Abstract
The nuclear pore complex (NPC) is the sole mediator of bidirectional nucleo-cytoplasmic transport and is also an important scaffold for chromatin organization and transcriptional regulation. Proteomic studies of numerous diverse eukaryotic species initially characterized the NPC as built with a number of remarkably similar structural features, suggesting its status as an ancient and conserved eukaryotic cell component. However, further detailed analyses now suggest that several key specific NPC features have a more convoluted evolutionary history than initially assumed. Recently we reported on TbNup92, a component in trypanosomes of one such conserved structural feature, a basket-like structure on the nuclear face of the NPC. We showed that TbNup92 has similar roles to nuclear basket proteins from yeasts and animals (Mlp and Tpr, respectively) in interacting with both the NPC and the mitotic spindle. However, comparative genomics suggests that TbNup92 and Mlp/Tpr may be products of distinct evolutionary histories, raising the possibility that these gene products are analogs rather than direct orthologs. Taken together with recent evidence for divergence in the nuclear lamina and kinetochores, it is apparent that the trypanosome nucleus functions by employing several novel or highly divergent protein complexes in parallel with conserved elements. These findings have major implications for how the trypanosomatid nucleus operates and the evolution of hierarchical nuclear organization.
Keywords: Trypanosoma brucei, nuclear pore complex, nuclear basket, mitotic spindle, chromosome segregation, mitosis, Mlp, nuclear evolution
Introduction
The nucleus and its associated machineries represent the major innovation differentiating eukaryotic from prokaryotic cells. Every nucleus is delimited by a double lipid membrane nuclear envelope (NE) into which NPCs (large structures containing ~30 distinct nucleoporins or Nups) are inserted.1,2 At the heart of each NPC is a well conserved core scaffold, composed of proteins with a protocoatomer architecture shared by many membrane coating proteins,3 that surrounds a central transport channel occupied by a subclass of FG-repeat containing nucleoporins (FG-Nups) (Fig. 1). Much of the bulk of FG-Nups is comprised of natively disordered domains rich in phenylalanine-glycine (FG) repeats, which function to selectively gate transport through the NPC by interacting with cargo-carrying transport factors.4 We previously speculated that the FG-Nup/transport factor recognition system is a mechanism designed to accommodate sequence evolution by the thousands of cargo molecules requiring selective transport.5,6 Here, we consider the question of whether other major component structures similarly allow the NPC to adapt to demands for evolutionary flexibility. In particular, we focus on recent work by ourselves and others on one such major NPC-associated structure, the nuclear basket, composed of filaments that project into the nucleoplasm (Fig. 1).
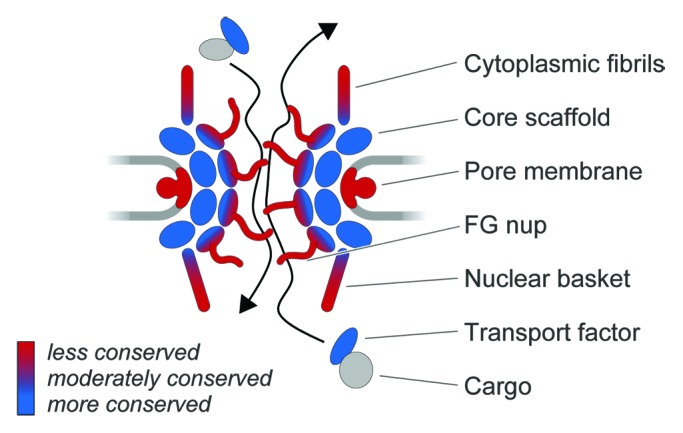
Figure 1. Evolvability, conservation and structural units of the nuclear pore complex. A schematic of the NPC is shown, with the cytoplasm at the top and nucleoplasm at the bottom. The nuclear envelope is drawn as two light gray lines. Units of the NPC are the core scaffold (blue), which are related to the protocoatomer complexes and act to deform membranes, the FG-repeat Nups (blue to red gradient), which act as the selectivity and gating activities, and the cytoplasmic fibrils and nuclear basket (blue to red gradient) which interface with cargo, chromatin, transcription and mRNA maturation pathways. The trans-membrane Nups are shown in red, reflecting our current understanding of significant variations in the identity of these Nups. The bar indicates low, intermediate and high conservation, as keyed by the blue to red gradient. Cargo is shown in gray to indicate the great variability and range in protein and RNA-protein complexes that interact with transport factors that are considerably well conserved in eukaryotes.
In the opisthokonts (animals and fungi, including yeast) the nuclear basket is composed of characteristic large (~200 kDa) proteins, termed Mlp1 and Mlp2 (Myosin-like protein 1 and 2) in yeast, and Tpr (Translocated promoter region) in mammals. NPCs use the basket proteins to associate with several nuclear peripheral processes, which extends NPC functionality.2,7 In both animal and yeast cells, ample evidence indicates that chromatin residing proximal to the NPC is rendered transcriptionally active by association with the nuclear basket, differentiating these regions from bulk peripheral heterochromatin, although this is complicated by observations that NPCs can also mediate chromatin silencing.2,8,9 Furthermore, there is evidence for specific interactions with promoter and other gene regions of particular subsets of regulated genes, that confers a “memory” of their recent expression state.10 Moreover, the nuclear basket is a key component for recruitment of RNA processing factors, linking RNA splicing, maturation, proofreading, and export through interactions with the Transcription-Export complex 2 (TREX-2) and transcription by the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex.2,7 However, it appears that not all members of these processing machineries are actually found in all eukaryotes, suggesting that alternate or highly divergent machineries are in place in many lineages.11 This raises the possibility that the basket itself might differ fundamentally between these same lineages.
Mlp/Tpr Proteins are Broadly Distributed among Eukaryotes
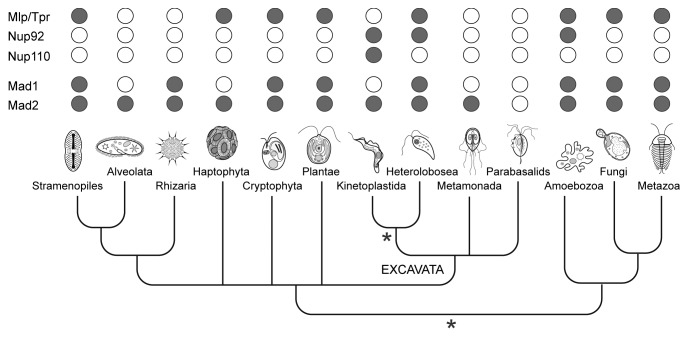
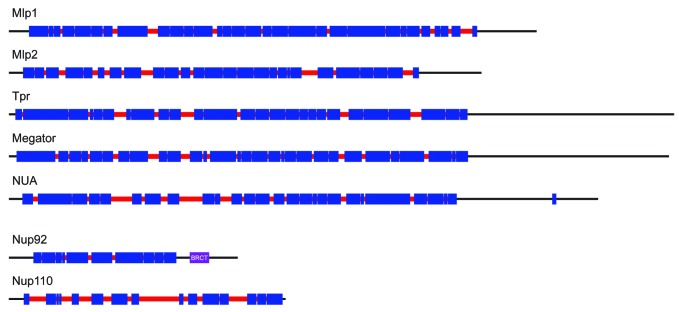
Although the Mlp/Tpr proteins have been best characterized in yeast and animals (both members of the Opisthokonta supergroup), similar proteins are present in other eukaryotic lineages (Fig. 2). An example is the plant nuclear basket protein NUA (NUclear pore Anchor), which has structural and functional similarity to Mlp/Tpr, suggesting conservation over great evolutionary distance.12,13 However, despite the broad taxonomic distribution, no Mlp/Tpr homologs have been found by sequence searches in several of the major eukaryotic groups, such as Alveolates and Kinetoplastids (supergroup Excavata), though poor primary sequence conservation complicates interpretation6,14 (Fig. 2). The Mlp/Tpr proteins are distinct from the members of the core scaffold or FG-Nup families in terms of domain architecture, being comprised almost entirely of long stretches of coiled-coil likely facilitating their oligomerization15 (Fig. 3). Most taxa possess a single Mlp/Tpr homolog per genome, though some lineages have duplicated their genes, so that for example Mlp1/Mlp2 of Saccharomyces cerevisiae arose from a relatively recent (~90 Myr ago) gene duplication event.6 Similarly, trypanosomes have two nuclear basket proteins, TbNup92 and TbNup110 in Trypanosoma brucei, that appear to be structural and functional analogs of Mlp1/Mlp2 (discussed below), but with clear distinctions.16,17 Specifically, TbNup92 (92kDa) and TbNup110 (110 kDa) are significantly smaller than Mlp/Tpr/NUA and the related Drosophila Megator basket protein, all of which are ~200 kDa (Fig. 3). Additionally, TbNup92 contains a breast cancer 1 susceptibility (BRCA1) C-terminus (BRCT) domain (Fig. 3), typically found in proteins involved in DNA repair and cell cycle checkpoints.18 Despite these differences, the functional similarities of these proteins are strongly suggestive of an ancestral origin, likely in the last eukaryotic common ancestor (LECA).6,14 Indeed, while TbNup110 appears to be a kinetoplastid-specific protein,17 orthologs of TbNup92 are present in two other eukaryotic lineages, the Amoebozoa and Heterolobosea (Fig. 2). Remarkably, both these lineages also possess a clear homolog of Mlp/Tpr, such that in these lineages, both types of nuclear basket protein are present.17
Figure 2. Distribution of the nuclear-basket components and Mad1/Mad2 proteins among eukaryotes. Homologs of the known constituents of the nuclear basket of animals, fungi and Trypanosomes as well as Mad1 and Mad2 proteins were searched in various eukaryotic groups using the BLAST and HMMER software. Positive hits (in gray) were verified by reverse BLAST. Two most preferred positions of the root within the phylogenetic tree of eukaryotes are marked with a star. While the root is traditionally placed between the “unikonts” (animals, fungi, and amoebozoa) and the remaining taxa, an alternative hypothesis suggests the root of eukaryotes lies within the group Excavata close to kinetoplastids.
Figure 3. Comparison of coiled-coil patterns of nuclear-basket proteins from different eukaryotes. The coiled-coil regions (in blue) were predicted using the Coils/Pcoils tool (http://toolkit.tuebingen.mpg.de/pcoils). Gaps in the coiled-coils are shown in red and mark the regions where either the probabilities of the coiled-coil conformation were lower than 50% for all three scanning windows (14, 21, and 28 residues) or there was a discontinuity in the heptad-repeat pattern.
Two possible explanations exist. The first is that these two classes of proteins at the basket—the TbNup92 type and Mlp/Tpr type—arose through independent events in a pre-LECA organism, forming an elaborate nuclear basket and ultimately sharing functionalities that each type retained as post-LECA sculpting removed one or the other type in most major lineages. Alternatively, both types arose by duplication from a distant, pre-LECA ancestor; post-duplication sculpting then altered the resulting two types sufficiently to obscure any obvious homology, though basic features of form and function were retained, explaining the remaining similarities noted above. Pre-LECA duplication and divergence of many other NPC components has already been proposed,1 with evidence being presented that some at least have retained similar functionalities,19 consistent with this possibility.
To further compare the nuclear basket proteins of various eukaryotes, we looked at their predicted coiled-coil pattern in greater detail (Fig. 3). Discontinuities in, and gaps between, the coiled-coil regions play important structural roles in coiled-coil proteins especially intermediate filaments such as nuclear lamins (another major family of coiled-coil proteins at the nuclear envelope).20 While only a small number of breaks in the coiled-coil pattern have been described for lamins, these features are well conserved between animals and amoebae.21 By contrast, the coiled-coils of Mlp/Tpr proteins appear more interrupted and the overall pattern of these interruptions is quite variable between various eukaryotes (Fig. 3), suggestive of evolutionary plasticity. Interestingly, the gaps between the coiled coil regions in TbNup110 are quite large and the protein apparently lacks the non-coiled C-terminal region present in other nuclear basket proteins, suggesting a distinct conformation (Fig. 3).
The Nuclear Basket and Spindle Checkpoint in Trypanosomes
As well as being NPC components there is good evidence that nuclear basket proteins play additional roles in cell cycle control and spindle morphogenesis. Thus, one of the two paralogs in both trypanosomes and yeast (TbNup92 and Mlp2 respectively), Tpr and Megator all exhibit cell cycle dependent positioning and have spindle-specific roles16,22-24 At mitosis, TbNup92 localizes along the length of the polymerizing spindle microtubules before accumulating at spindle attachment sites (SAS).17 Interestingly, a small but significant population of TbNup110 was also observed at the SAS, suggesting that TbNup92 shares some of its mitotic functionality with TbNup110.17 By contrast, at interphase both localized at the NPC. However, high-resolution imaging reveals that TbNup92 is juxtaposed to TbNup110 and extends into the nuclear interior, suggestive of a peripheral association of TbNup92 with the NPC via interaction with TbNup110.17 This is consistent with observations that TbNup92 interacts mainly with TbNup110 at the NPC, while TbNup110 has more extensive Nup contacts.17
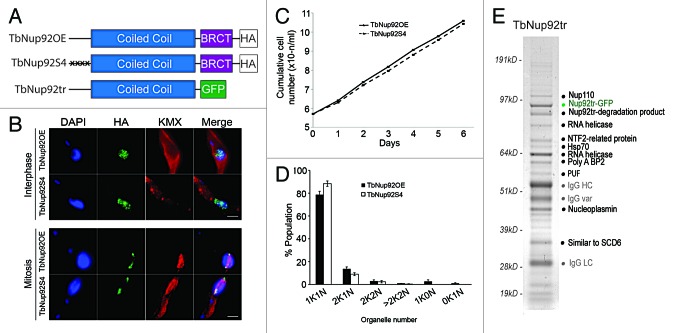
Since TbNup92 is unique among known nuclear basket proteins in possessing a BRCT domain, we assessed the role of this domain in cell cycle-dependent positioning of TbNup92, especially at mitosis. By eliminating the BRCT domain from one allele of TbNup92 (TbNup92tr), we recorded decreased proliferation combined with delays in G1/S and mitosis progression.17 Concurrently, deletion of the BRCT domain prevented the resulting TbNup92tr protein from distributing around the nuclear periphery and delayed migration to the SAS at mitosis,17 though this truncation did not prevent its interaction with TbNup110 (see below, Fig. 4A and E). Moreover, the localization of TbNup110 does not rely on TbNup92 and is unaffected in cells expressing a single copy of TbNup92tr with the second allele knocked out (TbNup92tr-KO) or in a TbNup92 null cell line (TbNup92Δ) which surprisingly, are viable.17 The TbNup92Δ cells exhibited severe proliferative defects, which mirrored an RNA interference knockdown phenotype in impacting progression through G1/S phase and chromosome segregation, possibly through improper formation of the spindle microtubules or the SAS.17 Intranuclear spindle pole bodies (SPBs) accumulate in Mlp2 knockouts,22 whereas Tpr directly influences chromosome migration to the spindle poles, through interaction with dynein and dynactin.25 Tpr and Mlps are required for the recruitment of the checkpoint proteins Mad1 and Mad2 to the kinetochores at mitosis,26-28 but in trypanosomes, only a Mad2 ortholog exists and surprisingly localizes to the basal body rather than inside the nucleus, a further example of the distinct nature of the biology at the trypanosome nuclear basket.29 Taken together, our data suggest that TbNup92 and Mlp/Tpr/Megator likely share roles in the correct construction of the mitotic spindle/SPB and are required for the correct migration of chromosomes to the poles at anaphase, but not as anchors for canonical mitotic checkpoint proteins. Delays in telomere separation to the poles point toward a role for TbNup92 at the metaphase-anaphase checkpoint. Unfortunately, the absence of cytological markers for the SAS/SPB in trypanosomes currently prevents a detailed analysis of the SAS itself. In contrast to BRCT domain-dependent interphase positioning, the coiled-coil domain of TbNup92 is important for SAS targeting, indicating that in addition to a likely structural role, the coiled-coil region contains sites for the regulated association of this protein to the spindle.
Figure 4. Mutation of known phosphorylation sites does not alter TbNup92 cell cycle-dependent targeting and the identification of interactions with RNA processing factors. (A) Schematic of TbNup92 constructs discussed here. The four known phosphorylated serine residues in the N-terminal portion of TbNup92 (S57, S58, S61, S62) were substituted with alanine (TbNup92S4) and overexpressed in procyclic cells. Overexpression of wild type TbNup92 (TbNup92OE) was used as a control. A C-terminal truncated version was also created that lacks the BRCT domain (TbNup92tr). (B) Cells expressing TbNup92S4 and TbNup92OE were probed with anti-HA and anti-β tubulin antibodies to visualize TbNup92 constructs and spindle microtubules respectively. Localizations of TbNup92S4 and overexpressed TbNup92 are indistinguishable throughout the cell cycle. DNA was visualized with DAPI (blue). Scale bars are 2 μm. (C, D) Mutation of phosphorylation sites does not impact cell growth or progression through the cell cycle. (E) TbNup92tr interacts with a number of RNA binding proteins as well as TbNup110, a heat shock protein (Hsp70) and a protein of unknown function (PUF), when affinity purified under mild buffer conditions. The TbNup92tr complexes were fractionated by SDS-PAGE and bands excised and proteins identified by electrospray ionization mass spectrometry (ESI-MS). IgG HC (heavy chain), IgG var (variant), and IgG LC (light chain) are llama anti-GFP antibodies used in the affinity isolation.
Phosphorylation is one key regulator of protein localization and several metazoan Nups are preferentially phosphorylated at mitosis to facilitate NPC and NE disassembly.30,31 Four serine phosphorylation sites (S57, S58, S61, S62) in the N-terminal portion of TbNup92 have been detected by mass spectrometry in a recent study of the T. brucei phosphoproteome.32 We tested the importance of phosphorylation by serine to alanine conversion on all four sites (TbNup92S4) (Fig. 4A). These mutations had no impact on the targeting of TbNup92 to the SAS or NPC, nor did they affect proliferation or chromosome ploidy (Fig. 4B-D), indicating that we have yet to find the regulatory mechanism for its site-specific functions or that associations with wild type TbNup92 is sufficient to faithfully target TbNup92S4.
A Minor Role for TbNup92 in RNA Processing
Trypanosomes have rather unusual mechanisms for controlling gene expression, possibly a reflection of early divergence that places them close to the eukaryotic root.33,34 The trypanosome genome contains ~8000 intron-free protein-coding genes, organized into directional polycistronic transcription units (PTUs).35-37 Each gene lacks an individual promoter but transcription start and stop sites are present for each PTU.38 Most PTUs are transcribed by RNA polymerase II into long polycistronic transcripts of functionally unrelated genes, and resolution of individual mRNAs occurs by trans-splicing and subsequent polyadenylation. Regulation of trypanosome gene expression relies heavily on mRNA turnover, as reflected by the expansion of genes encoding for RNA binding in the trypanosome genome.35,36 It is widely held that the majority of gene expression control is mediated via RNA-binding proteins acting on mRNA,39 although the gene order in PTUs contributes somewhat to regulation during the trypanosome cell cycle.40 With these considerations, we initiated a structure-function analysis of trypanosome nuclear basket proteins, because of their possible roles in mRNA surveillance, preventing unspliced or partially spliced polyadenylated RNA molecules from export to the cytoplasm.2
Significantly, TbNup92Δ cells differentially expressed genes with GO terms associated with RNA turnover, raising the possibility that TbNup92 could associate with a subset of RNA-binding proteins.17 Interestingly, a group of RNA helicases and splicing proteins co-purify with the TbNup92tr (lacking the BRCT domain) in an affinity isolation experiment performed under mild buffer conditions (Fig. 4E). Although the redistribution of TbNup92tr to the NPC is impaired, it remains localized to the nuclear periphery in a single focal point.17 It is unclear why relocalization should stabilize the interaction of TbNup92 with RNA processing proteins; nevertheless, this complex is potentially consistent with a role in mRNA processing at the nuclear basket, despite the highly divergent mechanisms of transcriptional control in trypanosomatids.
Distant Connections through Ancient Evolution
It seems that the functions of TbNup92 and its orthologs show many of the characteristics associated with Mlp/Tpr in Opisthokonts. However, the present evidence highlights the possibility that the mechanisms underpinning these functions may be somewhat distinct. Specifically, TbNup92 is more of an analog of Mlp/Tpr, rather than an ortholog, indicating a distinct evolutionary history that may either be quite separate or connected only by a very ancient common ancestor. There are additional examples of analogs functioning in otherwise conserved nuclear processes in trypanosomes. These include the lamina, which in mammals and amoeba is comprised of lamins but in trypanosomes has the NUP-1 protein as a major constituent.41 Similarly, kinetochores consist of 19 proteins in trypanosomes with no apparent similarity to kinetochore systems of non-trypanosomatid taxa.42 Do these examples represent distinct systems that evolved in parallel in LECA, with one or the other analog being subsequently lost in a lineage-specific manner? Or, does sequence divergence simply mask the deep underlying structural, evolutionary, and functional relationships? It is too early to discriminate between these possibilities, but the coincidence of baroque examples in trypanosomes provides a compelling case for digging deeply into these divergent organisms, to unravel the origins and scope of eukaryotic diversity and shed light on the mechanistic principles underlying the cellular machinery.
Animals and plants distinguish themselves from other eukaryotic kingdoms during mitosis by completely disassembling their NE. In these organisms, only one nuclear basket protein exists (Tpr/Megator/NUA). Most other eukaryotes retain their NE during mitosis. For them, the spindle is to some degree associated with the NE, and so may requires some NE associated structure, perhaps explaining the presence of a second spindle associated nuclear basket protein. We see many examples of proteins that at different times are in the NPC, the spindle, or both. Nuclear basket proteins shuttle between the spindle and NPC, including other Nups that also associate with the spindle organizer,43 and spindle checkpoint proteins are found at the NPC during interphase.44 Perhaps the NPC and SPB have a common evolutionary origin, and have retained the coordinated structural and functional interplay, though now only “touching from a distance.”
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
The work described on TbNup92 and related aspects of the trypanosome NPC was supported by the Wellcome Trust (program grant 082813 to M.C.F.), the MRC (studentship to J.M.H.), the Marie Curie fund (fellowship 300013 to L.K.), and the NIH (R21 AI096069, U54 GM103511 to B.T.C., J.A., and M.P.R., U01 GM098256 to M.P.R., P50 GM076547 to J.A. and P41 GM103314 to B.T.C.). S.K. is a Leverhulme Trust early career Fellow.
References
- 1.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 2.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 3.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8:1814–27. doi: 10.1128/EC.00225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Reilly AJ, Dacks JB, Field MC. Evolution of the karyopherin-β family of nucleocytoplasmic transport factors; ancient origins and continued specialization. PLoS One. 2011;6:e19308. doi: 10.1371/journal.pone.0019308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Field MC, Koreny L, Rout MP. Enriching the pore: splendid complexity from humble origins. Traffic. 2014;15:141–56. doi: 10.1111/tra.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ptak C, Aitchison JD, Wozniak RW. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol. 2014;28:46–53. doi: 10.1016/j.ceb.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J. 2010;29:1659–73. doi: 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell. 2013;24:3920–38. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brickner JH. Transcriptional memory at the nuclear periphery. Curr Opin Cell Biol. 2009;21:127–33. doi: 10.1016/j.ceb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erben E, Chakraborty C, Clayton C. The CAF1-NOT complex of trypanosomes. Front Genet. 2014;4:299. doi: 10.3389/fgene.2013.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–48. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding D, Muthuswamy S, Meier I. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol Biol. 2012;79:203–16. doi: 10.1007/s11103-012-9903-4. [DOI] [PubMed] [Google Scholar]
- 14.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hase ME, Kuznetsov NV, Cordes VC. Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell. 2001;12:2433–52. doi: 10.1091/mbc.12.8.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–30. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holden JM, Koreny L, Obado S, Ratushny AV, Chen WM, Chiang JH, Kelly S, Chait BT, Aitchison JD, Rout MP, et al. Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol Biol Cell. 2014;25:1421–36. doi: 10.1091/mbc.E13-12-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10:2461–70. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. J Cell Biol. 2012;196:419–34. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–85. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger A, Batsios P, Baumann O, Luckert E, Schwarz H, Stick R, Meyer I, Gräf R. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell. 2012;23:360–70. doi: 10.1091/mbc.E11-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, Rout MP. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005;170:225–35. doi: 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi H, Rath U, Wang D, Xu YZ, Ding Y, Zhang W, Blacketer MJ, Paddy MR, Girton J, Johansen J, et al. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol Biol Cell. 2004;15:4854–65. doi: 10.1091/mbc.E04-07-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lince-Faria M, Maffini S, Orr B, Ding Y, Cláudia Florindo, Sunkel CE, Tavares A, Johansen J, Johansen KM, Maiato H. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J Cell Biol. 2009;184:647–57. doi: 10.1083/jcb.200811012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano H, Funasaka T, Hashizume C, Wong RW. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem. 2010;285:10841–9. doi: 10.1074/jbc.M110.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–19. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4362–74. doi: 10.1091/mbc.E05-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22:2926–31. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akiyoshi B, Gull K. Evolutionary cell biology of chromosome segregation: insights from trypanosomes. Open Biol. 2013;3:130023. doi: 10.1098/rsob.130023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favreau C, Worman HJ, Wozniak RW, Frappier T, Courvalin JC. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 1996;35:8035–44. doi: 10.1021/bi9600660. [DOI] [PubMed] [Google Scholar]
- 31.Laurell E, Beck K, Krupina K, Theerthagiri G, Bodenmiller B, Horvath P, Aebersold R, Antonin W, Kutay U. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 2011;144:539–50. doi: 10.1016/j.cell.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Nett IR, Martin DM, Miranda-Saavedra D, Lamont D, Barber JD, Mehlert A, Ferguson MA. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol Cell Proteomics. 2009;8:1527–38. doi: 10.1074/mcp.M800556-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 2010;6:342–5. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He D, Fiz-Palacios O, Fu CJ, Fehling J, Tsai CC, Baldauf SL. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24:465–70. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 35.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–22. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 36.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–42. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6:e1001090. doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel TN, Hekstra DR, Kemp LE, Figueiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Cross GA. Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev. 2009;23:1063–76. doi: 10.1101/gad.1790409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clayton CE. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–8. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly S, Kramer S, Schwede A, Maini PK, Gull K, Carrington M. Genome organization is a major component of gene expression control in response to stress and during the cell division cycle in trypanosomes. Open Biol. 2012;2:120033. doi: 10.1098/rsob.120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DuBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, et al. NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akiyoshi B, Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–58. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatel G, Fahrenkrog B. Nucleoporins: leaving the nuclear pore complex for a successful mitosis. Cell Signal. 2011;23:1555–62. doi: 10.1016/j.cellsig.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 44.Wozniak R, Burke B, Doye V. Nuclear transport and the mitotic apparatus: an evolving relationship. Cell Mol Life Sci. 2010;67:2215–30. doi: 10.1007/s00018-010-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]