Abstract
Anoxygenic and oxygenic bacteria directly convert solar energy into biomass using photosynthesis. The formation and composition of photosynthetic complexes has to be tightly controlled in response to environmental conditions, as exposure to sunlight can be harmful due to the generation of reactive oxygen species and the damaging effects of UV irradiation. Therefore, photosynthetic bacteria are exposed to a particular set of regulatory challenges in addition to those that also affect other bacteria, requiring sophisticated regulatory systems. Indeed, hundreds of potential regulatory RNAs have been identified in photosynthetic model bacteria as well as antisense RNAs (asRNAs) of up to several kb in length that protect certain mRNAs from degradation. The trans-acting small non-coding RNAs (sRNAs), PcrZ and PsrR1, control pigment and photosystem biogenesis in Rhodobacter sphaeroides and cyanobacteria, respectively. The asRNAs IsrR and As1_flv4 act as negative regulators and the asRNAs PsbA2R and PsbA3R as positive effectors of photosynthesis gene expression in Synechocystis 6803.
Keywords: antisense transcription, cyanobacteria, differential RNA-seq, gene expression regulation, photosynthesis, α-proteobacteria
Introduction
Photosynthetic bacteria are exposed to a particular set of regulatory challenges that set them apart from other bacteria. During recent years, extensive transcriptome analyses have identified hundreds of regulatory RNA candidates in photosynthetic model bacteria. The expression of several of these RNA molecules is tightly linked to the presence of certain oxygen species; additionally, their expression may be regulated in concert with light intensity or factors such as the availability of nutrients, such as iron, that affect photosynthetic electron transfer processes. Moreover, several cis-antisense RNAs have been found that are co-regulated or inversely regulated with their photosynthetic host genes. Such patterns of gene expression suggest that these RNA molecules might play a role in the control of photosynthetic gene expression. Indeed, the functions of several regulatory RNA candidates have recently been elucidated.
In this review, we provide an overview of these regulatory RNA molecules. However, we also aim to give some sense of how many more regulatory RNA candidates might exist and still await detailed characterization.
Photosynthetic bacteria that serve as model organisms
Several photosynthetic bacteria have been established as model organisms due to their environmental importance and the availability of genome information and sophisticated genetic systems. In the frame of this review, we will focus on four photosynthetic model bacteria for which extensive transcriptome information has been elucidated.
Anoxygenic phototrophic bacteria are mainly found in the anoxic zone of many water bodies. Photosynthetic proteobacteria are also called purple bacteria, due to their color, which originates from their high carotenoid content. They contain bacteriochlorophylls a or b, are able to fix dinitrogen, and use a type II reaction center for photosynthesis.1 Among bacteria capable of anoxygenic photosynthesis, Rhodobacter sphaeroides 2.4.1 forms photosynthetic complexes in response to environmental stimuli, i.e., it is facultatively phototrophic. Under high oxygen tension, R. sphaeroides performs aerobic respiration and the expression of photosynthesis genes is repressed. However, at low oxygen tension or in the absence of oxygen, photosynthesis genes become transcribed and the organism switches to a photosynthesis-driven physiology.2
Cyanobacteria perform oxygenic photosynthesis. They are considered to be promising resources for the sustainable production of biofuels and other products, but many species belonging to this group are also important primary producers. Their habitat is widespread, encompassing nearly all aquatic and soil habitats. In addition to oxygenic photosynthesis and photosynthetic carbon fixation, many cyanobacteria are also able to fix atmospheric dinitrogen in an oxic environment. Because the key enzyme of dinitrogen fixation, nitrogenase, is sensitive to oxygen, these cyanobacteria either are capable of differentiating heterocysts, a cell type specialized for dinitrogen fixation, or have evolved a temporal separation of photosynthesis and dinitrogen fixation.3,4
The unicellular Synechocystis sp. PCC 6803 (from here on Synechocystis 6803) is one of the most popular cyanobacterial model organisms as it is amenable to straightforward genetic manipulation. Moreover, Synechocystis 6803 was the first phototrophic organism and the third organism ever for which a complete genome sequence was determined.5 In contrast, Prochlorococcus is a unicellular marine cyanobacterium without an established genetic system. However, Prochlorococcus numerically dominates the phytoplankton of the oligotrophic open oceans, typically between 40°N and 40°S. With up to 105 cells per ml, it contributes a significant fraction of the photosynthetic biomass and primary productivity, and thus, has a tremendous impact on the oceanic ecosystem and biogeochemical cycles.6 Therefore, Prochlorococcus has become a useful model of an ecologically important cyanobacterium that may be used to study the nature and regulation of biological diversity across all scales of complexity.7 Prochlorococcus exists in several ecotypes; the most important of these have been defined according their ability to adapt to high-light (e.g., the laboratory model strain MED4) or low-light regimes (e.g., the strain MIT9313).8 Prochlorococcus typically possesses very compact and streamlined genomes of 1.6–2.4 Mbp,9 with only very few annotated genes encoding protein regulators, which raised the question of whether the regulatory network relies mainly on regulatory RNA.10
Anabaena sp. PCC 7120 (also known as Nostoc sp. PCC 7120, from here on Anabaena 7120) differentiates heterocysts.3 Therefore, Anabaena 7120 is a model organism for those cyanobacteria that have true multi-cellularity, differentiate specialized cell types, and perform dinitrogen fixation.
Regulation of photosynthesis and photooxidative stress
Multiple functions of sRNAs in the regulation of bacterial gene expression are well established. It is not surprising that many sRNAs were also identified in photosynthetic bacteria, which have special requirements for balanced gene expression. Photosynthetic complexes on one hand confer the ability to use light as energy source; but on the other hand, exposure to light can be harmful due to the damaging effect of UV and the generation of reactive oxygen species. While respiratory electron transfer favors superoxide (O2‒) and hydrogen peroxide (H2O2) generation in a process known as autoxidation of flavoenzymes, the presence of (bacterio)chlorophylls in bacterial photosynthetic membranes gives rise to singlet oxygen (1O2) through photo-sensitized reactions.11,12 To avoid photooxidative stress, photosynthetic bacteria adjust the amount and/or the composition of the photosynthetic complexes (especially antennae) based on the existing light conditions. Many photosynthetic species possess alternative pathways for ATP production and can regulate the formation of photosynthetic complexes in response to other environmental conditions. For example, anoxygenic photosynthetic bacteria of the genus Rhodobacter do not form photosynthetic complexes at high oxygen concentrations. In contrast, cyanobacteria evolve oxygen during photosynthesis. Thus, all photosynthetic bacteria have evolved special strategies to protect photosynthetic complexes from damage and to defend against photooxidative stress. In addition, optimization of the photosynthetic apparatus according to the light conditions ensures efficient light driven electron transfer reactions, exemplified by cyanobacteria, which change the composition of their antennae system, the phycobilisome, in response to different light qualities to absorb a maximal amount of photons of different wavelengths.13 Moreover, several cyanobacteria are able to move toward favorable or away from harmful light conditions.14,15 Unique among prokaryotes, cyanobacteria possess a true circadian clock that is important for anticipating light and dark phases.16
Review
Methods for studying the transcriptomes of photosynthetic bacteria and for identifying sRNAs
Several technically different approaches have been utilized to identify the non-coding transcriptome of photosynthetic bacteria. Computational predictions in conjunction with experimental verification experiments were performed for model cyanobacteria including Synechocystis 6803,17,18 marine Synechococcus,19 and Prochlorococcus.20
Tiling or high-resolution microarrays were employed for R. sphaeroides,21 Synechocystis 6803, and Prochlorococcus MED4.10,22,23
High-throughput RNA sequencing based on 454 pyrosequencing was applied to study the transcriptomes of R. sphaeroides24 and of the cyanobacteria Synechocystis 6803,23 Prochlorococcus MED4,25 and Nodularia spumigena,26 which led to the genome-wide identification of transcriptional start sites (TSS) and non-coding transcripts. In an extension of this approach, comparative analysis of the primary and secondary transcriptomes was performed for Anabaena 7120.27
Exploring the intergenic regions using microarrays
Classic approaches for the global detection and quantification of transcripts use gene expression microarrays, which are based on hybridization of RNA or corresponding cDNA samples to DNA probes. However, microarrays are usually blind to transcripts derived from intergenic regions (IGRs), because most microarray designs merely rely on the coding information of genomes. Adding probes for IGRs in both strand orientations to the microarray setup therefore represents a suitable strategy for the identification of sRNAs and was successfully applied to the model cyanobacteria Synechocystis 6803 and to Prochlorococcus MED4 and MIT9313.10,22,23 Because the genome coverage and resolution of a given microarray are determined by the probe density, the most comprehensive results can be achieved by overlapping probes all over the genome in both strand orientations. The use of tiling microarrays successfully identified sRNAs in IGRs and explored massive antisense transcription in the model cyanobacterium Synechocystis 6803.22 Furthermore, high-resolution tiling arrays identified sRNAs in the cell division model bacterium Caulobacter crescentus and the legume symbiont Rhizobium etli.28,29 The Caulobacter microarray used a tiling probe spacing of 5 bp that facilitated the accurate determination of sRNA 5′ and 3′ ends. However, none of these examples achieved resolution of sRNAs on the single nucleotide level.
Using high-resolution Affymetrix microarrays to study the transcriptional landscape of Prochlorococcus MED4, 14 novel sRNAs and 24 asRNAs were identified.10 The absence of an efficient genetic system for Prochlorococcus makes the functional characterization of these sRNAs difficult, but expression profiling suggested several of these sRNAs to be involved in the adaptation to light stress, based on their regulated expression. Moreover, a role of some of these sRNAs in stress adaptation processes would be consistent with the preferred location of their genes in hypervariable genomic islands.10 It is interesting to note that the proportion of sRNA genes in Prochlorococcus MED4 is comparable with that found in model bacteria such as Escherichia coli, i.e., with 1–2% of the genes encoding trans-acting sRNAs. This is in contrast to the number of protein regulators, which is considerably smaller in Prochlorococcus (e.g., six vs. 32 two-component response regulators present in E. coli). Apparently, sRNA regulators require less coding capacity and are less costly to make than protein factors. Therefore, regulation of gene expression through sRNAs may play a relatively more important role in Prochlorococcus than in E. coli, and would be consistent with its extreme adaptation to the low nutrient conditions of the open oceans and the associated genome streamlining process.
Differential RNA sequencing to explore the primary transcriptome of photosynthetic bacteria
To overcome the limitations of microarray-based approaches, RNA sequencing (RNA-seq) has been applied to identify sRNAs and to map exact transcript boundaries in many bacterial species. In particular, differential RNA-seq (dRNA-seq) emerged as a powerful tool for exploring the primary transcriptome by treating RNA samples with Terminator 5′-Phosphate-Dependent Exonuclease (TEX). Primary transcripts typically possess a 5′ triphosphate, while processed transcripts have a 5′ monophosphate or 5′ OH group instead. When treating total RNA samples with TEX, processed transcripts are depleted and primary transcripts are therefore indirectly enriched. The comparison of RNA-seq results from TEX-treated and untreated RNA both identifies sRNAs and determines the TSS of virtually all genes. dRNA-seq was developed to investigate the primary transcriptome of the human pathogen Helicobacter pylori,30 but has been successfully applied to plenty of other model prokaryotes, leading to the identification of manifold sRNAs in these organisms.31-38
Rhodobacter sphaeroides
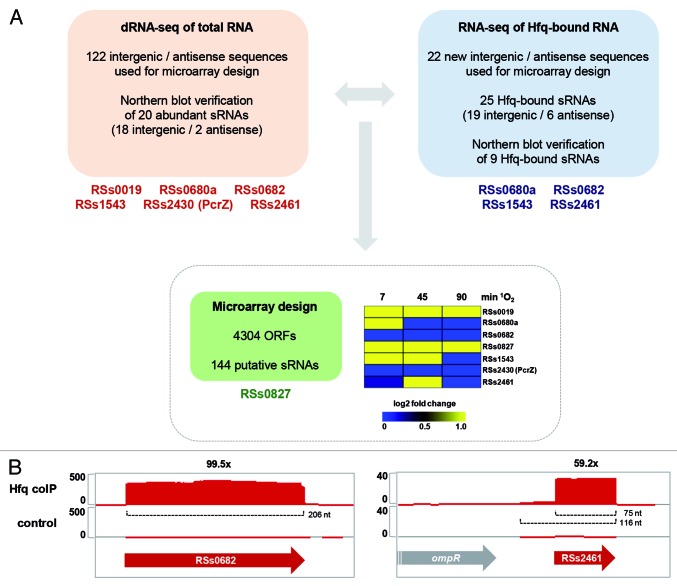
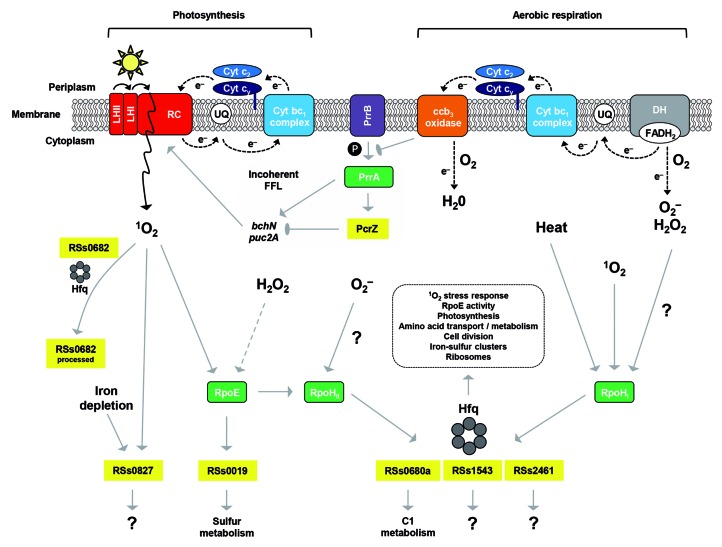
In R. sphaeroides, dRNA-seq identified more than 122 transcripts that are from IGRs or that are antisense to annotated genes, representing at least 107 putative sRNAs and 15 asRNAs (Fig. 1A). The 18 most abundant sRNAs were verified by northern blot analysis and named RSs (RNAs for R. sphaeroides sRNAs).24 This initial study focused on the stress factor 1O2, and dRNA-seq was therefore performed for samples from 1O2 stress experiments. In addition, cDNA libraries from both unstressed and O2‒ conditions served as references for the detection of 1O2-specific events. Intriguingly, the fraction of intergenic transcripts increased from ~10% in the unstressed control library to ~21% and ~16% in libraries from 1O2 and O2‒ experiments, respectively. This indicates that transcription of sRNAs is considerably induced under stress conditions, underscoring their functional role in the oxidative stress response of R. sphaeroides. Two sRNAs, namely RSs0680a and RSs2461, were induced by both 1O2 and O2‒, while RSs0019 was 1O2-specific (Fig. 2). Another sRNA, RSs0682, was the most abundant transcript in all cDNA libraries and exhibited strong accumulation of a processed product when exposed to 1O2, suggesting that processing serves an important function. RSs1543 and RSs2430, although initially identified in the dRNA-seq approach, were not recognized as oxygen-responsive sRNAs until subsequent screens were performed. RSs1543 was found together with RSs0680a and RSs2461 to depend on the RNA polymerase sigma factors RpoHI and RpoHII.39 The RpoHI/RpoHII promoter is responsible for their induction by H2O2 and 1O2 through the RpoE-RpoHII signaling cascade and by heat stress via RpoHI (Fig. 2). In this example, integration of different stress signals is mediated by alternative sigma factors, and obviously involves post-transcriptional regulation by downstream sRNAs. Finally, RSs2430 (now renamed PcrZ, see below) was revealed as a regulation factor balancing the production of photosynthetic components in an incoherent feed-forward loop under low oxygen tension.40 For a quick reference on the regulatory RNAs discussed here in detail, see Table 1.
Figure 1. Global analyses for the identification and characterization of sRNAs in R. sphaeroides. (A) dRNA-seq and RNA-seq from Hfq coIP experiments identified intergenic and antisense transcripts representing putative sRNAs and asRNAs, respectively.24,82 Newly identified transcripts and ORF annotations were used for microarray design. The sRNAs given below the colored boxes indicate their identification in the respective analysis and were chosen due to their oxygen-responsive expression (see main text and Fig. 2 for details). The heat map illustrates results for these sRNAs from a microarray study conducted for the singlet oxygen (1O2) stress response.21 Expression levels after treatment with methylene blue in light are compared with expression levels before singlet oxygen stress. (B) Integrated Genome Browser (IGB) screenshots for RNA-seq results from Hfq coIP experiments. Mapping of cDNA reads is shown for the coIP with 3xFLAG-tagged Hfq (Hfq coIP) and an unspecific control experiment (control). The scale on the left hand side of each screenshot displays relative read numbers. Red and gray arrows represent sRNA genes and ORFs, respectively. The numbers at the top indicate enrichment factors calculated for sRNAs. Transcript lengths are given for each sRNA.
Figure 2. Regulation model for oxygen responsive sRNAs in R. sphaeroides. Reactive oxygen species are either generated by energy transfer during photosynthesis or due to electron transfer mediated by flavoenzymes during aerobic respiration. Several alternative sigma factors (RpoE, RpoHI, RpoHII) are activated, which subsequently induce sRNAs. During aerobic respiration, phosphorylation of the response regulator PrrA by the sensor kinase PrrB is inhibited due to electron flow through the ccb3 oxidase. Under low oxygen tension, PrrA is phosphorylated and triggers PcrZ transcription, which in turn, represses photosynthesis gene expression. Interaction of some sRNAs with Hfq is indicated. (LH, light harvesting complex; RC, reaction center; UQ, ubiquinone pool; Cyt, cytochrome; P, phosphate, DH, dehydrogenase; FADH2, reduced flavin adenine dinucleotide; FFL, feed-forward loop; 1O2, singlet oxygen; O2‒, superoxide radical; H2O2, hydrogen peroxide).
Table 1. Individual sRNA and asRNA regulators discussed in this review.
| Name | Organism | Targets | Comments | Ref. |
|---|---|---|---|---|
| RSs0680a | R. sphaeroides | likely C1 metabolism genes | inducible by 1O2, O2‒ and heat | 24 , 39 |
| RSs1543 | R. sphaeroides | n.k. | inducible by 1O2, O2‒ and heat | 39 |
| RSs2461 | R. sphaeroides | n.k. | inducible by 1O2, O2‒ and heat | 24 , 39 |
| RSs0019 | R. sphaeroides | likely sulfur metabolism genes | inducible by 1O2 | 24 |
| RSs0682 | R. sphaeroides | n.k. | most abundant sRNA | 24 |
| RSs2430, PcrZ | R. sphaeroides | bchN mRNA (light independent protochlorophyllide reductase) and puc2A mRNA (pigment binding protein) | induced under low oxygen tension | 40 |
| SyR1 or PsrR1 | Synechocystis 6803 | likely several targets in the photosynthetic apparatus | strongly inducible by high light | 23 |
| IsrR | Synechocystis 6803 | isiA negative regulator | involved in iron stress response, codegradation with target mRNA | 61 , 62 |
| As_flv4 | Synechocystis 6803 | flv4, flv4-flv2 operon negative regulator | involved in the response to shifts in inorganic carbon supply | 65 |
| PsbA2R, PsbA3R | Synechocystis 6803 | positive regulator of psbA2 and psbA3 mRNA stability | expression is co-stimulated with target mRNAs by light | 66 |
| α-furA | Anabaena 7120, Microcystis aeruginosa PCC7806 | furA negative regulator | involved in the integration of iron and redox stress responses | 77 , 78 |
| NsiR1 | Anabaena 7120 | n.k. | inducible by nitrogen depletion in developing heterocysts | 43 |
| Yfr1 | Prochlorococcus MED4, Synechococcus elongatus PCC 6301, many other | outer membrane proteins PMM1119 and PMM1121 (P. MED4); sodium-dependent bicarbonate transporter SbtA (S. elongatus) | very abundant sRNA | 20 , 57 , 58 , 60 |
The first column gives the name of the respective sRNA or asRNA, the second the organisms in which it has been found, the third the regulatory targets (n.k., not known), followed by additional relevant observations and respective references (Ref.).
Synechocystis 6803
In Synechocystis 6803, dRNA-seq identified more than 400 TSS driving the transcription of putative non-coding RNAs from IGRs (nTSS) and more than 1000 TSS antisense to annotated genes (aTSS) in the chromosome and plasmids pSYSA, G, M, and X.23 Details of the TSS numbers and classification are summarized in Table 2. Interestingly, sRNAs derived from the three clustered regularly interspaced short palindromic repeats (CRISPR) systems on plasmid pSYSA (crRNAs) were among the most abundant transcripts in the cell.41 Complementary microarray analyses demonstrated a significant expression level of asRNAs (404 of 537) and sRNAs (166 of 194) for which probes were available on the used microarrays. However, most importantly, a similar percentage of putative trans-acting sRNAs (46.4%) and of mRNAs (43%) showed a significantly reduced or enhanced expression level when cyanobacterial cultures were exposed to three conditions considered relevant for photosynthetic growth (high light, CO2 depletion, or darkness).23 Thus, in a “guilty-by-association” approach, the identified sRNAs appeared as likely to be functionally relevant. Indeed, the sRNA SyR1 (now re-named PsrR1), which was most strongly upregulated under high light in this experiment eventually turned out to be a regulator of photosynthesis.
Table 2. Overview on the number and types of putative TSS (see Fig. 3 for explanation of TSS types) mapped for photosynthetic model bacteria by dRNA-seq.
Anabaena 7120 and Nodularia spumigena CCY9414
Anabaena 7120 and N. spumigena are filamentous diazotrophic cyanobacteria that differentiate heterocysts. Whereas Anabaena 7120 is a well-established model system for bacterial developmental biology and for the study of dinitrogen fixation (for reviews, see refs. 3 and 42), N. spumigena is of ecological interest as it frequently dominates the annual late summer cyanobacterial blooms in brackish water ecosystems such as the Baltic Sea. Their dRNA-seq-based transcriptome analysis provided genome-wide maps of more than 6000 and 10 000 TSSs (Table 2), driving the transcription of coding and non-coding regions, respectively.26,27 From these, orphan TSSs located in intergenic regions led to the prediction of 764 sRNAs in N. spumigena, among them, 70 copies of a possible retrotransposon and several potential RNA regulators, some of which are also present in other dinitrogen-fixing cyanobacteria.26 The comparison of dRNA-seq data from three different cyanobacteria shows consistently that only 25–33% of all TSS drive the transcription of annotated genes, whereas the vast majority of the transcriptional output is likely non-coding (Table 2).
In an extension of the dRNA-seq approach, a comparative analysis was performed on the primary and secondary transcriptomes of the wild-type Anabaena 7120 strain and of a hetR inactivation strain that shows no sign of heterocyst differentiation, both under nitrogen-replete conditions and 8 h after nitrogen step-down.27 This study led to the identification of a high number of previously unknown asRNAs and sRNAs whose expression was strongly regulated in response to nitrogen stress. The similar expression dynamics to those of known regulators of cell differentiation and dinitrogen fixation, combined with the fact that several of these asRNAs are located antisense to known heterocyst-related genes, suggested that many of these transcripts might, in fact, be involved in these processes.27 An example of a particularly interesting sRNA is NsiR1 (nitrogen stress inducible RNA1), which is transcribed from a tandem array of direct repeats upstream of hetF,43 one of the known regulators of heterocyst differentiation.44 There are two interesting aspects linked to NsiR1. First, NsiR1 is transcribed from a series of tandem repeats, a genetic element that has seldom been reported in bacteria.45 Except for the CRISPR repeats, functions of bacterial tandem repeats have largely remained known.46 Second, the transcription of NsiR1 depends on the two transcription factors critical for dinitrogen fixation and heterocyst development, NtcA and HetR, and thus is restricted to developing heterocysts, which was demonstrated in promoter-GFP fusion experiments.43 With these features, NsiR1 is the only known bacterial sRNA that is developmentally regulated, whereas the parallel but regulated expression of up to 12 almost identical repeats suggests its very high concentration at a certain points of time is critical for its function. See Table 1 for an overview on the sRNAs discussed in detail in this review.
Adaptation to Alternating Light and Oxygen Regimes Requires sRNAs in Rhodobacter sphaeroides
An sRNA as a regulator of photosynthesis genes: PcrZ
The first dRNA-seq approach to identify sRNAs in R. sphaeroides revealed RSs2430 (now renamed PcrZ for photosynthesis control RNA Z,40) as a highly abundant sRNA in this bacterium.24 Interestingly, this sRNA showed a similar expression pattern as genes encoding proteins for pigment binding and pigment synthesis: its expression increased upon reduction of oxygen tension. Indeed, PcrZ is under control of the two component system PrrB/PrrA,47,48 which activates photosynthesis genes when oxygen tension drops. Interestingly, growth conditions do not only affect transcription of the pcrZ gene but also affect processing of the PcrZ RNA generating stable fragments approximately 50 nt from the 5′ end of the 136 nt primary transcript. A strain overexpressing PcrZ verified the hypothesis that PcrZ may be involved in regulation of photosynthesis gene expression. The strain with increased PcrZ expression showed a significantly lighter color due to reduced amounts of pigment protein complexes, indicating that PcrZ is counteracting the induction of photosynthesis genes at low oxygen. This effect was not observed when a 51 nt PcrZ processing fragment was overexpressed, indicating that processing renders PcrZ functionally inactive. Microarray analyses revealed that PcrZ affects the expression of a very limited number of R. sphaeroides genes, the majority of which were photosynthesis genes. A newly established reporter gene system for alphaproteobacteria confirmed the bchN mRNA (for light independent protochlorophyllide reductase) and the puc2A mRNA (pigment binding protein) as direct targets of PcrZ (Table 1 and Fig. 2).
Transcription of PcrZ is induced together with photosynthesis genes under low oxygen tension by the PrrA response regulator, but PcrZ itself downregulates photosynthesis gene expression, indicating that it is part of an incoherent feed-forward loop (Fig. 249). The kinetics of induction of the pucBA genes for pigment binding proteins and of PcrZ were followed after reduction of oxygen tension in a wild-type strain and a strain with constitutively high levels of PcrZ. While pucBA mRNA levels in wild-type increased rapidly upon reduction of oxygen and then slightly decreased, constant high levels of PcrZ led to delayed induction of pucBA, which steadily increased over the 5 h of the experiment. PcrZ thus helps to balance the response of photosynthesis genes to oxygen conditions, thereby avoiding a steady accumulation of photosynthetic complexes that may consequently lead to increased photooxidative stress.
sRNAs with a role in oxidative stress responses
The initial dRNA-seq study by Berghoff and colleagues24 detected sRNAs not only under aerobic growth conditions but also after applying superoxide or 1O2 stress. As mentioned above, further analyses revealed that some sRNAs are induced by a variety of stresses. Those sRNAs were mostly under the control of RpoHI/RpoHII sigma factors.39 Genes for both alternative sigma factors show increased expression in response to heat or oxidative stress. While rpoHI expression shows the strongest response to heat stress, rpoHII expression shows a stronger increase in response to 1O2.50 RSs0680a, RSs2416, and RSs1543 are among the sRNAs triggered by various stresses (Fig. 2). While all other stress-induced sRNAs are present in single copy on the chromosome, RSs0680a is present as a repeat of four redundant sRNAs.24 All four individual sRNAs, called RSs0680a-d, contain a CCUCCUCCC motif within two stem-loop structures. RSs0680a-d are transcribed from an RpoHI/RpoHII promoter together with the upstream gene RSP_6037, which encodes a protein with a domain of unknown function (DUF) 1127. Interestingly, homologs of RSs0680a-d genes are also found in other alphaproteobacteria, in at least two adjacent copies and either upstream or downstream of genes encoding DUF1127 proteins.
The exact mechanism of RSs0680a-d function is still under investigation. Bioinformatic target predictions, as well as microarray and proteome analyses with a strain overexpressing this sRNA repeat, strongly support a role in C1-metabolism, and consequently, an effect on the glutathione pool (unpublished data). Because glutathione provides the reducing power to reverse the oxidation of cellular proteins by reactive oxygen species, it has a central role in oxidative stress defense.
There are indications that glutathione levels are influenced by yet another sRNA through a completely different pathway. The essential component of glutathione is the sulfur-containing amino acid cysteine, which switches between a reduced and oxidized state to carry out its antioxidant function. However, in proteins, cysteine residues are the primary target of singlet oxygen, leading to the formation of numerous hydroperoxides as secondary damage.51 The sRNA RSs0019 was found to be induced by both singlet oxygen and organic hydroperoxides (ref. 24 and unpublished). RSs0019 is under the control of the alternative sigma factor RpoE, which has a major role in the singlet oxygen response in R. sphaeroides.11,52,53 Overexpression of RSs0019 combined with microarray analysis strongly points to genes of sulfur metabolism as being predominantly repressed by this particular sRNA. As in the case of PcrZ and photosynthesis genes, RSs0019 might balance the uptake of sulfate and synthesis of sulfur-containing amino acids to guarantee appropriate cysteine and glutathione levels under the given environmental conditions.
Even more stress-related sRNAs?
The results obtained for RSs0680a-d and RSs0019 strongly emphasize their functional role in the (photo)oxidative stress response. Thus, resistance phenotypes are the expected outcome when manipulating their expression levels. This is consistent with observations that altered expression of RSs2461 or RSs1543 sRNAs affects the resistance of R. sphaeroides to photooxidative stress (unpublished data), strongly indicating their role in this response. However, their mechanisms of action have not been elucidated to date. This begs the question, how many sRNAs can be found that respond to a single stress? The information derived from dRNA-seq and an Hfq coIP approach (see below) was subsequently used for microarray design. The R. sphaeroides custom gene expression microarrays from Agilent Technologies (8x15K; ID: 027061) contain oligodeoxynucleotide probes (60-mers) for 4304 ORFs and 144 putative sRNAs and asRNAs (Fig. 1A). In a survey of the R. sphaeroides 1O2 response, the microarrays were applied to reveal global expression dynamics. For RSs0019, RSs0680a, RSs1543, and RSs2461, induction patterns similar to those observed in northern blot analyses were apparent. RSs0682 and PcrZ did not show major changes in expression levels, as expected (Fig. 1A). These data confirmed the reliability of the microarray approach for assessing sRNA expression levels and additionally identified a new 1O2-dependent sRNA (RSs082721), which was formerly described as being induced under iron limitation.54 Hence, the combination of global gene expression approaches highlighted at least five sRNAs that are induced upon 1O2 stress in R. sphaeroides.
Regulation of Photosynthesis-Relevant Genes with Regulatory RNAs in Cyanobacteria
The trans-acting sRNA PsrR1 and oxygenic photosynthesis
Biocomputational prediction,18 tiling microarrays,22 and pyrosequencing of Synechocystis 6803 transcripts23 revealed the existence of an sRNA, initially called SyR1 (for Synechocystis RNA 1) but now re-named PsrR1 (for Photosynthesis regulatory RNA 1; Table 2). At the time, a homolog of PsrR1 was computationally predicted in Synechococcus sp. PCC 6301 and two were predicted in Microcystis aeruginosa sp. PCC 7806, whereas no putative homolog was predicted in the more distantly related Thermosynechococcus sp. BP1.18 In Synechocystis 6803, transcription of the 131 nt-long PsrR1 was demonstrated from the IGR between the genes fabF (slr1332) and hoxH (sll1226).18 More detailed analyses of its expression revealed that PsrR1 is strongly upregulated under high light treatment and CO2 depletion.22 The overexpression of PsrR1 from an inducible promoter led to a bleaching phenotype with considerably reduced amounts of photosynthetic pigments,23 suggesting this sRNA is a key regulator of photosynthetic gene functions. Indeed, when subjected to a recently developed advanced computational target prediction approach,55 genes for photosystem I and the light harvesting apparatus appear among the top-scoring candidates. However, as computational target predictions are known not to be overly accurate,56 these results still need to be experimentally verified.
The sRNA Yfr1 in cyanobacteria
Yfr1 (for cyanobacterial functional RNA 1) is an abundant sRNA that was found initially in three species of marine cyanobacteria on the basis of computational predictions.20 With a length of only 54–57 nt, it is one of the shorter sRNAs. Using RNAmotif to find homologs in additional bacterial families revealed the ubiquitous and exclusive appearance of Yfr1 in all lineages of cyanobacteria except for two Prochlorococcus strains that belong to the low-light adapted clade.57 The presence of an ultra-conserved and likely unpaired undecanucleotide sequence 5′-ACUCCUCACA C-3′ resembling an anti-Shine-Dalgarno motif suggested Yfr1 to function as a trans-acting sRNA. However, another study reported that Yfr1 is constitutively expressed and accumulates up to 18 000 copies per cell in Synechococcus elongatus PCC 6301.58 Both findings, the high copy number as well as the constitutive expression, are rather unusual for a base-pairing sRNA.
Nevertheless, the analysis of a Yfr1-knockout mutant by microarrays revealed upregulation of sbtA, encoding the sodium-dependent bicarbonate transporter SbtA, which therefore appeared as a possible target.58 SbtA plays a crucial role in cyanobacteria in the acquisition of HCO3- that is needed for photosynthetic carbon fixation.59 Membrane proteins also appeared to be likely targets of Yfr1 in Prochlorococcus MED4, when one of the first successful computational searches for the targets of a bacterial sRNA was performed.60 In that case, the IntaRNA algorithm was applied. From the three top-ranking predicted targets, two, PMM1119 and PMM1121, encoding outer membrane proteins (soms or porins), were experimentally verified using a heterologous test system.60 These target mRNAs also belong to the most abundant mRNA species in the cell, which might, in turn, explain why Yfr1 is present at such a high copy number. The fact that Yfr1 is not further regulated appears contradictory to a regulatory function, but possible mechanisms might include the controlled sequestration by an RNA-binding protein or another sRNA, making only a subfraction of the Yfr1 molecules actually available for base-pairing interactions with its targets.
Modulation of cyanobacterial gene expression with antisense RNAs
The asRNA IsrR: The paradigm of an asRNA-mediated threshold-linear control mechanism
Several cis-antisense RNAs have been identified in cyanobacteria, which have an impact on the photosynthetic apparatus. The asRNA IsrR in Synechocystis 6803 controls the expression of the isiA gene, and thereby, of the iron stress-induced protein A (IsiA) as well as the IsiA-chlorophyll complexes in cyanobacterial cells under conditions of iron limitation and redox stress.61 Under iron deplete conditions, the IsiA protein forms an 18-mer antenna ring around the photosystem I trimer, which under continuing stress may later on evolve into an energy-dissipating and protective structure. These modifications include drastic changes in the composition and structural arrangement of the photosynthetic machinery that need to be tightly controlled. One of these control mechanisms utilizes IsrR. Tiling microarray studies have shown that under non-stress conditions IsrR is one of the most strongly accumulating transcripts.22 IsrR represses the accumulation of isiA mRNA under early stress conditions by co-degradation. When the inducing stress continues, the number of isiA mRNA molecules starts to out-compete the number of IsrR molecules. Thus, only a lasting strong stress condition can trigger IsiA protein synthesis. In this context, IsrR provides a unique element of gene expression control that cannot be easily conferred by protein-based regulation. The initial concentration of IsrR molecules is directly proportional to the threshold, above which isiA mRNA accumulation can occur.62 As a consequence, IsrR prevents the premature synthesis of IsiA and the re-organization of the photosynthetic apparatus. While the control of isiA expression by IsrR appears elegant and isiA genes are widely distributed within the cyanobacterial phylum, IsrR has thus far only been found in the two Synechocystis strains 6803 and PCC 6714.
The asRNA As1_flv4 establishes a transient threshold for the expression of its target mRNA during the early phase of the response to low carbon
A mechanism similar to the one used by IsrR may also be used by the Synechocystis 6803 asRNA As1_flv4. This asRNA was identified as one of three asRNAs (and one sRNA) that are associated with the flv4-2 operon in Synechocystis 6803. These transcripts were initially found by tiling microarray analysis, and then confirmed by dRNA-seq.22,27 The Flv2 and Flv4 proteins play a crucial role in photoprotection of photosystem II under low carbon conditions by accepting electrons from photosystem II.63,64 However, despite numerous studies, the understanding of low carbon control of gene expression in cyanobacteria is not complete. Here, the asRNA As1_flv4 was identified as a novel regulatory element. The As1_flv4 transcript accumulates in a reverse manner to the mRNA of its target gene flv4. In contrast to flv4, the As1_flv4 transcript accumulates already under high carbon conditions. After the shift to low carbon, it increases transiently in abundance and then starts to decline.65 The AbrB-like protein Sll0822 was identified as a direct or indirect negative transcriptional regulator for the promoter of as1_flv4, whereas the flv4-2 operon itself is controlled by another transcriptional regulator, NdhR.
Detailed mutational analyses suggest that As1_flv4 functions in preventing the premature expression of the flv4-2 flavoprotein operon upon a shift in inorganic carbon supply.65
PsbA2R and PsbA3R: low abundance asRNAs may have functions, too
These two asRNAs illustrate that among the hundreds or even thousands of low abundance natural antisense transcripts of a bacterial cell, some have functional relevance even when they are short-lived and occur in low abundances. At first, the stoichiometry of PsbA2R and PsbA3R was puzzling, e.g., the steady-state ratio of PsbA2R to its target, the psbA2 mRNA, is only 1:300.66 Another surprise was the observation that PsbA2R has a very short half-life of less than 1 min. Finally, the fact that these asRNAs appear to be co-regulated with their target mRNAs66 was surprising because this is very different from other examples of regulatory asRNAs such as IsrR or As-flv4.
PsbA2R and PsbA3R are associated with the genes psbA2 and psbA3 of Synechocystis 6803. These genes encode the D1 reaction center protein of water-splitting photosystem II and are extremely conserved from cyanobacteria to higher plants (e.g., 84% of the amino acid residues are identical between the Synechocystis 6803 and the Arabidopsis thaliana D1 protein). The highly oxidizing chemistry of photosystem II and the concomitant formation of reactive oxygen species induce lethal damage to the D1 protein. Therefore, this protein needs to be replaced rapidly, particularly under conditions when cells are exposed to excess light. Consequently, the D1 protein shows faster turnover than any other thylakoid protein, and under high-light conditions, it becomes the most highly expressed gene of the cyanobacterial cell. Accordingly, psbA genes possess very strong promoters, exhibit a codon usage poised for efficient translation and exist in many cyanobacteria in the form of a small gene family with up to five members. The major control mechanism of psbA gene expression in cyanobacteria is at the level of transcription.67,68 However, the asRNAs PsbA2R and PsbA3R affect a post-transcriptional control mechanism that involves the RNase E-mediated destabilization of the mRNA by cleavage at the “AU box” and the ribosome-binding site, both located in the 5′ UTR.69 These cis-elements mediate the destabilization of psbA mRNAs under darkness;69,70 however, it was not known why the very same elements would not be targeted by endonucleolytic attack under light. The paper by Sakurai et al.66 showed that PsbA2R and PsbA3R are involved in the protection of psbA 5′UTRs from cleavage by RNase E. The TSSs of these asRNAs are located within the 5′UTR, just upstream of the ribosome binding site but on the complementary strand. Therefore, they cover the “AU box,” effectively protecting it from cleavage by RNase E.66 As the abundance of PsbA2R and PsbA3R is co-regulated with the abundance of their mRNA targets, their concentration increases when cells are shifted to higher light intensities, whereas PsbA2R and PsbA3 rapidly disappear when cells are shifted to darkness. Interestingly, a physiological impact of PsbA2R was noted. When its expression level was reduced, colonies on plates grew slower and the abundance of psbA2 mRNA was clearly decreased, which, based on both in vivo and in vitro experiments, was interpreted to result from a decrease in psbA2 mRNA stability. This decrease led to physiological consequences, such as a lowered amount of the D1 protein and of maximal photosystem II activity.66
These results suggested that PsbA2R and PsbA3R act as positive post-transcriptional regulators of gene expression. The experimental evidence indicated these asRNAs support a high expression of the psbA genes in Synechocystis 6803, which helps to maintain photosynthesis at a high level, especially at high light intensities. Of general interest is the finding that low-abundance and short-lived asRNAs cannot be regarded merely as transcriptional noise but can have important physiological functions, too.
CfrI, an asRNA linking a gene encoding a homing endonuclease with the 3′ end of psbA in a lytic cyanophage
The very idea of asRNA control of gene expression gained support early on from studying the biology of bacteriophages. Bacteriophage lambda expresses the 77-nt OOP asRNA overlapping the 3′ end of the cII-repressor gene. Overexpression of OOP led to the RNase III-dependent codegradation of the cII mRNA,71 setting a paradigm for the likely related mechanisms used by IsrR and As_flv4. It is therefore not surprising to find asRNAs originating from bacteriophages infecting cyanobacteria, the so-called cyanophages. An asRNA at the 3′ end of psbA was discovered in the cyanophage S-PM2, belonging to the myoviridae. This asRNA was called Cyanophage Functional RNA I (CfrI).72 Similar to the observations made for PsbA2R and PsbA3R asRNAs that cover the 5′ ends of chromosomal psbA genes in Synechocystis 6803,66 CfrI is co-expressed with its predicted target gene. However, in contrast to PsbA2R and PsbA3R, CfrI overlaps the 3′ end of psbA and originates from a TSS in the following gene.72 Another unique feature of the S-PM2 psbA gene is the fact that it is interrupted by a group I intron and that the following gene encodes the homing endonuclease for this intron.73 Therefore, CfrI links the gene for the homing endonuclease with psbA, the host gene for the group I intron to which this endonuclease belongs to. Although not addressed experimentally, the authors speculated that CfrI also plays a positive role in protecting the psbA mRNA; more precisely, the 3′ exon might not be associated with ribosomes for some time, due to the slow splicing process of the group I intron.72
RNA metabolism during phage infection
In a microarray study on whole-genome expression of the cyanophage P-SSP7 and its host Prochlorococcus MED4, it was shown that transcript abundance of 41 host genes, including RNase E, increased during lytic infection, while the vast majority of host mRNAs declined in abundance.74 The authors hypothesized that the induction of host RNase E transcript may help the phage by generating a source of nucleotides that are utilized by the phage for its own replication. Those results raised the question of how these host mRNAs are protected from degradation in light of the enhanced transcription of RNase E. Stazic et al. (2011)75 found evidence that extraordinarily long asRNAs of 3.5 and 7 kb protect some of these mRNAs from RNase E degradation by masking single-stranded recognition sites of RNase E, leading to increased stability of the mRNAs. The phage transcriptome itself is invulnerable to RNase E degradation due to the complete coverage of the mRNA pool with long asRNAs (unpublished data). The existence of long asRNAs and their function in stabilization of RNA adds a further facette to the possible functions of asRNAs, which are otherwise often involved in the degradation of mRNAs.
In Figure 3, a summary of cyanobacterial asRNA functions is presented. These asRNAs can act as repressors of gene expression, illustrated by IsrR,61 As1_flv4,65 and the asRNAs of the furA gene encoding the ferric uptake regulator,76-78 or support a high level of gene expression as speculated for phage CfrI72 and shown for PsbA2R and PsbA3R66 and can protect mRNAs in the form of long non-coding asRNAs.75
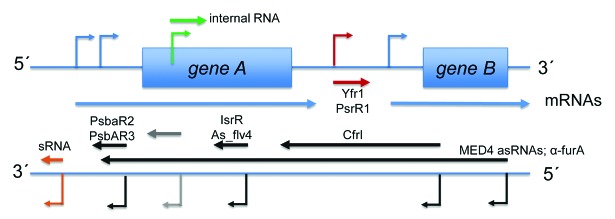
Figure 3. Summary of demonstrated asRNA and sRNA functions in cyanobacteria. TSS driving the transcription of annotated genes (gTSS, blue), putative sRNAs (nTSS, red), and asRNAs (aTSS, black), together with intragenic TSS sequences (iTSS, green) are visualized by the bent arrows. Details of the TSS numbers and classification mapped in dRNA-seq studies in different cyanobacteria are summarized in Table 2. The sRNAs with an at least partially characterized function (symbolized by straight arrows in dark red) are PsrR1 and Yfr1. These are specific trans-acting regulators of gene expression but there are also hundreds of additional sRNAs awaiting more detailed characterization (light red arrows). Relatively short intragenically located asRNAs can act as repressors of gene expression, illustrated by IsrR and As1_flv4 in Synechocystis 6803 (black arrows).61,65 Another type of antisense repressor is represented by α-furA, which in Anabaena 7120 controls the expression of the furA gene. This 2.2 kb-long asRNA originates by read-through from the downstream located gene alr1690.76-78 Longer and shorter asRNAs may also support a high level of gene expression as speculated for the asRNA CfrI of cyanophage S-PM2,72 infecting marine Synechococcus, demonstrated for PsbA2R and PsbA3R in Synechocystis 680366 or, in the form of several kb long asRNAs in Prochlorococcus MED4, may even protect mRNAs from the RNase E activity enhanced upon infection by the cyanophage P-SSP7.75 Last but not least, there are myriads of intragenic transcripts (green) and asRNAs (gray) whose functions are still entirely unknown.
The Role of the RNA Chaperone Hfq in Photosynthetic Bacteria
Hfq in R. sphaeroides
In many bacteria, the RNA chaperone Hfq is crucial for sRNA functions and strongly binds RNA via several distinct binding surfaces.79,80 The plethora of Hfq-bound transcripts is revealed in co-immunoprecipitation (coIP) experiments. For example, Hfq coIP combined with RNA-seq uncovered Hfq-bound transcripts on a global scale and identified several new sRNAs in Salmonella.81 Using a similar strategy, the R. sphaeroides Hfq homolog was C-terminally fused to a 3xFLAG-tag and applied to coIP after 1O2 stress experiments. Subsequent RNA-seq identified Hfq-bound transcripts in vivo.82 In total, 19 sRNAs and six asRNAs were enriched in the Hfq coIP experiment when compared with a non-specific control library. Hfq binding to nine sRNAs was validated by northern blot analysis, among them the well-known sRNA candidates RSs0680a, RSs0682, RSs1543, and RSs2461 (Fig. 1A). In the case of RSs0682, the full-length sRNA was enriched, while for RSs2461, a 75 nt processing fragment strongly accumulated in the coIP library (Fig. 1B). Because Hfq supports sRNA functions, the processed fragment of RSs2461 might represent the functional unit, which binds to mRNA targets with the aid of Hfq. Regulation of multiple targets is regularly observed for many trans-acting sRNAs, and Hfq is thought to facilitate the sRNA–mRNA interactions that rely on imperfect base-pairing. Therefore, it is not surprising to find sRNAs to be predominant among the Hfq-bound transcripts (64% of all transcripts in the R. sphaeroides Hfq coIP).82 In contrast, the enrichment of R. sphaeroides asRNAs during Hfq coIP seems enigmatic because cis-antisense transcripts have perfect complementarity to their target mRNAs and are therefore not expected to rely on Hfq for base pairing. However, in a SELEX approach to identify Hfq-binding RNAs in E. coli, many sequences antisense to translation start sites or intervening sequences between ORFs in operons were demonstrated to be Hfq-bound as well.83 There are several possible explanations for these findings. First, Hfq might be essential for the interaction between the asRNA–mRNA complex due to multifaceted secondary structures in one or both RNAs that have to be solved. Furthermore, it is a feasible scenario that asRNAs, when transcribed in excess of the mRNA, might bind to targets in trans and rely on Hfq for these secondary interactions. Finally, asRNAs may just randomly bind to Hfq when their concentration is increased within the cell, an idea which is in line with the “active cycling” model and a general high affinity of Hfq for any RNA.80,84 Whether binding of asRNAs to Hfq has a functional meaning or is just an unavoidable side-effect will be an exciting topic for future studies.
A R. sphaeroides strain lacking Hfq had a lighter color than the isogenic wild-type due to reduced amounts of photosynthetic complexes. This could be attributed to reduced half-lives of mRNAs encoding pigment binding proteins, implicating a stabilizing effect of Hfq on those mRNAs.82 As described above, the sRNA PcrZ has an important function in the regulation of photosynthesis genes, but PcrZ was not found to be associated with Hfq. Thus, the mechanisms behind the action of Hfq on photosynthetic complex formation include the direct effect on photosynthesis mRNAs and possibly other interactions, which need further investigation. In addition to a reduced pigmentation, inhibition assays revealed that the Hfq mutant has a higher sensitivity to 1O2, which could be correlated with reduced RpoE activity and a disordered induction of RpoHII-dependent genes.82 Here, a complex regulatory defect is obvious and might include dysfunction of the singlet oxygen inducible and Hfq-bound sRNAs as well as other Hfq-dependent factors.
Hfq in Synechocystis 6803
Most cyanobacteria harbor an hfq gene whose sequence is very distinct from those of other bacteria. However, crystal structures of two cyanobacterial Hfq proteins revealed that the structure and the organization of the homohexamer are well conserved in comparison to other bacteria.85 The two faces for the potential interaction with nucleic acids are defined as proximal and distal sides. Amino acid residues important for RNA binding on both faces are not conserved among cyanobacterial Hfq proteins. Moreover, the Synechocystis hfq is not able to complement an E. coli hfq mutant strain.85 So far, there is no evidence for the specific binding of cyanobacterial sRNAs to the Hfq protein. Compared with the wide-ranging effects of inactivation of hfq in other bacteria, mutational analysis revealed rather specific functions of Hfq in cyanobacteria. Inactivation of hfq in the filamentous cyanobacterium Anabaena 7120 led to a lower accumulation of transcripts encoding gene products of the nitrate assimilation pathway. In contrast to the wild-type, hfq mutant cells differentiated into heterocysts even in the presence of nitrate.86 Synechocystis 6803 mutants lacking the hfq gene lost their type IV pili. The wild-type strain is able to move over surfaces by a process called twitching motility using type IV pili. Accordingly, the hfq mutant showed no movement toward a light source.87 As type IV pili are also important for the uptake of DNA, hfq mutants lost their natural competence. Microarray analyses revealed several mRNAs that were downregulated in the Synechocystis hfq mutant strain. Many of them are under the control of a cAMP receptor protein (SyCRP1) that is involved in motility.88 Although some Hfq functions in cyanobacteria have been discovered, the mechanistic aspects remain unclear. The lack of conservation on the sequence level, an unfavorable surface charge and the low affinity for RNA binding85 make the known RNA-binding pockets of cyanobacterial Hfq proteins an unlikely candidate for RNA-binding. Considering the divergent properties of cyanobacterial Hfq homologs compared with other bacteria, we speculate that this special subfamily of Sm proteins controls gene expression by an as of yet unknown mechanism. Studies on the identification of protein partners of Hfq in E. coli revealed physical associations of the RNA chaperone with several proteins and protein complexes that modify RNA. RNase E is one of the most prominent partners of Hfq in E. coli.89 Hfq was also found in a complex with polynucleotide phosphorylase and poly(A) polymerase, thereby regulating polyadenylation-dependent RNA decay.90 Furthermore, a large-scale network analysis predicted that Hfq might interact with 34 proteins and subunits of multiprotein complexes, including ribosomal proteins, RNA polymerase, and proteins such as DnaK and DnaJ.91 Protein-protein interaction analyses are required to identify potential binding partners of cyanobacterial Hfq proteins. Thus, the function of Hfq in cyanobacteria might not be based on direct RNA interaction, but rather on binding of protein partners with a function in RNA metabolism or regulation.
Hfq in Prochlorococcus
The preceding sections suggested different roles and possibly different mechanisms of Hfq in R. sphaeroides compared with Synechocystis 6803. It is interesting to note that some cyanobacteria do not harbor Hfq at all. Hfq genes appear to have been lost during evolution of the Prochlorococcus genus and the genome streamlining process associated with it. Strains with relatively larger genome sizes (Prochlorococcus MIT9303 and 9313) possess an hfq gene, located within the dapF-leuS IGR.20 In contrast, all other Prochlorococcus isolates, including the MED4 strain, are devoid of a homolog. Nevertheless, no obvious differences were observed in the composition of the non-coding transcriptome from a strain bearing an hfq gene (MIT 9313) and a strain lacking it (MED4). This observation is in line with the lack of any evidence that cyanobacterial Hfq is an RNA binding protein at all.
Conclusions
Photosynthetic bacteria inhabit a myriad of environments and constitute some of the most ecophysiologically diverse phyla found in nature. Hence, their ability for adaptation suggests the existence of sophisticated regulatory mechanisms, implying that various types of regulatory RNAs might be integral elements within their signal processing machinery. Indeed, as we have summarized in this review, there are hundreds to thousands of non-coding RNAs that were identified in photosynthetic bacteria based on different and complementary technical approaches.
In view of these numbers, the most important tasks in the near future appear to be to filter out the most relevant regulatory RNAs and to identify their exact molecular functions and functional principles. Focused and efficient screening methods are a top priority in this endeavor, either by focusing on a particular physiological process such as the response to redox stress,40 or by using new technological advances such as the recently developed CopraRNA algorithm.55 From a more ecophysiological perspective, it will be interesting to see how the results from studies on regulatory and modulatory functional RNAs in model laboratory organisms compare with findings in microbial populations in the environment. Unique microbial sRNAs have been identified through metatranscriptomic analyses,92 and such approaches still bear substantial potential. Another aspect is the evolution of eukaryotic phototrophs. There is increasing evidence that potentially regulatory RNAs are expressed not only in the nucleus of plants but also in chloroplasts.93-95 It will be important to discover to what extent RNA-based regulatory mechanisms evolved from the prokaryotic ancestor of chloroplasts or are involved in the genetic interactions between nucleus and chloroplast, which was suggested more than 30 y ago96 and is still an enigmatic possibility.
Further developments will take place in two fields tightly linked to regulatory RNA. These are the characterization of protein factors modulating the activity of sRNAs and asRNAs, and the identification of genes that are modulated by these non-coding RNAs. Work has begun to characterize interactions with Hfq, RNase E, and RNase III, but there are many more relevant protein factors also in photosynthetic bacteria. Last but not least, there are a small but growing number of regulatory RNAs in bacteria that derive from coding transcripts, and of regulatory RNAs that are dual-function RNAs and also encode peptides or short proteins. The characterization of these hybrid or chimeric RNA molecules will constitute another exciting field of research.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft Focus program “Sensory and regulatory RNAs in Prokaryotes” SPP1258, grants HE 2544/4-1 and 4-2, KL563/23-1, WI 2014/3-1 and 3-2, STE 1119/2-1 and 2-2, as well as grant KL563/20-2.
Glossary
Abbreviations:
- asRNA
cis-acting antisense RNA
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- dRNA-seq
differential RNA-seq
- DUF
domain of unknown function
- IGR
intergenic region
- sRNA
small noncoding RNA
- TSS
transcriptional start site (aTSS, of asRNAs, nTSS, of non-coding sRNAs)
References
- 1.Imhoff JF. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blankenship RE, Madigan MT, Bauer CE, eds. Anoxygenic photosynthetic bacteria Dordrecht, The Netherlands: Kluwer Academic Publishers, 1995:1-15. [Google Scholar]
- 2.Zeilstra-Ryalls J, Gomelsky M, Eraso JM, Yeliseev A, O’Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–9. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flores E, Herrero A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat Rev Microbiol. 2010;8:39–50. doi: 10.1038/nrmicro2242. [DOI] [PubMed] [Google Scholar]
- 4.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2:a000315. doi: 10.1101/cshperspect.a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 6.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–27. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman ML, Chisholm SW. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15:398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–7. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 9.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–7. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 10.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaeser J, Nuss AM, Berghoff BA, Klug G. Singlet oxygen stress in microorganisms. Adv Microb Physiol. 2011;58:141–73. doi: 10.1016/B978-0-12-381043-4.00004-0. [DOI] [PubMed] [Google Scholar]
- 12.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–54. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossman AR, Schaefer MR, Chiang GG, Collier JL. The phycobilisome, a light-harvesting complex responsive to environmental conditions. Microbiol Rev. 1993;57:725–49. doi: 10.1128/mr.57.3.725-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi JS, Chung YH, Moon YJ, Kim C, Watanabe M, Song PS, Joe CO, Bogorad L, Park YM. Photomovement of the gliding cyanobacterium Synechocystis sp. PCC 6803. Photochem Photobiol. 1999;70:95–102. doi: 10.1111/j.1751-1097.1999.tb01954.x. [DOI] [PubMed] [Google Scholar]
- 15.Nultsch W, Schuchart H, Höhl M. Investigations on the phototactic orientation of Anabaena variabilis. Arch Microbiol. 1979;122:85–91. doi: 10.1007/BF00408050. [DOI] [Google Scholar]
- 16.Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 1993;90:5672–6. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gierga G, Voss B, Hess WR. The Yfr2 ncRNA family, a group of abundant RNA molecules widely conserved in cyanobacteria. RNA Biol. 2009;6:222–7. doi: 10.4161/rna.6.3.8921. [DOI] [PubMed] [Google Scholar]
- 18.Voss B, Georg J, Schön V, Ude S, Hess WR. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics. 2009;10:123. doi: 10.1186/1471-2164-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierga G, Voss B, Hess WR. Non-coding RNAs in marine Synechococcus and their regulation under environmentally relevant stress conditions. ISME J. 2012;6:1544–57. doi: 10.1038/ismej.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berghoff BA, Konzer A, Mank NN, Looso M, Rische T, Förstner KU, Krüger M, Klug G. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLoS Genet. 2013;9:e1003576. doi: 10.1371/journal.pgen.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A. 2011;108:2124–9. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol. 2009;74:1497–512. doi: 10.1111/j.1365-2958.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 25.Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss B, Bolhuis H, Fewer DP, Kopf M, Möke F, Haas F, El-Shehawy R, Hayes P, Bergman B, Sivonen K, et al. Insights into the physiology and ecology of the brackish-water-adapted Cyanobacterium Nodularia spumigena CCY9414 based on a genome-transcriptome analysis. PLoS One. 2013;8:e60224. doi: 10.1371/journal.pone.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A. 2011;108:20130–5. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L. Small non-coding RNAs in Caulobacter crescentus. Mol Microbiol. 2008;68:600–14. doi: 10.1111/j.1365-2958.2008.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vercruysse M, Fauvart M, Cloots L, Engelen K, Thijs IM, Marchal K, Michiels J. Genome-wide detection of predicted non-coding RNAs in Rhizobium etli expressed during free-living and host-associated growth using a high-resolution tiling array. BMC Genomics. 2010;11:53. doi: 10.1186/1471-2164-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 31.Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci U S A. 2009;106:21878–82. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–36. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–51. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–77. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albrecht M, Sharma CM, Dittrich MT, Müller T, Reinhardt R, Vogel J, Rudel T. The transcriptional landscape of Chlamydia pneumoniae. Genome Biol. 2011;12:R98. doi: 10.1186/gb-2011-12-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidtke C, Findeiss S, Sharma CM, Kuhfuss J, Hoffmann S, Vogel J, Stadler PF, Bonas U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020–31. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilms I, Overlöper A, Nowrousian M, Sharma CM, Narberhaus F. Deep sequencing uncovers numerous small RNAs on all four replicons of the plant pathogen Agrobacterium tumefaciens. RNA Biol. 2012;9:446–57. doi: 10.4161/rna.17212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madhugiri R, Pessi G, Voss B, Hahn J, Sharma CM, Reinhardt R, Vogel J, Hess WR, Fischer HM, Evguenieva-Hackenberg E. Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol. 2012;9:47–58. doi: 10.4161/rna.9.1.18008. [DOI] [PubMed] [Google Scholar]
- 39.Nuss AM, Glaeser J, Berghoff BA, Klug G. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J Bacteriol. 2010;192:2613–23. doi: 10.1128/JB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci U S A. 2012;109:16306–11. doi: 10.1073/pnas.1207067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholz I, Lange SJ, Hein S, Hess WR, Backofen R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving three Cas6 and a Cmr2 protein. PloS One. 2013;8 doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muro-Pastor AM, Hess WR. Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol. 2012;20:548–57. doi: 10.1016/j.tim.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Ionescu D, Voss B, Oren A, Hess WR, Muro-Pastor AM. Heterocyst-specific transcription of NsiR1, a non-coding RNA encoded in a tandem array of direct repeats in cyanobacteria. J Mol Biol. 2010;398:177–88. doi: 10.1016/j.jmb.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Wong FC, Meeks JC. The hetF gene product is essential to heterocyst differentiation and affects HetR function in the cyanobacterium Nostoc punctiforme. J Bacteriol. 2001;183:2654–61. doi: 10.1128/JB.183.8.2654-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–93. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 47.Eraso JM, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eraso JM, Kaplan S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1996;178:7037–46. doi: 10.1128/jb.178.24.7037-7046.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mank NN, Berghoff BA, Klug G. A mixed incoherent feed-forward loop contributes to the regulation of bacterial photosynthesis genes. RNA Biol. 2013;10:347–52. doi: 10.4161/rna.23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuss AM, Glaeser J, Klug G. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J Bacteriol. 2009;191:220–30. doi: 10.1128/JB.00925-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattison DI, Rahmanto AS, Davies MJ. Photo-oxidation of proteins. Photochem Photobiol Sci. 2012;11:38–53. doi: 10.1039/c1pp05164d. [DOI] [PubMed] [Google Scholar]
- 52.Glaeser J, Zobawa M, Lottspeich F, Klug G. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J Proteome Res. 2007;6:2460–71. doi: 10.1021/pr060624p. [DOI] [PubMed] [Google Scholar]
- 53.Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A. 2005;102:6502–7. doi: 10.1073/pnas.0502225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peuser V, Remes B, Klug G. Role of the Irr protein in the regulation of iron metabolism in Rhodobacter sphaeroides. PLoS One. 2012;7:e42231. doi: 10.1371/journal.pone.0042231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright PR, Richter AS, Papenfort K, Mann M, Vogel J, Hess WR, Backofen R, Georg J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc Natl Acad Sci U S A. 2013;110:E3487–96. doi: 10.1073/pnas.1303248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Backofen R, Hess WR. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 2010;7:33–42. doi: 10.4161/rna.7.1.10655. [DOI] [PubMed] [Google Scholar]
- 57.Voss B, Gierga G, Axmann IM, Hess WR. A motif-based search in bacterial genomes identifies the ortholog of the small RNA Yfr1 in all lineages of cyanobacteria. BMC Genomics. 2007;8:375. doi: 10.1186/1471-2164-8-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura T, Naito K, Yokota N, Sugita C, Sugita M. A cyanobacterial non-coding RNA, Yfr1, is required for growth under multiple stress conditions. Plant Cell Physiol. 2007;48:1309–18. doi: 10.1093/pcp/pcm098. [DOI] [PubMed] [Google Scholar]
- 59.Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria: function and phylogenetic analysis. J Biol Chem. 2002;277:18658–64. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 60.Richter AS, Schleberger C, Backofen R, Steglich C. Seed-based INTARNA prediction combined with GFP-reporter system identifies mRNA targets of the small RNA Yfr1. Bioinformatics. 2010;26:1–5. doi: 10.1093/bioinformatics/btp609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci U S A. 2006;103:7054–8. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legewie S, Dienst D, Wilde A, Herzel H, Axmann IM. Small RNAs establish delays and temporal thresholds in gene expression. Biophys J. 2008;95:3232–8. doi: 10.1529/biophysj.108.133819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang P, Eisenhut M, Brandt AM, Carmel D, Silén HM, Vass I, Allahverdiyeva Y, Salminen TA, Aro EM. Operon flv4-flv2 provides cyanobacterial photosystem II with flexibility of electron transfer. Plant Cell. 2012;24:1952–71. doi: 10.1105/tpc.111.094417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM. Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS One. 2009;4:e5331. doi: 10.1371/journal.pone.0005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eisenhut M, Georg J, Klähn S, Sakurai I, Mustila H, Zhang P, Hess WR, Aro EM. The antisense RNA As1_flv4 in the Cyanobacterium Synechocystis sp. PCC 6803 prevents premature expression of the flv4-2 operon upon shift in inorganic carbon supply. J Biol Chem. 2012;287:33153–62. doi: 10.1074/jbc.M112.391755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sakurai I, Stazic D, Eisenhut M, Vuorio E, Steglich C, Hess WR, Aro EM. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012;160:1000–10. doi: 10.1104/pp.112.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Golden SS, Brusslan J, Haselkorn R. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 1986;5:2789–98. doi: 10.1002/j.1460-2075.1986.tb04569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulo P, Sakurai I, Aro EM. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta. 2012;1817:247–57. doi: 10.1016/j.bbabio.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 69.Horie Y, Ito Y, Ono M, Moriwaki N, Kato H, Hamakubo Y, Amano T, Wachi M, Shirai M, Asayama M. Dark-induced mRNA instability involves RNase E/G-type endoribonuclease cleavage at the AU-box and SD sequences in cyanobacteria. Mol Genet Genomics. 2007;278:331–46. doi: 10.1007/s00438-007-0254-9. [DOI] [PubMed] [Google Scholar]
- 70.Agrawal GK, Kato H, Asayama M, Shirai M. An AU-box motif upstream of the SD sequence of light-dependent psbA transcripts confers mRNA instability in darkness in cyanobacteria. Nucleic Acids Res. 2001;29:1835–43. doi: 10.1093/nar/29.9.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krinke L, Wulff DL. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987;1:1005–13. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- 72.Millard AD, Gierga G, Clokie MR, Evans DJ, Hess WR, Scanlan DJ. An antisense RNA in a lytic cyanophage links psbA to a gene encoding a homing endonuclease. ISME J. 2010;4:1121–35. doi: 10.1038/ismej.2010.43. [DOI] [PubMed] [Google Scholar]
- 73.Zeng Q, Bonocora RP, Shub DA. A free-standing homing endonuclease targets an intron insertion site in the psbA gene of cyanophages. Curr Biol. 2009;19:218–22. doi: 10.1016/j.cub.2008.11.069. [DOI] [PubMed] [Google Scholar]
- 74.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, Kettler G, Sullivan MB, Steen R, Hess WR, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. 2007;449:83–6. doi: 10.1038/nature06130. [DOI] [PubMed] [Google Scholar]
- 75.Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–9. doi: 10.1093/nar/gkr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hernández JA, Alonso I, Pellicer S, Luisa Peleato M, Cases R, Strasser RJ, Barja F, Fillat MF. Mutants of Anabaena sp. PCC 7120 lacking alr1690 and alpha-furA antisense RNA show a pleiotropic phenotype and altered photosynthetic machinery. J Plant Physiol. 2010;167:430–7. doi: 10.1016/j.jplph.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 77.Hernández JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. J Mol Biol. 2006;355:325–34. doi: 10.1016/j.jmb.2005.10.079. [DOI] [PubMed] [Google Scholar]
- 78.Martin-Luna B, Sevilla E, Gonzalez A, Bes MT, Fillat MF, Peleato ML. Expression of fur and its antisense α-fur from Microcystis aeruginosa PCC7806 as response to light and oxidative stress. J Plant Physiol. 2011;168:2244–50. doi: 10.1016/j.jplph.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 79.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner EG. Cycling of RNAs on Hfq. RNA Biol. 2013;10:619–26. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol. 2011;80:1479–95. doi: 10.1111/j.1365-2958.2011.07658.x. [DOI] [PubMed] [Google Scholar]
- 83.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–6. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bøggild A, Overgaard M, Valentin-Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA-binding properties. FEBS J. 2009;276:3904–15. doi: 10.1111/j.1742-4658.2009.07104.x. [DOI] [PubMed] [Google Scholar]
- 86.Puerta-Fernández E, Vioque A. Hfq is required for optimal nitrate assimilation in the Cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2011;193:3546–55. doi: 10.1128/JB.00254-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dienst D, Dühring U, Mollenkopf H, Vogel J, Golecki J, Hess WR, Wilde A. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 2008;154:3134–43. doi: 10.1099/mic.0.2008/020222-0. [DOI] [PubMed] [Google Scholar]
- 88.Yoshimura H, Yanagisawa S, Kanehisa M, Ohmori M. Screening for the target gene of cyanobacterial cAMP receptor protein SYCRP1. Mol Microbiol. 2002;43:843–53. doi: 10.1046/j.1365-2958.2002.02790.x. [DOI] [PubMed] [Google Scholar]
- 89.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–86. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–20. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 91.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–7. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 92.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature. 2009;459:266–9. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 93.Hotto AM, Germain A, Stern DB. Plastid non-coding RNAs: emerging candidates for gene regulation. Trends Plant Sci. 2012;17:737–44. doi: 10.1016/j.tplants.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 94.Hotto AM, Schmitz RJ, Fei Z, Ecker JR, Stern DB. Unexpected Diversity of Chloroplast Noncoding RNAs as Revealed by Deep Sequencing of the Arabidopsis Transcriptome. G3 (Bethesda) 2011;1:559–70. doi: 10.1534/g3.111.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24:123–36. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bradbeer JW, Atkinson YE, Börner T, Hagemann R. Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesised RNA. Nature. 1979;279:816–7. doi: 10.1038/279816a0. [DOI] [Google Scholar]