Abstract
Streptomycetes are Gram-positive, GC-rich, soil dwelling bacteria, occurring ubiquitary throughout nature. They undergo extensive morphological changes from spores to filamentous mycelia and produce a plethora of secondary metabolites. Owing to their complex life cycle, streptomycetes require efficient regulatory machinery for the control of gene expression. Therefore, they possess a large diversity of regulators. Within this review we summarize the current knowledge about the importance of small non-coding RNA for the control of gene expression in these organisms.
Keywords: Streptomyces, S. coelicolor, small non-coding RNA, sRNA, antisense RNA, asRNA
Streptomycetes grow in a complex life cycle undergoing extensive morphological changes from spores to filamentous mycelia. Their ability to degrade chitin and cellulose makes them environmentally important in their habitat.1,2 Furthermore, streptomycetes produce a variety of secondary metabolites. Not only two-thirds of all known antibiotics are produced by these bacteria, but also immunosuppressants, antivirals, and herbicides, making them highly relevant for pharmaceutical and industrial purposes.1,3,4
The life cycle of streptomycetes is a complex succession of morphological changes in response to altered environmental conditions (Fig. 1). In general, bacteria have to adapt constantly to their environment. Still, changes in gene expression and morphology as grave as shown by streptomycetes are rarely observed among other microorganisms. Their life cycle starts with the germination of a monogenomic spore that prolongs, mainly by growing from the tip, into filamentous tubes.5 Those grow into the substrate and form a highly branched mycelium, the so called vegetative or primary mycelium. When nutrition becomes scarce in the vegetative colonies, part of the mycelium undergoes programmed cell death to serve as substrate for aerial hyphae growing out of the soil.6 Each hyphal filament of this aerial or secondary mycelium contains several copies of the chromosome. Eventually, the single-layered septa in the filaments from vegetative growths are replaced by thick, pigmented, double-layered septa forming monogenomic spores. The single chromosome is well protected by the outlasting mantle and the spores can be reactivated even after decades.7-10
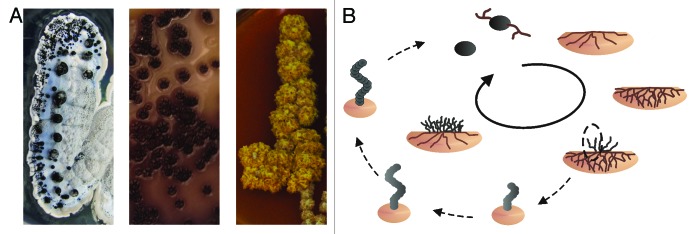
Figure 1. Complex life cycle of streptomycetes. (A) Morphological diversity of different Streptomyces strains. (B) The life cycle of S. coelicolor. Monogenomic spores of S. coelicolor germinate and grow into the soil. When nutrition becomes limited the vegetative mycelia digest themselves to serve as substrate for aerial mycelia growing out of the soil. Those aerial hyphae differentiate into spores again through insertion of septa.
Streptomyces coelicolor, the Model Organism for Gram-Positive, GC-Rich Bacteria
S. coelicolor is considered the model organism for streptomycetes. It has a linear chromosome of nearly 8.7 Mb with almost 8000 annotated open reading frames (ORFs). The linear chromosome is kept in a circularized manner by proteins on the free 5′ends. Replication takes place bidirectionally from a central origin of replication. The distribution of genes throughout the chromosome shows a distinction between the core region and the distal arms located on the left and right. The core region contains most housekeeping genes, such as those for DNA replication, cell division, and translation, whereas the arm regions mostly code for less essential functions as secondary metabolites or exoenzymes.1 Due to their complex life cycle, streptomycetes are in need of efficient regulatory machinery. Therefore, it is not surprising that S. coelicolor possesses a large diversity of such regulators. More than 12% of their proteins are predicted to have a regulatory function and hundreds of transcription factors are described.11 In addition to that, nearly 70 different sigma factors for the direct influence of the promotor activity are known or predicted. Compared with only seven sigma factors in E. coli, the amount of regulators mirrors not only the need for a strong regulation in organisms with a larger genome, but also their ability to adapt to constantly changing environmental conditions.12
Small Non-Coding RNAs—Novel Regulators in Bacteria
Within the last years it has become obvious that small non-coding RNAs (sRNAs) represent a further level of regulation in bacteria. These usually untranslated sRNAs are characterized by their small size of ~50–300 nucleotides (nt) and commonly are encoded in the intergenic regions (IGR) of the genome. Generally, they are transcribed from their own promoters, which often are induced by specific stress conditions or environmental and morphological changes. This leads to a very efficient and defined regulation of sRNA expression.13 Besides some examples of sRNAs binding to and thereby modifying the function of proteins, the majority of these regulators directly target mRNAs. They do so by imperfect basepairing within or close to the translation initiation region, thus influencing initial steps of translation. The imperfect basepairing, in Gram-negative bacteria often mediated by the chaperone Hfq, allows them to recognize and regulate the expression of multiple target mRNAs. Those targets rarely are encoded in close proximity to the sRNA gene itself, but rather are dispersed all over the genome. Thereby, the functions of sRNAs are tremendously versatile. In contrast to other RNA transcripts in prokaryotes, termination of sRNA transcription mainly occurs Rho-factor independent. Instead, intrinsic stem-loops serve as terminator structures.14,15 sRNAs not only form their own terminator structures, but also fold themselves into highly structured conformations, which are crucial for their function.
It is still unclear if sRNAs are—in evolutionary terms—quite old or rather recent inventions. When comparing the conservation however, known protein binding RNAs like 6S or tmRNA are generally more widely conserved than the ones targeting mRNAs. Often the genetic neighborhood helps to overcome the lack of sequence conservation in an sRNA to identify homologs in other species by pointing the researcher to the right region of the genome. The sRNA scr5239 of S. coelicolor, for example, has—up to now—always been found encoded up- or downstream of the same histidine kinase. A functional connection between the sRNA and this kinase remains to be elucidated. A comprehensive review of sRNA function, evolution, and conservation has been presented recently by Susan Gottesmann.16
Up to now, the majority of knowledge about sRNAs was gathered from Gram-negative bacteria such as E. coli and Salmonella. Little is still known about their abundance and function in Gram-positive bacteria. Due to their size, their interesting morphological development, and the ability to produce a multitude of secondary metabolites, we assumed that sRNAs are important regulators in streptomycetes, too. Therefore, we performed a bioinformatic analysis and a deep sequencing approach of the primary transcriptome of S. coelicolor M145. Within this review we will summarize the current knowledge of sRNAs in streptomycetes obtained by our lab and others, what is known about their occurrence and function, and what is similar and different to other bacteria.
Approaches to Identify sRNAs in Streptomycetes
The first attempt to identify sRNAs in streptomycetes was done by Vohradský and co-workers.17 They performed a BLAST search of conserved intergenic regions between S. coelicolor and S. avermitilis. In the next step, the co-localization of the identified conserved sequences with Rho-independent terminators was analyzed. The resulting 32 IGR were proposed to contain sRNAs. Microarray analysis revealed the expression of 20 sRNAs within these predicted 32. Expression of nine of them was further validated by RT-PCR. In addition, the grade of conservation of the secondary structure was also taken in consideration, which was generally high among the predicted sRNAs. The authors also performed a search for functional homologs of already known sRNAs of other species, like spot42, micF, or 6S RNA. Interestingly, only one sRNA, with similarities to the 6S RNA of E. coli, was identified.17
A further, bioinformatic approach came from the Elliot lab.18 Notably, they concentrated their search on the IGR of the core regions of the chromosome. The combination of conserved sequences found by BLAST search in different Streptomyces species combined with the results of sRNA Finder, a program that considers secondary structural conservation, resulted in 114 IGR with potential sRNA transcripts. Twenty of those sequences were chosen for further analysis. Expression of six sRNAs could then be validated by northern blot.
Further experiments were performed in several bld and whi mutant strains, which are impaired in development of aerial hyphae or spore formation, respectively. sRNA expression was analyzed under several conditions in these strains. For at least one sRNA it was shown that it might be dependent on a certain sigma factor. Interestingly, only two of the sRNAs identified in this study had been found by the Vohradský group.18
In 2011, we performed a first deep sequencing approach. RNA of S. coelicolor cultures harvested at the end of the exponential phase (where streptomycetes start to switch on their secondary metabolism) was sequenced using the 454 technology, resulting in about 40 000 reads. The majority of the reads corresponded to known open reading frames (ORFs), rRNAs, and tRNAs. Besides these, sequencing revealed over one thousand transcripts located in IGR, most of them located in the core region of the chromosome. For further analysis, stringent filtering criteria were applied. Those required minimum length of 80 nt, no overlap with other transcripts, and a minimum distance of 60 nt to known ORFs, respectively. This left 24 putative sRNAs, which were tested for expression in northern blot analysis. For these, RNA prepared from three different time points during growth was used. That way 11 sRNAs were validated, nine of them—all encoded in the genomic core—showed a strong growth phase dependency.
Examination of the degree of conservation of these sRNAs among all available microbial genomes showed that all 11 sRNAs could only be found in Streptomyces species. The grade of conservation inside of the genus of Streptomyces is highly variable, ranging from abundance in all available streptomycete genomes analyzed to being present in S. coelicolor only. It has to be mentioned though, that the high GC-content of streptomycetes might inhibit sequence-based search tools such as BLAST from finding homologs in lower GC species. The comparison of our work to previous sRNA predictions could solely yield an overlap of one sRNA: scr5676, which has not been further characterized, yet.19
Another streptomycete species, S. griseus, was analyzed by the Horinouchi group in 2009.20 They used a bioinformatic analysis in combination with experimental validation via northern blot and RT-PCR. Originally, they identified 321 transcripts in IGR by looking for sequence homologies between S. griseus, S. coelicolor,and S. avermitilis. They further reduced this number by excluding known RNA sequences like those for rRNAs, tRNAs, and established ribowitches. Furthermore, they required a minimum length of 80 nt and a minimum distance of 10 nt of a conserved sequence to the next ORF. In the end, 12 of 54 predicted sRNAs were validated by northern blot analysis and RT-PCR experiments. The identified sRNAs showed a growth phase-dependent expression. Similar to Swiercz et al.,18 they also detected differences in sRNA expression in strains with developmental defects. However, no distinct function could be identified. Interestingly, only two sRNAs were found in this study that had been identified by the Vohradský group,17 and only one sRNA from the work of Swiercz et al.18 Taken into account that different approaches and parameters were used in all studies, such a small overlap is not too big a surprise.20
A recent RNA sequencing study of the Elliot lab compared the transcriptome of S. coelicolor, S. avermitilis,and S. venezulae.21 The bacteria were grown under similar conditions and the RNA was sequenced at different developmental steps. This time the authors did not only screen for sRNAs but also for non-coding antisense transcripts (asRNAs).
asRNAs are encoded on the complementary strand of their target genes, which guarantees perfect basepairing to its mRNA. As a result, they usually have only one target each. Fifty-nine new asRNAs were detected in S. avermitilis, 79 in S. venezuelae, and 99 in S. coelicolor. Most of these new asRNAs were found to be species specific, only 11 of them were conserved in other Streptomyces species.
Using an adjusted set of parameters that had been successfully employed in earlier studies, Moody et al.21 identified 90 sRNAs in S. coelicolor, with 71 of them being novel. The number of newly found sRNAs in S. avermitilis (199) and S. venezuelae (176) were distinctly higher than in S. coelicolor. This came with little surprise, as this was the first screen for non-coding RNAs in these two species. Conservation of the 90 newly identified transcripts in the three used species corresponds to the fact of sRNAs generally showing a higher degree of conservation than asRNAs. In all three Streptomyces strains, the expression of conserved sRNAs showed a strong dependency on the growth phase. A similar pattern was observed in non-conserved sRNAs within the respective species, though. This is little surprising, as a life cycle as complex as the one being used by streptomycetes offers a wide field for regulation. Therefore, the 105 sRNAs discovered in S. coelicolor today may likely be only the tip of the iceberg. Swiercz et al.18 already showed growth phase dependency for sRNA transcripts, most often in connection to the offered nutrition source, which we could also observe for many transcripts in own experiments (unpublished data).
In a recent publication, they tried to unravel the biological function of the transcript scr4677, which was identified in their transcriptomics approach.11 The work led to insights into the general behavior of this small RNA, such as differential expression in different media; however, it did not fully reveal a clear target or function for scr4677. The proposed potential of the sRNA to regulate the neighboring operon SCO4676–SCO4677 has to be further examined. The sRNA also shows alternate processing under different nutrition conditions. Interestingly, one of the sRNAs under survey in our lab, scr4632, also shows a similar pattern of potentially processed RNA transcripts (unpublished data). In both cases, an involvement of procession regulation has not been entirely understood at present. To what extend this is important for sRNA mediated regulation in streptomycetes has to be determined, still.
Nomenclature of Small Non-Coding RNAs in Streptomycetes
A nomenclature for sRNAs has been introduced by the Elliot lab.18 scrXXXX stands for Streptomyces coelicolor small RNA, with XXXX as the SCO number of the gene downstream of the sRNA. As an example, scr5239 is a small RNA in S. coelicolor located upstream of the gene SCO5239. Consequently, sRNAs from S. griseus are named sgrXXXX, those from S. venezuelae svrXXXX, and of S. avermitilis sarXXXX, respectively. Up to now, the nomenclature of antisense RNA is not uniform yet with cncXXXX used by the Takano lab and asRNA by the Elliot lab.
What’s Same, What’s Different—sRNA Characteristics of Streptomyces
The sRNAs discovered in streptomycetes so far all are exclusively found in the genus of Streptomyces. Even conservation to closely related actinobacteria has not been determined. This may be due to the high GC-content of streptomycetes, which makes a sequence-based discovery of sRNAs difficult in other species. However, it is also possible that sRNAs from streptomycetes are highly species-specific and related to the production of secondary metabolites or developmental changes, and therefore, simply cannot be found elsewhere.
The conservation of sRNAs within the genus of Streptomyces exists to different extents, ranging from abundance in all available streptomycete genomes to being present in S. coelicolor only. Relating to the distribution across the genome the majority of sRNAs is located in the core region of the linear chromosome (about 80%), with only a lesser amount being encoded in the arm regions. sRNAs have been identified in all developmental stages and have even been found in spores of S. coelicolor, some to quite a high degree (unpublished data).19
Another insight that several groups share is the presence of stems with C-rich loops within the sRNA structure.19,21 Those are assumed to act as alternative termination structures to known Rho-independent termination.17 Besides that, C-rich loops have also been shown to act as an initial binding site to the mRNA target followed by the melting of the stem, thereby leading to a conformational change enabling the binding of the target.22,23 These studies, however, were done in Gram-negative bacteria with a significantly lower GC-content. If a C-rich loop yields enough target specificity in a high-GC organism, it remains to be experimentally validated.
Within the last years, it came into focus that sRNAs might be encoded even within a 5′ or 3′UTR of an mRNA.24 Moody et al. detected a putative sRNA in S. venezuelae that is highly expressed out of an internal 62 nt-long region of the mRNA; in Salmonella, the derivation of sRNAs out of the 3′UTR was shown by Chao et al., whether there is a specific functional role of this mechanism remains unclear.21,25 Regarding the predicted structure of the 3′UTR arisen sRNA in S. venezuelae, it seems plausible that this stem-loop-like structure could be a terminator, whose stable stem resists RNase digestion for a bit longer than the rest of its mRNA, and therefore, is found to be enriched in RNA sequencing.
A similarity of streptomycetes to other organisms is their usage of RNase III for sRNA-mediated regulation. RNase III degrades RNA by cleaving at the double stranded basepairing region of mRNA–sRNA complexes. A homolog to the E. coli RNase III has been identified in S. coelicolor.26 It shows endonucleolytic activity specific for double strands. Gatewood et al. used RNaseq to compare the transcriptome of S. coelicolor M145 and an RNase III deletion mutant in order to find substrates for the RNase III homolog in S. coelicolor. They identified several sRNAs from earlier studies both in the wild-type strain and the deletion mutant. For most of them, the expression was more or less the same in both strains. Yet, two sRNAs (scr6925 and scr2101) showed a higher expression in the deletion strain: scr6925 2-fold and scr2101 even 7-fold. A direct interaction with RNase III has not been shown so far, because the targets of both sRNAs are not known, yet.
sRNA With Known Function in S. coelicolor
One of the currently best-characterized sRNAs in Streptomyces is probably scr5239. It was initially identified in a deep sequencing approach of S. coelicolor and is encoded between the genes SCO5238, a TetR family regulator of unknown function, and SCO5239, a highly conserved histidine kinase.19 The sRNA sequence and structure is well conserved and can be found in silico in 19 of the 31 Streptomyces genomes currently available, its expression has been shown in five of them by northern blot.27 Interestingly, and as already discussed above, the sRNA could not be detected outside the genus of Streptomyces by bioinformatic means.
Expressional analysis showed that scr5239 is constitutively expressed at a basal level during the whole developmental cycle under most conditions tested. Expression level of scr5239 was only decreased when the cells reached stationary phase under nitrogen-limiting conditions.
The secondary structure of the sRNA was analyzed by Inline and enzymatic probing. It folds into five consecutive stem-loops, which share an even higher degree of conservation among streptomycetes than the sequence alone. Most notably, the interaction site with at least one of its target-mRNAs, the agarase dagA, is buried in stem P4 that has to open up before an interaction can take place. This makes the interaction, in vitro, temperature dependent.
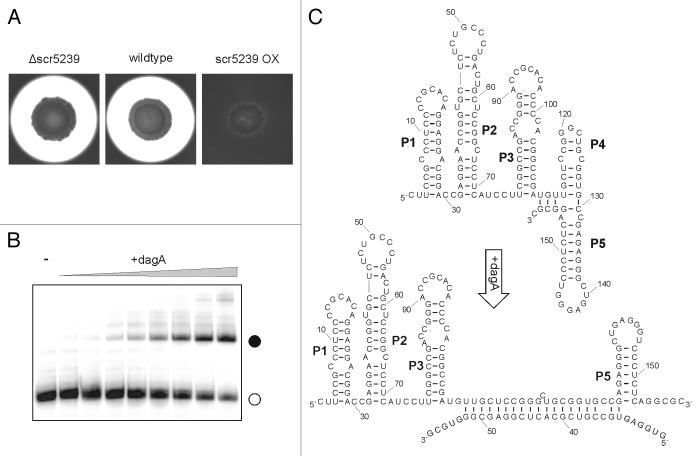
The aforementioned target of scr5239, dagA, was determined by phenotype analysis of scr5239 overexpression and deletion strains.27 The scr5239 overexpression strain did not sink into the medium as the wild-type and deletion strains do, a phenomenon that is caused by the utilization of agar as a carbon source (Fig. 2A). The agarase DagA is responsible for the first step of agar decomposition, the cleavage of agar into the disaccharide neoagarobiose.
Figure 2. Agarase DagA controlled by scr5239. (A) Agarase assay of S. coelicolor wild-type, scr5239 deletion, and overexpression strain. Agarase expression is visualized as a pale halo around the colony. (B) Electrophoretic mobility shift of radiolabelled scr5239 (open circle) incubated with an increasing amount of a 100 nt mRNA fragment containing the target site of dagA. Complex formation (closed circle) was visualized by gelelectrophoresis. (C) Secondary structure of scr5239 alone (top) and in complex with the mRNA target site (bottom). The structure was calculated using RNAfold29 and confirmed by structural probing. Stems (pedesand = P) are numbered, the 5′ and 3′ end of each RNA is indicated.
The interaction site of scr5239 with dagA was found to 30 nt within the coding region of the mRNA. Surprisingly, northern blot analysis showed that while scr5239 overexpression leads to a repression of agarase expression, it has no influence on the steady-state level of the agarase mRNA (Fig. 2B).
A second sRNA with a known function in S. coelicolor is the cis-encoded antisense RNA cnc2198.1.28 It is encoded antisense the glutamine synthase gene I glnA. Overexpression of cnc2198.1 led to a ~40% reduction of GlnA as seen in western blot analysis. Another of the currently identified asRNAs is proposed to be involved in the regulation of complex I formation, an important subunit of the respiratory transport chain. Still, this assumption has yet to be verified.21 Further functions of sRNAs or asRNAs have not been determined so far.
Conclusion
Clearly, more work needs to be done on the identification of sRNAs under different conditions, such as the variation of nutrient conditions, the application of stress situations, and most definitely on the growth phase dependency. All of the previous works stated that the found sRNAs are connected to their experimental setting and that another setup of the conditions may alter the findings enormously. Besides that, it will be very interesting to determine where the “standard” binding site of sRNAs on their target mRNA is located, as all sRNAs characterized so far bind their target mRNA within its ORF. In addition, the constantly increasing number of accessible Streptomyces genomes will provide an even greater tool for sRNA and target search in the future.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work has been supported by grants of the Deutsche Forschungsgemeinschaft (SU402/2-2 within the priority program “Regulatory RNAs in Bacteria”).
References
- 1.Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 2.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bérdy J. Bioactive microbial metabolites. J Antibiot (Tokyo) 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 4.Kekuda TRP. Fascinating diversity and Potent biological activities of Actinomycete metabolites. J Pharm Biomed Anal. 2010;3:250. [Google Scholar]
- 5.Ruban-Ośmiałowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwińska J. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J Bacteriol. 2006;188:7311–6. doi: 10.1128/JB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagüe P, Rodríguez-García A, López-García MT, Martín JF, Rioseras B, Sánchez J, Manteca A. Transcriptomic analysis of Streptomyces coelicolor differentiation in solid sporulating cultures: first compartmentalized and second multinucleated mycelia have different and distinctive transcriptomes. PLoS One. 2013;8:e60665. doi: 10.1371/journal.pone.0060665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak J, Dharmatilake AJ, Jiang H, Kendrick KE. Differential regulation of ftsZ transcription during septation of Streptomyces griseus. J Bacteriol. 2001;183:5092–101. doi: 10.1128/JB.183.17.5092-5101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flärdh K, Buttner MJ. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 9.McCormick JR, Flärdh K. Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev. 2012;36:206–31. doi: 10.1111/j.1574-6976.2011.00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakimowicz D, van Wezel GP. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol Microbiol. 2012;85:393–404. doi: 10.1111/j.1365-2958.2012.08107.x. [DOI] [PubMed] [Google Scholar]
- 11.Hindra, Moody MJ, Jones SE, Elliot MA. Complex Intra-Operonic Dynamics Mediated by a Small RNA in Streptomyces coelicolor. PLoS One. 2014;9:e85856. doi: 10.1371/journal.pone.0085856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strakova E, Zikova A, Vohradsky J. Inference of sigma factor controlled networks by using numerical modeling applied to microarray time series data of the germinating prokaryote. Nucleic Acids Res. 2014;42:748–63. doi: 10.1093/nar/gkt917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol Chem. 2005;386:1219–38. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 15.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol. 2007;10:262–70. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pánek J, Bobek J, Mikulík K, Basler M, Vohradský J. Biocomputational prediction of small non-coding RNAs in Streptomyces. BMC Genomics. 2008;9:217. doi: 10.1186/1471-2164-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiercz JP, Hindra, Bobek J, Bobek J, Haiser HJ, Di Berardo C, Tjaden B, Elliot MA. Small non-coding RNAs in Streptomyces coelicolor. Nucleic Acids Res. 2008;36:7240–51. doi: 10.1093/nar/gkn898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vockenhuber M-P, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 2011;8:468–77. doi: 10.4161/rna.8.3.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tezuka T, Hara H, Ohnishi Y, Horinouchi S. Identification and gene disruption of small noncoding RNAs in Streptomyces griseus. J Bacteriol. 2009;191:4896–904. doi: 10.1128/JB.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moody MJ, Young RA, Jones SE, Elliot MA. Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics. 2013;14:558. doi: 10.1186/1471-2164-14-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 24.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–9. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 25.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatewood ML, Bralley P, Weil MR, Jones GH. RNA-Seq and RNA immunoprecipitation analyses of the transcriptome of Streptomyces coelicolor identify substrates for RNase III. J Bacteriol. 2012;194:2228–37. doi: 10.1128/JB.06541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vockenhuber MP, Suess B. Streptomyces coelicolor sRNA scr5239 inhibits agarase expression by direct base pairing to the dagA coding region. Microbiology. 2012;158:424–35. doi: 10.1099/mic.0.054205-0. [DOI] [PubMed] [Google Scholar]
- 28.D’Alia D, Nieselt K, Steigele S, Müller J, Verburg I, Takano E. Noncoding RNA of glutamine synthetase I modulates antibiotic production in Streptomyces coelicolor A3(2) J Bacteriol. 2010;192:1160–4. doi: 10.1128/JB.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofacker IL, Stadler PF. Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics. 2006;22:1172–6. doi: 10.1093/bioinformatics/btl023. [DOI] [PubMed] [Google Scholar]