Abstract
Small regulatory RNAs (sRNAs) are universally distributed in all three domains of life, Archaea, Bacteria, and Eukaryotes. In bacteria, sRNAs typically function by binding near the translation start site of their target mRNAs and thereby inhibit or activate translation. In eukaryotes, miRNAs and siRNAs typically bind to the 3′-untranslated region (3′-UTR) of their target mRNAs and influence translation efficiency and/or mRNA stability. In archaea, sRNAs have been identified in all species investigated using bioinformatic approaches, RNomics, and RNA-Seq. Their size can vary significantly between less than 50 to more than 500 nucleotides. Differential expression of sRNA genes has been studied using northern blot analysis, microarrays, and RNA-Seq. In addition, biological functions have been unraveled by genetic approaches, i.e., by characterization of designed mutants. As in bacteria, it was revealed that archaeal sRNAs are involved in many biological processes, including metabolic regulation, adaptation to extreme conditions, stress responses, and even in regulation of morphology and cellular behavior. Recently, the first target mRNAs were identified in archaea, including one sRNA that binds to the 5′-region of two mRNAs in Methanosarcina mazei Gö1 and a few sRNAs that bind to 3′-UTRs in Sulfolobus solfataricus, three Pyrobaculum species, and Haloferax volcanii, indicating that archaeal sRNAs appear to be able to target both the 5′-UTR or the 3′-UTRs of their respective target mRNAs. In addition, archaea contain tRNA-derived fragments (tRFs), and one tRF has been identified as a major ribosome-binding sRNA in H. volcanii, which downregulates translation in response to stress. Besides regulatory sRNAs, archaea contain further classes of sRNAs, e.g., CRISPR RNAs (crRNAs) and snoRNAs.
Keywords: archaea, small regulatory RNAs, tRNA-derived fragments, translation, Methanosarcina mazei, Haloferax volcanii, Sulfolobus solfataricus, Nanoarchaeum equitans
Introduction
About 35 y ago it has been proposed that archaea form a third domain of life, phylogenetically separated from the bacteria and eukaryotes.1 This proposal was based on partial 16S/18S rRNA sequences of very few methanogenic archaea, bacteria, and lower eukaryotes, but has been proven to be true after the analysis of thousands additional species and sequencing of hundreds of archaeal and bacterial genomes.2-4 Like bacteria, archaea do not contain a nucleus and are thus prokaryotes. In addition, archaea and bacteria share a much higher metabolic diversity compared with eukaryotes, including a large variety of anaerobic energy yielding pathways. On the other hand, archaea and eukaryotes share homologous proteins in many central biological processes to the exclusion of the bacteria, including DNA packaging, replication, transcription, translation, and cell cycle. Given this dichotomy, we find it attractive to compare the results obtained about small sRNAs in archaea with the knowledge on sRNAs in bacteria and eukaryotes.
The sRNAs are universally distributed in all three domains of life: archaea, bacteria, and eukaryotes. In 1984, the first sRNA was identified in the genome of Escherichia coli,5 but it was long thought to be exceptional. However, in recent years it became clear that bacteria typically contain 200–400 sRNAs that can target more than 1000 mRNAs. Typically, bacterial sRNAs are 50–300 nucleotides in length and fulfill numerous important regulatory functions, including stress response, and regulation of virulence genes, carbon source uptake, and metabolism (for reviews see refs. 6–10). Bacterial sRNAs are often encoded in intergenic regions in trans to their target genes and typically function by imperfect base-pairing interactions with their target mRNAs near the ribosomal binding site (RBS). This interaction can repress translation by masking the RBS or can induce translation by making the RBS accessible. Also, additional molecular mechanisms of sRNA functions in bacteria have been described, including destabilization or stabilization of the target mRNAs or the action via specific binding to a protein. Another class of sRNAs are antisense sRNAs (asRNAs), which are encoded in cis on the opposite strand of their target gene, and thus, have full complementarity to their target mRNA. The fraction of genes regulated by asRNAs varies widely in different bacterial species and can be higher than 40%.11
It has been suggested that bacterial sRNAs are responsible for fine-tuning of gene regulation due to the fact that sRNA gene deletion mutants often have no obvious phenotype compared with the parent strain or the phenotype is very mild (reviewed in refs. 8 and 12). Many sRNAs in gram-negative bacteria need the Hfq protein for function, which is required for target mRNA recognition and complex formation (reviewed in ref. 13).
The best-studied eukaryotic sRNAs are microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-associated RNAs (piRNAs), which are approximately 20–30 nt in length and are thus much smaller than bacterial sRNAs. Currently, it is thought that in higher eukaryotes, e.g., humans, more than half of all mRNAs is regulated by these sRNAs in their translation and/or stability.14 In animals, miRNAs typically repress translation of their target mRNAs via imperfect base pairing between miRNAs and the 3′-UTRs. In eukaryotes, the lack of a miRNA often leads to severe phenotypic defects. In humans, miRNA dysfunction or deficiency is associated with a variety of diseases, developmental defects, and cancer formation (reviewed in refs. 15–18). A second class of eukaryotic sRNAs, siRNAs, is often processed from exogenous RNAs that enter the cell. Typically, siRNAs bind to the 3′-UTRs of target mRNAs, have the ability to form perfect hybrids, and binding results in degradation of the target mRNA. Thus, siRNAs are thought to have evolved as a defense system against RNA viruses (reviewed in refs. 19 and 20). The piwi-interacting RNAs (piRNAs) form complexes with Piwi proteins and are involved in the silencing of transposons during spermatogenesis (reviewed in ref. 21). A different class of eukaryotic sRNAs, the tRNA-derived fragments (tRFs), is generated via the processing of mature tRNAs or precursor tRNAs. Recently, it became clear that tRFs are not only degradation intermediates, but can have important regulatory functions (reviewed in ref. 22). An additional class of eukaryotic sRNAs comprises small nucleolar RNAs (snoRNAs), which are localized in the nucleolus and guide site-specific modifications of the rRNAs. Based on specific sequence motifs, they are divided into the two groups of C/D box and H/ACA box snoRNAs (for detailed reviews, see refs. 23–25).
Surprisingly, the first sRNAs identified in archaea were C/D box snoRNAs, which had not been expected in a prokaryote lacking a nucleolus. Based on the conserved function, these sRNAs are also called small “nucleolar” RNAs in archaea. C/D box snoRNAs guide ribose methylation at specific sites of the rRNAs. They were first predicted to exist in three Pyrococcus species, and subsequently, experimentally verified in Sulfolobus acidocaldarius (reviewed in refs. 23 and 26). Furthermore, H/ACA box snoRNAs have been identified in archaea. H/ACA box snoRNAs guide the conversion of uridine to pseudouridine at specific sites of the rRNA. Both types of snoRNAs are associated with at least four protein molecules in nucleolar ribonucleoprotein (snoRNP) complexes, which recognize target sites and catalyze the modification reaction. The protein content of the two snoRNP complexes differs (reviewed in ref. 27). One important C/D box snoRNA-associated protein is fibrillarin. Fibrillarin functions as a methyltransferase and the structures of several archaeal fibrillarins have been determined.28-30 The conserved presence and function of snoRNAs in archaea and eukaryotes substantiates that they are of ancient evolutionary origin. However, archaeal snoRNAs are not in the major focus of this article and we would like to refer to several recent reviews about archaeal snoRNAs and possible scenarios of snoRNA evolution.24,31,32
Subsequent to snoRNAs, additional groups of sRNAs have been identified in archaea, i.e., regulatory cis- and trans-encoded sRNAs, tRNA-derived fragments, and CRISPR RNAs (crRNAs). This review will focus mainly on regulatory cis- and trans-encoded sRNAs and summarizes the current knowledge of their identification, their participation in various biological processes, their interaction partners, and molecular mechanisms of action. At the end we will give an outlook on current technical challenges and foreseeable future trends.
Identification of sRNAs in Archaea
Shortly after the identification of snoRNAs in archaea, a new class of sRNAs was identified in several species using experimental and bioinformatic approaches. About 10 y ago, bioinformatic approaches led to the identification of sRNA genes in the genomes of Methanocaldococcus janaschii and Pyrococcus furiosus.33-35 These approaches predicted 18 putative sRNA genes in M. jannaschii and five sRNA genes in P. furiosus, in addition to the previously known snoRNAs. At the same time, small scale experimental RNomics approaches led to the identification of sRNAs in the euryarchaeon Archaeoglobus fulgidus,36 and later in Sulfolobus solfataricus37 and in Haloferax volcanii.38 While these initial approaches verified the existence of sRNAs in several archaeal species, in recent years it became clear that the number of sRNAs is much higher than anticipated in the beginning.
Whole genome transcriptome analysis via high-throughput sequencing of cDNA libraries, called RNA-Seq, became available a few years ago and enabled the qualitative analysis of the RNA inventory of species as well as the quantitative analysis of differential transcript levels under various conditions.39,40
The first RNA-Seq study with an archaeal species was performed for Methanosarcina mazei Gö1 under different nitrogen availabilities, leading to the identification of 242 intergenic and antisense sRNA, including six cis-antisense sRNAs overlapping with transposase genes and 40 sRNA candidates containing very short ORFs potentially encoding peptides smaller than 30 amino acids.41 The transcriptome of H. volcanii was characterized in exponentially growing and in stationary phase cultures grown under optimal conditions as well as, respectively, under reduced salt concentration and elevated temperature using a multiplexing RNA-Seq approach.42 One hundred and forty-five intergenic sRNAs and 45 antisense sRNAs were identified and it was revealed that the levels of many sRNAs differed depending on growth phase and/or external conditions. Sense sRNAs were also observed, but not further characterized, because they might correspond to degradation intermediates of full-length transcripts. Notably, also tRNA-derived fragments were observed, an indication that they might play regulatory roles, similar to their function in eukaryotes. The 190 intergenic and antisense sRNAs included all sRNAs identified in a previous small scale RNomics study,38 but not all sRNAs that had been predicted using genome comparisons in silico.43 Thus, it is very likely that not all sRNA genes were expressed under the three tested conditions and the actual number is higher than the 190 sRNAs observed in this RNA-Seq study.
A RNA-Seq study was also performed with four Pyrobaculum species, i.e., P. aerophilum, P. arsenaticum, P. calidifontis, and P. islandicum.44 However, this study focused on the characterization of snoRNAs and CRISPR-derived crRNAs. In all four species approx. 85 C/D box snoRNAs and 10 H/ACA snoRNAs were identified.
Using a combination of experimental and in silico approaches, small RNAs were also detected in Pyrococcus abyssi.45 Recently, the non-coding transcriptome of this hyperthermophilic archaeon was additionally analyzed by RNA-Seq, surprisingly demonstrating that several highly expressed or highly conserved sRNAs are AU-rich, suggesting RNA functions that do not require extensive secondary structure in the high-temperature environment of P. abyssi.46
Record numbers of 126 C/D box snoRNAs could be identified in the sRNA profile of the hyperthemophilic Methanopyrus kandleri.47 The high amount of these RNA species appears to correlate with the increased growth temperature of up to 110 °C, which would necessitate 2-O-methylation for the stabilization of rRNA folding.
In the subdomain of Crenarchaeota, two RNA-Seq studies have been performed with the species Sulfolobus solfataricus. The first study led to the identification of more than 300 sRNAs, including 13 snoRNAs, 18 crRNAs, and 28 cis-antisense sRNAs overlapping with transposase genes.48 Notably, the highest fraction was comprised of antisense sRNAs with 185 members, which exceed the fraction of intergenic sRNAs (125 members) by far, in contrast to the euryarchaeal species discussed above. The sRNAs identifed with the RNA-Seq approach included all sRNAs that had been identified earlier using small scale RNomics.36,49 As the genome of S. solfataricus is 2.99 Mbp and encodes less than 3000 protein-encoding genes, the fraction of sRNA genes is about 10% and higher than that in the euryarchaeal species characterized thus far. Recently, another RNA-Seq study concentrated on the identification of very small sRNAs in S. solfataricus.50 Total RNA was size-fractionated and RNA from about 18–30 nt was analyzed. A large number of sRNAs around 20 nt was found, verifying that very small sRNAs exist in Archaea.
Only few members of the phylum Nanoarchaeota have been identified, with Nanoarchaeum equitans being the single cultured organism.51,52 N. equitans contains a minimal and highly compacted genome and its RNA production relies on the import of nucleotides from an associated archaeon, Ignicoccus hospitalis. Small RNA profiling via RNA-Seq methodology was applied to identify sRNA molecules that are produced under these constraints.53 It was shown that C/D box snoRNAs and CRISPR RNAs are abundant in the cell, which underlines the importance of small RNA-guided RNA modification and viral defense mechanisms. In addition, several novel sense and antisense sRNAs were identified that could fulfil regulatory functions in the cell.
Until now, no sRNA investigation studies have been performed in species belonging to the kingdoms of Korarchaeota and Thaumarchaeota. Nevertheless, the occurrence of sRNAs in all species from the kingdoms Euryarchaeota, Crenarchaeota, and Nanoarchaeota that have been studied thus far underscores the wide-spread or even universal occurrence of sRNAs in archaea. Table 1 gives an overview of the results discussed above and lists several subclasses of sRNAs that have been experimentally verified by RNomics or RNA-Seq approaches.
Table 1. Trans-encoded intergenic sRNAs, cis-encoded antisense sRNAs (asRNAs), and tRNA-derived fragments (tRFs) experimentally identified in various archaea using RNA-Seq approaches.
| Number of genes for | ||||
|---|---|---|---|---|
| Intergenic sRNAs | asRNAs | tRFs | Reference | |
| Euryarchaeota | ||||
| M. mazei | 199 | 43 | n.d. | 41 |
| H. volcanii | 145 | 45 | 11 | 42 |
| P. abyssei | 107 | 215 | n.d.*1 | 46 |
| Crenarchaeota | ||||
| S. solfataricus | 43 | 185 | n.d. | 48 |
| Pyrobaculum (4 species) | present*2 | present*2 | present*2 | 44 |
| Nanoarchaeota | ||||
| N. equitans | present*2 | present*2 | present*2 | 53 |
*1Not detectable due to the method of library construction. *2Number of genes not included in the publication
It should be noted that most archaeal sRNAs are not well conserved; many sRNA genes are not even shared by species of the same genus. For example, only very few sRNAs that have been found in H. volcanii are also present in Haloferax mediteranii.54 In M. mazei, the majority of sRNAs genes are conserved in other species of methanogenic archaeea, but are not found outside of the methanogens.41 However, this observation is not confined to archaea, but similar findings have been reported for bacterial sRNA genes. For example, more than 90% of the 500 sRNAs identified in Pseudomonas aeruginosa have no homolog in any other bacterial species.55 In Salmonella, sRNAs were identified, which are not existent in E. coli.56 In addition, “highly conserved” sRNAs are typically confined to one taxonomic group, e.g., highly conserved cyanobacterial sRNAs are not found outside of the cyanobacteria.57 Therefore, it seems that in prokaryotes, both archaea and bacteria, the pace of evolution is much higher for sRNA genes than for protein-encoding genes. An interesting but as-yet-unresolved question is whether the species-specific sRNA genes were generated de novo during species evolution or whether the mutation rates of preexisting sRNA genes were so high that the similarity of homologous genes in different species has dropped below recognizable limits.
In Vivo Functions of sRNAs in Archaea
Although sRNAs have been identified in a variety of archaeal species, detailed analyses of the biological functions in vivo have until now only been performed with two species, M. mazei Gö1 and H. volcanii. In both species differential sRNA levels have been quantified using northern blot, DNA microarray, and RNA-Seq analyses, and mutants have been generated and characterized.
The methanogenic archaeon M. mazei is able to fix molecular nitrogen under conditions of nitrogen limitation and is genetically tractable, thus has been used for many years as a model to study nitrogen-dependent gene regulation in archaea.58-62 The recent discovery of regulatory sRNAs and their predicted role in nitrogen-dependent differential gene expression add another layer of regulation in addition to the well-studied transcriptional regulation.41 RNA-Seq analysis revealed that M. mazei contains nearly 250 sRNAs, 40 of which with coding capacity that might be oligopeptide-encoding mRNAs or dual function sRNAs.41 One hundred and thirty-five sRNAs were found to have differential levels in response to nitrogen availability, 36 were solely detected during nitrogen abundance, and 99 under nitrogen limitation. For several sRNAs, differential levels in response to the nitrogen availability were verified by northern blot analyses. Notably, in several cases, conserved motifs could be identified in the promoters of nitrogen-regulated sRNAs, indicating a network of coordinated transcriptional regulation.
One example, sRNA154, was further functionally characterized by a genetic approach.63 sRNA154 was found to be exclusively present under nitrogen-limiting conditions and has been shown to be under strict transcriptional control of the general nitrogen regulatory protein NrpR, indicating that it is relevant under nitrogen-fixating conditions. A deletion mutant of the sRNA154 gene had a severe growth defect under nitrogen-limiting conditions. However, under nitrogen sufficiency, it grew indistinguishable from the parent strain, emphasizing a crucial role of sRNA154 for nitrogen fixation.63 An in-depth characterization of a further sRNA, which led to the discovery of a first archaeal target mRNA, is discussed in the next chapter.
Differential sRNA levels under various conditions have also been analyzed in the halophilic archaeon H. volcanii, which has an optimal salt concentration of 2.1 M NaCl. For example, northern blot analyses revealed that sRNA194 is highly expressed during exponential phase, but absent or barely detectable in stationary phase cells.38 A 6-fold multiplexed RNA-Seq analysis yielded a genome-wide overview of sRNA levels in cultures grown under three different conditions to exponential phase and to stationary phase, respectively.42 The largest difference was found between optimal salt concentration and reduced salt concentration, 24 sRNAs had higher levels at the optimal salt concentration, while 19 sRNAs had higher levels at the reduced salt concentration. The six highest sRNA levels were found in cells grown at the reduced salt concentration, indicating that sRNAs have important regulatory functions for osmotic adaptation in H. volcanii.
For H. volcanii, a very efficient genetic system has been established and optimized.64-66 Generation of two deletion mutants revealed that phenotypic comparison of mutants and parent strain is an effective approach to unravel the biological roles of sRNAs in haloarchaea, deletion mutant of sRNA30 could not grow at 51 °C, in contrast to the parent strain, while deletion mutant of sRNA63 had a severe growth defect at low salt concentrations.38 Recently, growth of H. volcanii in microtiter plates was established, enabling the parallel comparison of growth parameters of many cultures.67 The efficient genetic system and the possibility of massive parallel cultivation together enabled the generation of a set of 27 deletion mutants of sRNA genes and their phenotyping under 11 different growth or stress conditions. In addition, cell morphology was checked and swarming was analyzed.54 Interestingly, 24 of the 27 sRNA gene deletion mutants exhibited a phenotype under at least one of the tested conditions, unraveling the broad importance of sRNAs in haloarchaea for many biological functions. They are involved in metabolic regulation, adaptation to extremes of temperature and osmolarity, stress response, growth phase adaptation, and even regulation of cellular morphology and behavior. Notably, seven of the 27 deletion mutants showed a gain-of-function phenotype, e.g., a shorter lag phase, a faster growth rate, a higher growth yield, or faster swarming than the parent strain. It is known that depletion of miRNAs in higher eukaryotes can result in a gain-of-function.68-70 Thus, it appears that in archaea and eukaryotes evolution of sRNAs did not result in the highest possible function of a single biological process, but in optimized flexibility and stability of the regulatory network.
Loss of sRNA function can also lead to a severe loss of function, both in archaea (see above) and in eukaryotes. For example, mutations in miRNA genes in C. elegans can impair developmental timing in larvae.71,72 In humans, miRNA dysfunction or deficiency induces strong disease patterns, cancer (reviewed in ref. 73), multiple sclerosis,74 Parkinson disease,75 or Alzheimers disease.76 In contrast, sRNA gene deletion mutants in bacteria often lack a detectable phenotypic difference to the parent strain or have only a very mild phenotype; therefore, it has sometimes been proposed that the major role of bacterial sRNAs is the “fine-tuning of gene expression.”77
Targets of Archaeal sRNAs
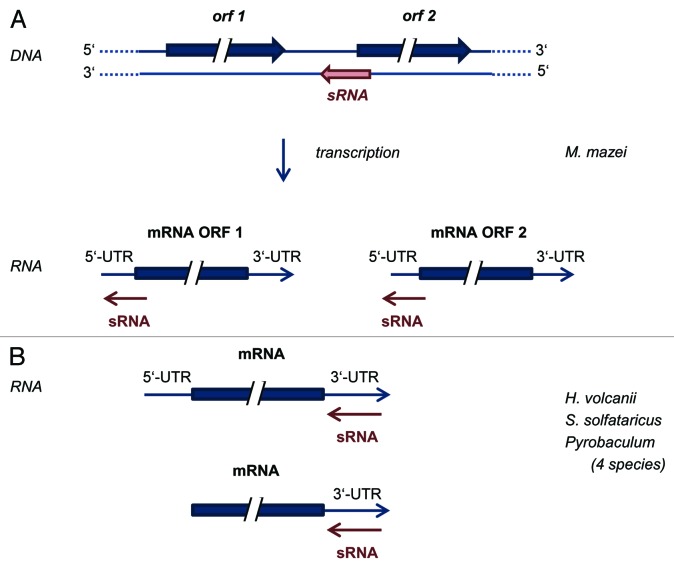
In bacteria, sRNAs typically interact with their target mRNAs near the 5′-end, often masking the ribosome binding site (RBS) or leading to an unfolding of the mRNA that makes the RBS accessible for ribosomes.78,79 In eukaryotes, mi/si/piRNAs typically bind to the 3′-UTRs of their target mRNAs, leading to translational inhibition and/or degradation of the mRNAs (reviewed in ref. 80). In both domains, a number of computational approaches have been established that allow a genome-wide in silico search for putative targets of a specific sRNA.81-83 However, these programs designed for and trained with bacterial and eukaryotic sRNAs, respectively, cannot necessarily be used for a genome-wide bioinformatic prediction of targets of archaeal sRNAs without specific adaptations. Early attempts to apply prediction programs for bacterial sRNA targets failed for archaeal sRNAs, indicating that the principles of interactions between sRNAs and target mRNAs might be different in archaea and bacteria. Thus, experimental approaches appear to be crucial to identify target mRNAs for archaeal sRNAs, subsequently verified targets could be used to guide bioinformatic predictions. Very recently, the group of Rolf Backofen optimized the target prediction program IntaRNA84 and integrated comparative genomics into the prediction pipeline,85 (see also contribution in this volume by Backofen, et al.) which was successfully used in combination with an experimental approach for the identification of the first target of an archaeal sRNA.86 This first target for an archaeal sRNA was identified in M. mazei combining genetic approaches, genome-wide transcriptome analysis of sRNA mutants, and computational target predictions using the tool IntaRNA.84,86,87 The bicistronic mRNA (MM2441-MM2442) was identified as the trans-encoded target mRNA of the respective sRNA (sRNA162). The predicted target interaction between the non-structured single-stranded linker region of sRNA162 and its target results in masking the RBS of MM2441 by base-pairing as well as the translational start codon, leading to a dis-coordinated expression of the bicistronic mRNA, and thus, to a depletion of the transcriptional regulator MM2441. The predicted interaction between sRNA162 and the RBS of MM2441 was verified in vitro (by EMSA) and in vivo (by ectopic expression of sRNA162 and various mutants). Further studies strongly indicated the involvement of sRNA162 in the expression regulation of soluble methyltransferases—most likely due to translation inhibition of the transcriptional regulator MM244—following the metabolic switch between growth on methanol vs. methylamine. Notably, it was also obtained that sRNA162 overlaps the 5UTR of the MM2442 transcript, and thus, is a also acting as a cis-encoded antisense RNA, in addition to regulating MM2441 expression as a trans-encoded sRNA (compare Fig. 1). Besides the recent report on an asRNA acting in trans in Staphylococcus aureus,88 an sRNA acting as an antisense RNA on cis-encoded as well as on trans-encoded mRNAs most likely via two distinct domains has not been shown in prokaryotes so far. Thus, the methanoarchaeal sRNA162 is the prototype of a novel class of “dual-mechanistic” sRNAs, which has the ability to act simultaneously in cis and in trans. The interaction of sRNA162 with the 5′-region of at least two genes, overlapping the RBS, resembles the molecular mechanism of typical bacterial sRNAs. Transcripts of methanoarchaea usually have long 5′-UTRs, thus interactions between sRNAs and 5′-UTRs are very likely. Further studies are required to unravel whether it is indeed typical for the action of methanoarchaeal sRNAs.
Figure 1. Modes of interactions between archaeal sRNAs and target mRNAs characterized until now. (A) A novel mode of interaction of a sRNA that acts as a cis-encoded antisense sRNA as well as a trans-encoded sRNA, in both cases interacting with the 5′-region of the target mRNA. (B) sRNAs that interact with the 3′-UTRs of target mRNAs in several species of archaea, both of leadered and of leaderless transcripts.
In contrast to Methanoarchaea, various archaeal species exist in which most or all transcripts do not contain 5′-UTRs, e.g., Pyrobaculum aerophilum,89 Sulfolobus solfataricus,48 Halobacterium salinarum, and H. volcanii.90 If differential regulation of translational efficiencies and/or stabilities of these leaderless transcripts would occur, respective motifs must be localized either in within the ORF or in the 3′-UTR. Genome-wide analyses of translational regulation (translatome analyses) in H. salinarum and H. volcanii revealed that translational regulation is not seldom in these two species. In fact 20% and 10%, respectively, of all transcripts have growth phase-specific differential translational efficienies.91 Further studies showed that 5′-UTRs and 3′-UTRs are sufficient to transfer translational regulation to a reporter transcript, and that the direction of translational regulation (up- or downregulation) is encoded in the 3′-UTR.92 To our knowledge, this was the first report that 3′-UTRs are involved in translational regulation in a prokaryotic species. However, it remained unclear whether sRNAs were involved in these examples. Recently, the transcriptomes of seven sRNA gene deletion mutants were compared with that of the parent strain with the aim to identify putative sRNA targets experimentally (Jaschinski, Babski, and Soppa, unpublished results). In each case, several or many genes differed in their transcript levels between mutant and parent strain. The numbers of transcripts with differential levels ranged from less than 10 to about 100, indicating that the regulatory role of haloarchaeal sRNAs can be very specific or rather broad, depending on the identity of the sRNA. Bioinformatic comparisons of the sequences of sRNAs and putative target mRNAs led to the discovery of several examples of putative interactions between sRNAs and the 3′-UTRs of mRNAs. Currently, experimental approaches to analyze whether these interactions occur and are relevant in vivo are under way, e.g., several UTRs of several transcripts have been transferred to a reporter transcript, and quantification of translational efficiencies in sRNA deletion mutants vs. parent strain indicate that the interaction between sRNA and the 3′-UTR can indeed guide translational efficiency (Jaschinski, Höfle, and Soppa, unpublished results).
Additional implication for an archaeal sRNA targeting a 3′UTR has been obtained from a recent comparative RNA sequencing approach studying four Pyrobaculum strains.93 Here, a 65 nucleotid-long antisense transcript, asR3, has been identified that is conserved in Pyrobaculum and binds to the 3′-end of the tpi gene (encoding triose-phosphate-isomerase), either overlapping the stop codon or binding to the 3′-UTR. Furthermore, a conserved structural element located close to the stop codon has been recognized in the 3′-region of the tpi gene (designated tpi element). Binding of asR3 to the tpi mRNA might be able to compete against the formation of the intramolecular tpi element structure, and consequently, might modulate the function of the highly conserved tpi element.
Recently, an interaction between a sRNA and the 3′-UTR of a target mRNA was reported for S. solfataricus.94 The sRNA257(1) was regulated in a phosphate-dependent manner and had a high level during phosphate sufficiency and a low level during phosphate starvation. The transcript of the gene Sso1183 was found to be inversely regulated in response to changing phosphate concentrations. A sequence comparison in silico revealed a very long stretch of high complementarity of about 60 nt between the sRNA and the 3′-UTR of the mRNA. The 3′-UTR and a mutated version of the 3′-UTR (with highly reduced complementarity) were fused to the reporter transcript lacS, and the transcript levels of the sRNA257(1) and the mRNA of the lacS variants were quantified in cultures grown in the presence and absence of phosphate. It could indeed be shown that the level of lacS containing the 3′-UTR of Sso1183 was inversely regulated to the sRNA level, in contrast to two lacS controls lacking a 3′-UTR or containing a 3′-UTR of an unrelated gene. Thus, first examples are accumulating that archaeal sRNAs can execute their regulatory role via interaction with the 3′-UTRs of target genes.
In summary, experimental approaches have led to the identification of first target mRNAs in three species of archaea. Already, these first examples reveal that the molecular mode of action of archaeal sRNAs is not universal in this domain of life and that the characterization of archaeal sRNAs leads to the identification of molecular mechanisms of sRNAs that have not been found for any bacterial sRNA. The modes of sRNA target mRNA interactions are schematically summarized in Figure 1.
tRNA-Derived Fragments
Recently, it has been discovered that eukaryotes contain small RNAs that are derived from tRNAs (tRNA-derived fragments, tRF). Initially, it was thought that tRFs are simple degradation products with increased stability, but now it is clear that they fulfill specific regulatory functions in many biological processes (reviewed in ref. 22), e.g., cell proliferation,95 in protein biosynthesis, and in the siRNA and miRNA pathway.96 In accordance with their regulatory roles, the processing of tRNAs to tRFs is itself regulated and is induced under specific conditions, e.g., stress application.97,98
Subsequently, tRFs were also identified in bacteria and archaea. In archaea, the existance of tRFs has been reported for several species of Pyrobaculum99 and H. volcanii.42,100 In H. volcanii, tRFs could be observed from 11 of the 51 tRNAs using RNA-Seq, and their existence could be verified using northern blot analyses.42 In another study, all small RNAs that could be co-isolated with ribosomes were identified by RNA-Seq.100 Twenty-six percent of the co-isolated sRNAs were tRFs, indicating that tRFs can directly or indirectly be associated with the ribosome. The tRFs of the tRNA-Valine (tRF[Val]) were the most prominent co-isolated small RNAs. It could be verified by density gradient centrifugation and in vitro binding studies that tRF(Val)directly binds to the small ribosomal subunit. Furthermore, using an in vitro translation system, it was shown that binding of tRF(Val) to the ribosome inhibits translation. In addition, it was revealed that the generation of tRF(Val) from the tRNA(Val) was highly induced under specific stress conditions, e.g., alkaline stress.100 Taken together, these results show that tRFs can potentially fulfill regulatory functions not only in eukaryotes, but also in archaea.
CRISPR RNAs in Archaea
In prokaryotes, one family of sRNAs, the CRISPR RNAs (crRNAs), play a key role in the defense against foreign DNA, e.g., viruses or conjugative plasmids. Genomic CRISPR arrays were discovered 1987 in E. coli.101 The term CRISPR is an acronym for “clustered regularly interspaced short palindromic repeats,”102 and as such, CRISPR loci are characterized by series of short repeated DNA sequences interspaced by unique spacer sequences. These spacers can represent foreign DNA elements that have been acquired at the promoter-proximal position of a CRISPR array. The CRISPR elements are transcribed and processed into small CRISPR RNAs, which contain a single spacer element flanked by repeat RNA fragments. These spacer sequences can guide a complex of CRISPR-associated (Cas) proteins to cDNA targets. This allows the detection of a repeated exposure to foreign DNA, which can subsequently be degraded by this interference ribonucleoprotein complex. Thus, CRISPR and Cas proteins represent an adaptive immune system, which follows rules of Lamarckian evolution.103 Different CRISPR-Cas systems exist with a plethora of Cas protein families that highlight functional differences (e.g., recognition of DNA or RNA) and the co-evolution of CRISPR and anti-CRISPR systems in prokaryotes and viruses.104,105 The diversification of Cas proteins is most obvious for crRNA maturation endonucleases and large subunits of the interference complex.106,107 The biogenesis of CRISPR RNA molecules and the mechanisms of CRISPR-Cas interference have been reviewed in detail before (e.g., refs. 108–110). A comparison of bacterial and archaeal CRISPR systems reveals that the latter contain (1) more and longer CRISPR systems, (2) a more diverse Cas protein landscape, and (3) more often a constitutive production of crRNAs. However, these observations also correlate with an increasingly hot environment, which is most often reserved for thermophilic or hyperthermophilic archaeal organisms. It remains to be analyzed if extreme growth conditions limit the diversity of host/virus interactions, which results in an accelerated evolution of defense and counter-defense measures.
Future Aspects
As summarized above, in recent years, a plethora of sRNAs have been found in any archaeal species that has been studied, and their importance for many different biological processes has been elucidated. Notably, first identifications of mRNA targets of sRNAs has led to the discovery of mechanisms of action unprecedented in prokaryotes, i.e., a dual-mechanistic sRNA that is a cis-antisense sRNA and a trans-acting sRNA at the same time, and a few sRNAs that act via binding to the 3′-UTR. Nevertheless, knowledge about archaeal sRNAs is lacking behind knowledge obtained with bacteria and eukaryotic sRNAs, and the following developments seem to be important for the near future.
Until now, the genome-wide prediction of sRNA targets has been successful for M. mazei in combination with genetic approaches, but not for other archaea. The experimentally identified targets should be used to train bioinformatic prediction programs with the aim to develop reliable high-quality predictions, possibly in an iterative exchange between experimental results and bioinformatic improvements. The results with M. mazei, H. volcanii, and S. solfataricus show that a domain-wide “archaeal” prediction program will not be possible, but several attempts for different groups of archaea are necessary.
Molecular analysis of a significant number of sRNA–target mRNA pairs will be necessary to understand the modes of actions and to enable generalization of principles. In E. coli and Salmonella, a dual plasmid system with a reporter gene (carrying UTRs and modified versions thereof) on one plasmid and the cognate sRNA gene on another plasmid has been proven to be extremely useful (e.g., refs. 111–114). Equivalent systems should be developed for several species of archaea to allow detailed analyses of sRNAs and motifs within UTRs of target mRNAs in vivo.
It is currently unknown whether and which proteins are required for the regulatory functions of sRNAs. In H. volcanii, the Lsm protein has been shown to bind a small subset of the existing sRNA pool, but it is unclear whether this is important for function or whether other sRNAs need different proteins or regulate solely at the RNA level. Methods for the affinity isolation of sRNAs and/or regulated mRNAs would be desirable to enable the identification of interaction partners.
It has been shown for many sRNAs that their levels are very different under various growth conditions. However, the regulation of sRNA generation and decay has not been studied yet, neither for sRNA genes with an own promoter, nor for sRNAs like tRFs that are processed from a longer precursor.
In summary, significant progress has been made in recent years concerning the presence and distribution of sRNAs in archaea, their biological functions, and initial successes have been made in target identification and characterization of sRNA–target RNA interactions, but it is also clear that considerable further work is needed to deepen our understanding of archaeal sRNAs and their regulatory networks.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The work in the groups of Marchfelder A, Schmitz RA, and Soppa J were funded by the German Research Council (Deutsche Forschungsgemeinschaft, DFG) through the Priority Program SPP 1258 “Sensory and Regulatory RNAs in Prokaryotes” (grants DFG Ma1538/11, Schm1052/9, So264/14). The group of Randau L was associated to the Priority Program and was funded by the Max-Planck-Society. Currently, the collaborative project of the groups of Marchfelder A and Soppa J are funded through the DFG grants Ma1538/18 and So264/21.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74:5088–90. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardanov AV, Ravin NV. The impact of genomics on research in diversity and evolution of archaea. Biochemistry (Mosc) 2012;77:799–812. doi: 10.1134/S0006297912080019. [DOI] [PubMed] [Google Scholar]
- 3.Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 2011;14:274–81. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 4.Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol. 2010;33:175–82. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 5.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci U S A. 1984;81:1966–70. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalaouna D, Simoneau-Roy M, Lafontaine D, Massé E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta. 2013;1829:742–7. doi: 10.1016/j.bbagrm.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 7.De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie. 2011;93:1943–9. doi: 10.1016/j.biochi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desnoyers G, Bouchard MP, Massé E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet. 2013;29:92–8. doi: 10.1016/j.tig.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi RU, Miyazaki H, Ochiya T. The role of microRNAs in the regulation of cancer stem cells. Front Genet. 2014;4:295. doi: 10.3389/fgene.2013.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulrane L, McGee SF, Gallagher WM, O’Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–62. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- 18.Kim WT, Kim WJ. MicroRNAs in prostate cancer. Prostate Int. 2013;1:3–9. doi: 10.12954/PI.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–8. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 20.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat Biotechnol. 2007;25:1435–43. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar MS, Chen KC. Evolution of animal Piwi-interacting RNAs and prokaryotic CRISPRs. Brief Funct Genomics. 2012;11:277–88. doi: 10.1093/bfgp/els016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10:1798–806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 24.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–8. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ. Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol. 2007;425:283–316. doi: 10.1016/S0076-6879(07)25013-X. [DOI] [PubMed] [Google Scholar]
- 26.Dennis PP, Omer A. Small non-coding RNAs in Archaea. Curr Opin Microbiol. 2005;8:685–94. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Reichow SL, Hamma T, Ferré-D’Amaré AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–64. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–23. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol. 2003;10:256–63. doi: 10.1038/nsb905. [DOI] [PubMed] [Google Scholar]
- 30.Deng L, Starostina NG, Liu ZJ, Rose JP, Terns RM, Terns MP, Wang BC. Structure determination of fibrillarin from the hyperthermophilic archaeon Pyrococcus furiosus. Biochem Biophys Res Commun. 2004;315:726–32. doi: 10.1016/j.bbrc.2004.01.114. [DOI] [PubMed] [Google Scholar]
- 31.Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 2013;54:53–77. doi: 10.1042/bse0540053. [DOI] [PubMed] [Google Scholar]
- 32.Lafontaine DL, Tollervey D. Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem Sci. 1998;23:383–8. doi: 10.1016/S0968-0004(98)01260-2. [DOI] [PubMed] [Google Scholar]
- 33.Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–40. doi: 10.1016/S0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 34.Schattner P. Searching for RNA genes using base-composition statistics. Nucleic Acids Res. 2002;30:2076–82. doi: 10.1093/nar/30.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein RJ, Misulovin Z, Eddy SR. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc Natl Acad Sci U S A. 2002;99:7542–7. doi: 10.1073/pnas.112063799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Hüttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–41. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Hüttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–81. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 38.Straub J, Brenneis M, Jellen-Ritter A, Heyer R, Soppa J, Marchfelder A. Small RNAs in haloarchaea: identification, differential expression and biological function. RNA Biol. 2009;6:281–92. doi: 10.4161/rna.6.3.8357. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536–46. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 41.Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci U S A. 2009;106:21878–82. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heyer R, Dörr M, Jellen-Ritter A, Späth B, Babski J, Jaschinski K, Soppa J, Marchfelder A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012;9:1011–8. doi: 10.4161/rna.20826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Babski J, Tjaden B, Voss B, Jellen-Ritter A, Marchfelder A, Hess WR, Soppa J. Bioinformatic prediction and experimental verification of sRNAs in the haloarchaeon Haloferax volcanii. RNA Biol. 2011;8:806–16. doi: 10.4161/rna.8.5.16039. [DOI] [PubMed] [Google Scholar]
- 44.Bernick DL, Cox CL, Dennis PP, Lowe TM. Comparative genomic and transcriptional analyses of CRISPR systems across the genus Pyrobaculum. Front Microbiol. 2012;3:251. doi: 10.3389/fmicb.2012.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phok K, Moisan A, Rinaldi D, Brucato N, Carpousis AJ, Gaspin C, Clouet-d’Orval B. Identification of CRISPR and riboswitch related RNAs among novel noncoding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genomics. 2011;12:312. doi: 10.1186/1471-2164-12-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toffano-Nioche C, Ott A, Crozat E, Nguyen AN, Zytnicki M, Leclerc F, Forterre P, Bouloc P, Gautheret D. RNA at 92 °C: the non-coding transcriptome of the hyperthermophilic archaeon Pyrococcus abyssi. RNA Biol. 2013;10:1211–20. doi: 10.4161/rna.25567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su AA, Tripp V, Randau L. RNA-Seq analyses reveal the order of tRNA processing events and the maturation of C/D box and CRISPR RNAs in the hyperthermophile Methanopyrus kandleri. Nucleic Acids Res. 2013;41:6250–8. doi: 10.1093/nar/gkt317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wurtzel O, Sapra R, Chen F, Zhu Y, Simmons BA, Sorek R. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–41. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zago MA, Dennis PP, Omer AD. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:1812–28. doi: 10.1111/j.1365-2958.2005.04505.x. [DOI] [PubMed] [Google Scholar]
- 50.Xu N, Li Y, Zhao YT, Guo L, Fang YY, Zhao JH, Wang XJ, Huang L, Guo HS. Identification and characterization of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. PLoS One. 2012;7:e35306. doi: 10.1371/journal.pone.0035306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podar M, Makarova KS, Graham DE, Wolf YI, Koonin EV, Reysenbach AL. Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct. 2013;8:9. doi: 10.1186/1745-6150-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–7. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 53.Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012;13:R63. doi: 10.1186/gb-2012-13-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaschinski K, Babski J, Lehr M, Burmester A, Benz J, Heyer R, Dörr M, Marchfelder A, Soppa J. Generation and Phenotyping of a collection of sRNA gene deletion mutants of the haloarchaeon Haloferax volcanii. PLoS ONE. 2014 doi: 10.1371/journal.pone.0090763. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gómez-Lozano M, Marvig RL, Molin S, Long KS. Genome-wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol. 2012;14:2006–16. doi: 10.1111/j.1462-2920.2012.02759.x. [DOI] [PubMed] [Google Scholar]
- 56.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 57.Georg J, Hess WR. Regulatory RNAs in cyanobacteria: developmental decisions, stress responses and a plethora of chromosomally encoded cis-antisense RNAs. Biol Chem. 2011;392:291–7. doi: 10.1515/bc.2011.046. [DOI] [PubMed] [Google Scholar]
- 58.Weidenbach K, Ehlers C, Kock J, Ehrenreich A, Schmitz RA. Insights into the NrpR regulon in Methanosarcina mazei Gö1. Arch Microbiol. 2008;190:319–32. doi: 10.1007/s00203-008-0369-3. [DOI] [PubMed] [Google Scholar]
- 59.Veit K, Ehlers C, Ehrenreich A, Salmon K, Hovey R, Gunsalus RP, Deppenmeier U, Schmitz RA. Global transcriptional analysis of Methanosarcina mazei strain Gö1 under different nitrogen availabilities. Mol Genet Genomics. 2006;276:41–55. doi: 10.1007/s00438-006-0117-9. [DOI] [PubMed] [Google Scholar]
- 60.Veit K, Ehlers C, Schmitz RA. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Go1. J Bacteriol. 2005;187:6147–54. doi: 10.1128/JB.187.17.6147-6154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ehlers C, Weidenbach K, Veit K, Deppenmeier U, Metcalf WW, Schmitz RA. Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol Genet Genomics. 2005;273:290–8. doi: 10.1007/s00438-005-1128-7. [DOI] [PubMed] [Google Scholar]
- 62.Ehlers C, Veit K, Gottschalk G, Schmitz RA. Functional organization of a single nif cluster in the mesophilic archaeon Methanosarcina mazei strain Gö1. Archaea. 2002;1:143–50. doi: 10.1155/2002/362813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ehlers C, Jäger D, Schmitz RA. Establishing a markerless genetic exchange system for Methanosarcina mazei strain Gö1 for constructing chromosomal mutants of small RNA genes. Archaea. 2011;2011:439608. doi: 10.1155/2011/439608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bitan-Banin G, Ortenberg R, Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J Bacteriol. 2003;185:772–8. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–53. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammelmann M, Soppa J. Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J Microbiol Methods. 2008;75:201–4. doi: 10.1016/j.mimet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 67.Jantzer K, Zerulla K, Soppa J. Phenotyping in the archaea: optimization of growth parameters and analysis of mutants of Haloferax volcanii. FEMS Microbiol Lett. 2011;322:123–30. doi: 10.1111/j.1574-6968.2011.02341.x. [DOI] [PubMed] [Google Scholar]
- 68.Boon RA, Dimmeler S. MicroRNAs and aneurysm formation. Trends Cardiovasc Med. 2011;21:172–7. doi: 10.1016/j.tcm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 69.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–66. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 71.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 72.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 73.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39:6219–25. doi: 10.1007/s11033-011-1441-7. [DOI] [PubMed] [Google Scholar]
- 75.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–4. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hébert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum Mol Genet. 2010;19:3959–69. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 77.Shimoni Y, Friedlander G, Hetzroni G, Niv G, Altuvia S, Biham O, Margalit H. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 79.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang Y, Zhang JL, Yu XL, Xu TS, Wang ZB, Cheng XC. Molecular functions of small regulatory noncoding RNA. Biochemistry (Mosc) 2013;78:221–30. doi: 10.1134/S0006297913030024. [DOI] [PubMed] [Google Scholar]
- 81.Pichon C, Felden B. Small RNA gene identification and mRNA target predictions in bacteria. Bioinformatics. 2008;24:2807–13. doi: 10.1093/bioinformatics/btn560. [DOI] [PubMed] [Google Scholar]
- 82.Tjaden B. Prediction of small, noncoding RNAs in bacteria using heterogeneous data. J Math Biol. 2008;56:183–200. doi: 10.1007/s00285-007-0079-5. [DOI] [PubMed] [Google Scholar]
- 83.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc Natl Acad Sci U S A. 2005;102:2454–9. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–56. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–7. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jäger D, Pernitzsch SR, Richter AS, Backofen R, Sharma CM, Schmitz RA. An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic Acids Res. 2012;40:10964–79. doi: 10.1093/nar/gks847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prasse D, Ehlers C, Backofen R, Schmitz RA. Regulatory RNAs in archaea: first target identification in Methanoarchaea. Biochem Soc Trans. 2013;41:344–9. doi: 10.1042/BST20120280. [DOI] [PubMed] [Google Scholar]
- 88.Sayed N, Jousselin A, Felden B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol. 2012;19:105–12. doi: 10.1038/nsmb.2193. [DOI] [PubMed] [Google Scholar]
- 89.Slupska MM, King AG, Fitz-Gibbon S, Besemer J, Borodovsky M, Miller JH. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J Mol Biol. 2001;309:347–60. doi: 10.1006/jmbi.2001.4669. [DOI] [PubMed] [Google Scholar]
- 90.Brenneis M, Hering O, Lange C, Soppa J. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 2007;3:e229. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lange C, Zaigler A, Hammelmann M, Twellmeyer J, Raddatz G, Schuster SC, Oesterhelt D, Soppa J. Genome-wide analysis of growth phase-dependent translational and transcriptional regulation in halophilic archaea. BMC Genomics. 2007;8:415. doi: 10.1186/1471-2164-8-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brenneis M, Soppa J. Regulation of translation in haloarchaea: 5′- and 3′-UTRs are essential and have to functionally interact in vivo. PLoS One. 2009;4:e4484. doi: 10.1371/journal.pone.0004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bernick DL, Dennis PP, Höchsmann M, Lowe TM. Discovery of Pyrobaculum small RNA families with atypical pseudouridine guide RNA features. RNA. 2012;18:402–11. doi: 10.1261/rna.031385.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Märtens B, Manoharadas S, Hasenöhrl D, Manica A, Bläsi U. Antisense regulation by transposon-derived RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO Rep. 2013;14:527–33. doi: 10.1038/embor.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–95. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–42. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 98.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bernick DL, Dennis PP, Lui LM, Lowe TM. Diversity of Antisense and Other Non-Coding RNAs in Archaea Revealed by Comparative Small RNA Sequencing in Four Pyrobaculum Species. Front Microbiol. 2012;3:231. doi: 10.3389/fmicb.2012.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–33. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 103.Koonin EV, Wolf YI. Is evolution Darwinian or/and Lamarckian? Biol Direct. 2009;4:42. doi: 10.1186/1745-6150-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–32. doi: 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Plagens A, Tripp V, Daume M, Sharma K, Klingl A, Hrle A, Conti E, Urlaub H, Randau L. In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku120. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brendel J, Stoll B, Lange SJ, Sharma K, Lenz C, Stachler A-E, Maier L-K, Richter H, Nickel L, Schmitz RA, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of crRNAs in Haloferax volcanii. J Biol Chem. 2014 doi: 10.1074/jbc.M113.508184. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–9. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 109.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 110.Marchfelder A, Fischer S, Brendel J, Stoll B, Maier LK, Jäger D, Prasse D, Plagens A, Schmitz RA, Randau L. Small RNAs for defence and regulation in archaea. Extremophiles. 2012;16:685–96. doi: 10.1007/s00792-012-0469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–37. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol. 2008;68:890–906. doi: 10.1111/j.1365-2958.2008.06189.x. [DOI] [PubMed] [Google Scholar]
- 114.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–37. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]