts and Schneider cells and was shown to be regulated in a DPE-dependent manner.17 We have generated flies containing either the natural enhancer of Antp P2 promoter (wt) or an Antp P2 promoter with a mutation in the DPE motif (mDPE). EGFP expression, which was detected by immunostaining of 2–16 h embryos with anti-GFP antibodies, is typical for the Antp genomic fragment used35,36 and is highly dependent on the DPE, as mDPE-Antp P2-EGFP flies do not express EGFP (Fig. 3). Hence, the DPE motif is necessary for transcription of the Antp gene in the developing Drosophila embryo.
Figure 3. Expression of Antp P2-EGFP in transgenic Drosophila embryos is dependent on the DPE motif. Homozygous transgenic flies expressing the EGFP reporter gene driven by the natural enhancer and downstream promoter of the DPE-dependent Hox gene Antennapedia (Antp P2) were generated using site-specific integration. The expression of the EGFP was detected by staining of 2–16 h embryos with anti-GFP antibodies followed by a secondary antibody labeled with a Cy2 fluorophore. DIC images of the same embryos are depicted on top of each fluorescent image. Transgenic embryos for EGFP driven by the wild type (wt) enhancer-promoter of Antp P2 display an expression pattern (marked by an arrowhead) that is typical for the Antp P2 genomic fragment used, whereas transgenic embryos for EGFP driven by the Antp P2 enhancer-promoter containing a mutated DPE (mDPE) do not express EGFP.
The next challenge would be to investigate the effects of mutations in different core promoter elements of multiple developmental regulators, in order to decipher the in vivo importance of the core promoter's composition. It would be exciting to compare the in vivo expression driven by promoters in which the TATA box could compensate for the loss of a DPE and examine whether in vivo DPE-driven transcription is different than TATA-driven transcription. Such analysis may also provide new insights into the spatial and temporal expression of different core promoter variants.
The Core Promoter Is a Pivotal Dimension of Gene Regulatory Networks
Overall, these findings, together with previously published data4,37,38 strongly advocate that the core promoter itself is a critical regulatory component for gene expression. We envision the core promoter composition as an additional component of the dorsal-ventral gene regulatory network, which contributes to the combinatorial transcriptional output (Fig. 4). The enrichment of the DPE in Dorsal target promoters, as compared with the TATA box motif, is most striking in the mesodermal genes, which respond to the highest Dorsal nuclear concentrations.24 The enrichment of the DPE motif in the promoters of Dorsal target genes is decreasing with Dorsal nuclear concentration. The correlation between Dorsal nuclear concentration and core promoter composition, as well as the regulation of multiple key mesodermal Dorsal target genes via the DPE motif, reinforce this model. Further support for this model is provided by the expression pattern of hybrid enhancer-promoter constructs that recapitulate the same pattern detected for the relevant natural core promoter, and not the enhancer.
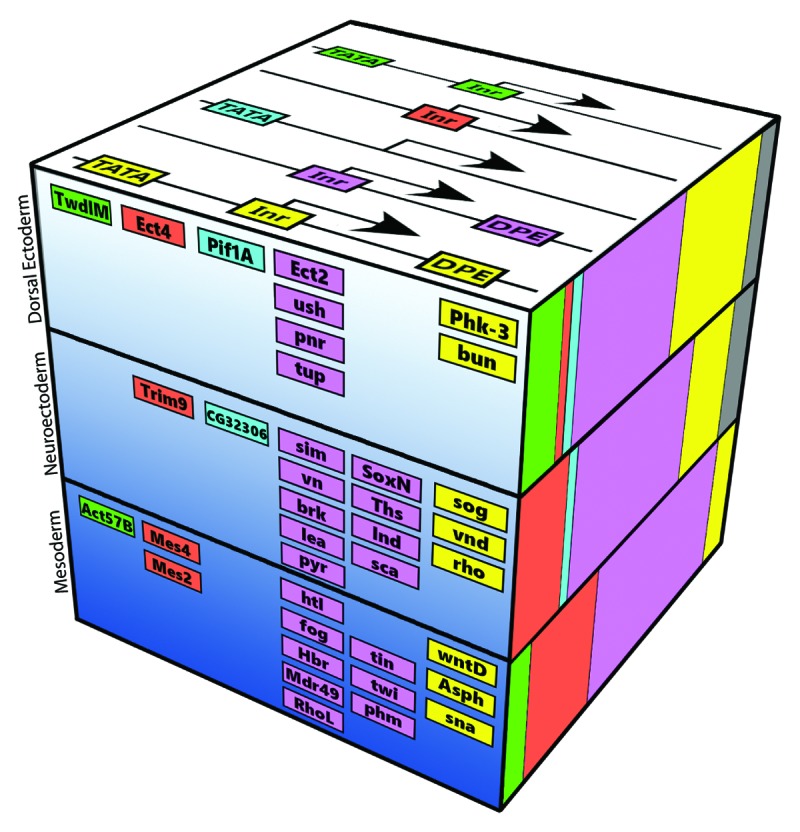
Figure 4. The core promoter composition establishes an additional dimension in the dorsal-ventral gene regulatory network. Dorsal target genes are classified according to their embryonic tissues: the mesoderm, neurogenic ectoderm, and dorsal ectoderm. The Dorsal nuclear gradient is represented by the depth of the blue color. The upper side of the cube displays the color coding of the possible combinations of the discussed core promoter elements. The front depicts selected Dorsal target genes with the corresponding color-coded core promoter composition. The relative frequency of each core promoter combination among all Dorsal target genes in each of the three tissue (using the same color code) is shown on the right.
Taken together, the core promoter composition is an integral part of transcriptional regulation. We believe that developmental gene regulatory networks that are assembled based on transcription factors and cis-regulatory modules should be revisited to incorporate the additional dimension, that of the core promoter composition.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Ron Wides for invaluable advice. We thank Dan and Ron Even for graphic design assistance. We thank Doron Ginsberg, Diana Ideses, Hila Shir, Adi Kedmi, Yehuda Danino, and Avital Ovadia-Shochat for critical reading of the manuscript. We thank Albert Courey and Norbert Perrimon for the generous gift of reagents. This research was supported by grants from the Israel Science Foundation to T.J-G. (no. 798/10) and the European Union Seventh Framework Programme (Marie Curie International Reintegration Grant) to T.J-G. (no. 256491).
References
- 1.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 2.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–51. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 3.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–6. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagha M, Bothma JP, Esposito E, Ng S, Stefanik L, Tsui C, Johnston J, Chen K, Gilmour DS, Zeitlinger J, et al. Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell. 2013;153:976–87. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 6.Ochoa-Espinosa A, Small S. Developmental mechanisms and cis-regulatory codes. Curr Opin Genet Dev. 2006;16:165–70. doi: 10.1016/j.gde.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–8. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 8.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 9.Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juven-Gershon T, Hsu J-Y, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–9. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339:225–9. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1:40–51. doi: 10.1002/wdev.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohler U, Wassarman DA. Promoting developmental transcription. Development. 2010;137:15–26. doi: 10.1242/dev.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parry TJ, Theisen JWM, Hsu J-Y, Wang Y-L, Corcoran DL, Eustice M, Ohler U, Kadonaga JT. The TCT motif, a key component of an RNA polymerase II transcription system for the translational machinery. Genes Dev. 2010;24:2013–8. doi: 10.1101/gad.1951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikstein R. The unexpected traits associated with core promoter elements. Transcription. 2011;2:201–6. doi: 10.4161/trns.2.5.17271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller F, Demény MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J Biol Chem. 2007;282:14685–9. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- 17.Juven-Gershon T, Hsu J-Y, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008;22:2823–30. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine M, Davidson EH. Gene regulatory networks for development. Proc Natl Acad Sci U S A. 2005;102:4936–42. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/S0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- 20.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–8. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson E, Levin M. Gene regulatory networks. Proc Natl Acad Sci U S A. 2005;102:4935. doi: 10.1073/pnas.0502024102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci U S A. 2008;105:20072–6. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves GT, Stathopoulos A. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehavi Y, Kuznetsov O, Ovadia-Shochat A, Juven-Gershon T. Core promoter functions in the regulation of gene expression of Drosophila dorsal target genes. J Biol Chem. 2014;289:11993–2004. doi: 10.1074/jbc.M114.550251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldberg ML. Ph.D. thesis. Stanford University 1979. [Google Scholar]
- 26.Reeve JN. Archaeal chromatin and transcription. Mol Microbiol. 2003;48:587–98. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- 27.Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–24. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 28.Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–31. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–5. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- 30.Hsu JY, Juven-Gershon T, Marr MT, 2nd, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–8. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–69. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rushlow CA, Han K, Manley JL, Levine M. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell. 1989;59:1165–77. doi: 10.1016/0092-8674(89)90772-1. [DOI] [PubMed] [Google Scholar]
- 33.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–83. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fish MP, Groth AC, Calos MP, Nusse R. Creating transgenic Drosophila by microinjecting the site-specific phiC31 integrase mRNA and a transgene-containing donor plasmid. Nat Protoc. 2007;2:2325–31. doi: 10.1038/nprot.2007.328. [DOI] [PubMed] [Google Scholar]
- 35.Boulet AM, Scott MP. Control elements of the P2 promoter of the Antennapedia gene. Genes Dev. 1988;2(12A):1600–14. doi: 10.1101/gad.2.12a.1600. [DOI] [PubMed] [Google Scholar]
- 36.Zink B, Engström Y, Gehring WJ, Paro R. Direct interaction of the Polycomb protein with Antennapedia regulatory sequences in polytene chromosomes of Drosophila melanogaster. EMBO J. 1991;10:153–62. doi: 10.1002/j.1460-2075.1991.tb07931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–56. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler JE, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–9. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]