Abstract
Whereas, the majority of bacterial non-coding RNAs and functional RNA elements regulate post-transcriptional processes, either by interacting with other RNAs via base-pairing or through binding of small ligands (riboswitches), 6S RNAs affect transcription itself by binding to the housekeeping holoenzyme of RNA polymerase (RNAP). Remarkably, 6S RNAs serve as RNA templates for bacterial RNAP, giving rise to the de novo synthesis of short transcripts, termed pRNAs (product RNAs). Hence, 6S RNAs prompt the enzyme to act as an RNA-dependent RNA polymerase (RdRP). Synthesis of pRNAs exceeding a certain length limit (~13 nt) persistently rearrange the 6S RNA structure, which in turn, disrupts the 6S RNA:RNAP complex. This pRNA synthesis-mediated “reanimation” of sequestered RNAP molecules represents the conceivably fastest mechanism for resuming transcription in cells that enter a new exponential growth phase. The many different 6S RNAs found in a wide variety of bacteria do not share strong sequence homology but have in common a conserved rod-shaped structure with a large internal loop, termed the central bulge; this architecture mediates specific binding to the active site of RNAP. In this article, we summarize the overall state of knowledge as well as very recent findings on the structure, function, and physiological effects of 6S RNA examples from the two model organisms, Escherichia coli and Bacillus subtilis. Comparison of the presently known properties of 6S RNAs in the two organisms highlights common principles as well as diverse features.
Keywords: 6S RNAs, structure, function, mechanism, transcriptional regulation, physiological role
Introduction
Bacterial 6S RNAs belong to the small group of regulatory RNA molecules that globally inhibit transcription by directly binding to RNA polymerase (RNAP). First identified in E. coli more than 40 years from now,1,2 6S RNAs have recently been identified in a wide variety of different bacteria using RNA structure-based annotation tools.3 It is worth noting that in some organisms paralogous 6S RNAs with probably non-redundant functions are expressed.3-7
A key toward understanding 6S RNA function(s) was the demonstration that the RNA is associated with the RNAP holoenzyme in E. coli cell extracts.8,9 Computational and experimental analyses then revealed that 6S RNAs are able to form a distinct secondary structure consisting of two irregular helical stem regions flanking a large internal loop, termed the central bulge (CB), which is reminiscent of an open promoter DNA.3,7 This rod-shaped structure with a central region in which the top and bottom strands interact only weakly or not at all was later shown to be a general feature of 6S RNAs (Fig. 1). The RNA’s open promoter mimicry is essential for recognition by RNAP and immediately explains 6S RNA-dependent transcriptional inhibition, which was demonstrated independently in different laboratories.9-13 Considerable progress has since been made toward unravelling the molecular details of the 6S RNA-RNAP interaction. In E. coli, the molecule shows a strong preference for binding to the E. coli σ70-housekeeping holoenzyme of RNAP (Eσ70) that is responsible for transcription during exponential growth phase.7,9,11 However, not all σ70-dependent promoters are equally regulated by 6S RNA. Evidence has been provided that promoters with an extended -10 motif or promoters lacking a -35 consensus sequence are primarily affected by 6S RNA. Since quite a number of exceptions to this rule have been observed,10-13 the structural criteria for the prediction of which promoters are affected to what extent need to be refined.
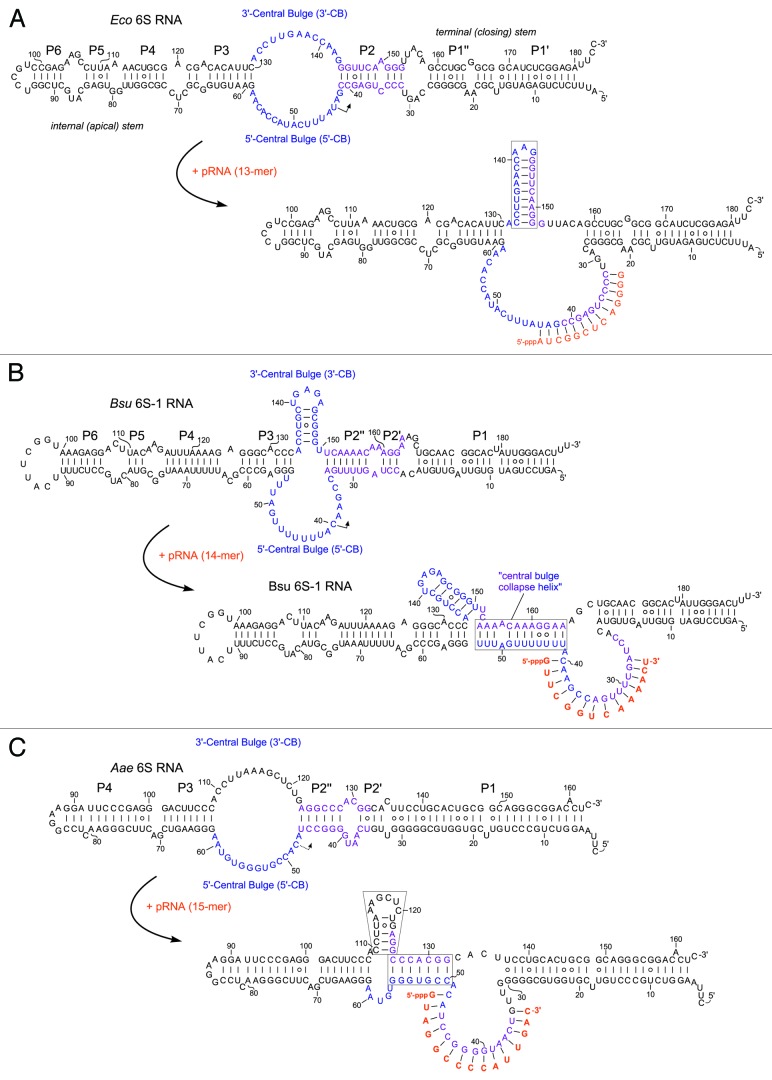
Figure 1. Ground state structures and predicted pRNA-induced structural rearrangements of 6S RNAs (E. coli 6S RNA, B. subtilis 6S-1 RNA and A. aeolicus 6S RNA). (A) In E. coli (Eco) representing the γ-proteobacteria, pRNA-induced disruption of 6S RNA structure triggers formation of an extended hairpin in the 3′-CB (boxed).21,47,50 The CB and P2 are thought to include a structural equivalent to extended -10 elements of DNA promoters.3 Unrau and co-workers termed the free structure at the top “S1 state,” the same secondary structure bound to RNAP “S2 state,” and the rearranged structure at the bottom “S4 state.”21 (B) B. subtilis (Bsu) 6S-1 RNA represents a group of 6S RNAs for which a hairpin can form (at least transiently) in the 3′-CB already in the free RNA; in this type of 6S RNA, the structural rearrangement is achieved by formation of a central bulge collapse helix (boxed).50 (C) A. aeolicus 6S RNA may involve both mechanistic components in its pRNA-induced rearrangement, hairpin formation in the 3′-CB and formation of a central bulge collapse helix (boxed elements).
As the cellular concentration of E. coli 6S RNA increases toward stationary phase, a general function of 6S RNA in transcriptional adaptation during the transition from exponential to stationary growth has been suggested.7 Likewise, highest 6S RNA expression was also seen in Legionella pneumophila (a γ-proteobacterium as E. coli), when cells reached the post-exponential growth phase.14 For the ε-proteobacterium Helicobater pylori, northern blot analyses indicated an increase of cellular 6S RNA levels from early to mid-exponential phase after which the levels remained rather constant up to late stationary phase.15 A distinct pattern was observed in the cyanobacterium Synechococcus sp. strain PCC6301, where 6Sa RNA levels largely decreased in stationary phase, suggesting a 6S RNA function in actively dividing cells.16 Several Firmicutes, including Bacillus subtilis, express two 6S RNAs, one with an expression profile comparable to that of E. coli 6S RNA, and the second one not accumulating or even decreasing toward stationary phase (see below).3,5,7,17 Hence, 6S RNA-mediated adaptations of the transcription machinery appear to be versatile and not necessarily focus on the stationary phase where cells reduce and reprogram transcription. The complexity of regulation by 6S RNAs is further illustrated by the outcome of genome-wide transcriptome analyses that have revealed a large number of genes to be under 6S RNA control, either directly or indirectly and including inhibition as well as activation effects.12
A milestone discovery was the finding that 6S RNA can act as template for transcription, thereby turning bacterial RNAP into an RNA-dependent RNA polymerase.11,18 This remarkable property is not restricted to 6S RNA from E. coli but has been observed for 6S RNAs in several other bacteria as well. 6S RNA-templated transcription results in the synthesis of small “product RNAs (pRNAs),” usually < 20 nt in length.5,15,19,20 Functional analyses in the E. coli system demonstrated that pRNA transcription reverts 6S RNA-dependent inhibition by disrupting the RNAP:6S RNA complex.18,21,22 Whether reversal of transcriptional inhibition is the only functional effect that can be attributed to this unusual reaction is a matter of current studies.
In the following sections, we summarize previous and recent findings on 6S RNAs that have contributed to a better functional understanding of this regulator in the two organisms E. coli and B. subtilis, where most of the studies have been performed so far. The molecular basis of several common and divergent regulatory features as well as the functional importance of 6S RNAs and its interdigitation with other regulatory pathways of transcription will be discussed.
Regulation of 6S RNA Expression in E. coli
E. coli 6S RNA (gene ssrS) is expressed as the first cistron of a bicistronic operon also including the ygfA gene coding for the enzyme 5-formyltetrahydrofolate cycloligase,23,24 which is involved in the formation of antibiotic-resistant persister cells and upregulated during biofilm formation.25,26 Two tandem promoters, P1 and P2, direct the transcription of the operon, but differ in their σ factor requirements; P1 is strictly σ70-specific while P2 is recognized by both Eσ70 and Eσ38.27,28 A set of nucleoid-associated proteins, including FIS, H-NS, StpA, and LRP bind to the upstream promoter region and thereby contribute to the RNA’s growth phase-dependent expression.29,30 A recent analysis has revealed a divergent autoregulatory effect of 6S RNA overexpression on promoters P1 (activation) and P2 (feedback inhibition).29 The mature 5′−ends of 6S RNA transcripts derived from P1 are generated in a well-established way by RNases G and E, while the longer transcripts from P2 are processed exclusively by RNase E.27 The details of 3′-end maturation are less clear. Several Rho-dependent terminators located downstream of the 6S RNA cistron lead to transcriptional termination within the ygfA cistron, followed by 3′-processing of 6S RNA likely via an exonucleolytic pathway. This indicates that the Rho factor regulates the expression of YgfA.31 During normal growth, 6S RNA was shown to be metabolically stable with a half-life longer than the generation time.22 As the result, the cellular concentration of 6S RNA in E. coli cells accumulates over the growth cycle and varies from roughly 1000 copies during early exponential growth to about 10 000 copies in late stationary phase.9
Binding of E. coli 6S RNA to RNAP
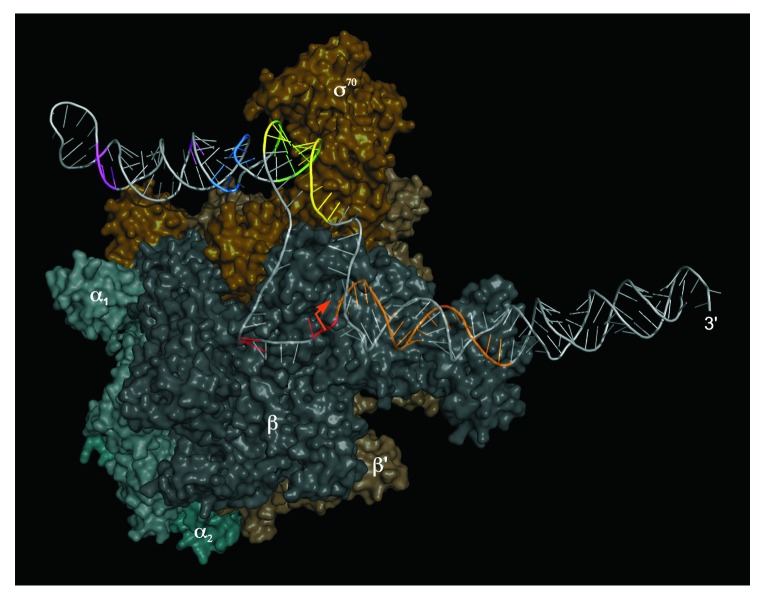
The binding of 6S RNA to RNAP was intensively studied in vitro and in vivo in several laboratories. 6S RNA binds with highest affinity to the Eσ70 (housekeeping) holoenzyme. In contrast, holoenzymes with the alternative specificity factors σ38, σ32, or the RNAP core enzyme are recognized only weakly or not at all. Furthermore, no binding to the free σ70 subunit was observed.7,9,11,13 UV crosslinking experiments have identified the β, β', and σ70 subunits of RNAP to be in contact with 6S RNA.9,11 Moreover, footprinting analyses indicated that sequence regions flanking the CB (Fig. 1) are involved in RNAP binding.11 Studies with σ70 mutants revealed the protein’s 4.2 region that recognizes the -35 element of DNA promoters to be crucial for the interaction with 6S RNA. In particular, a positively charged stretch of amino acids partially overlapping the helix-turn-helix DNA-binding domain of σ70 region 4.2 proved to be critical for 6S RNA binding.10,32 6S RNA:RNAP complexes were further probed using the chemical nuclease FeBABE.33 A selection of σ70 mutants harboring single cysteine residues, each positioned in a functionally defined region of the protein, were conjugated to the cleavage reagent FeBABE34 to generate hydroxyl radicals for RNA hydrolysis in the vicinity of the FeBABE moiety. A set of active RNAP holoenzymes was then reconstituted each with a different σ70 variant and complexed with 6S RNA. Sites of phosphodiester bond hydrolysis were identified on sequencing gels using 5′- and 3′-endlabeled 6S RNAs. In this way, a map of 6S RNA nucleotide positions in vicinity (within a range of 15 Å) to defined σ70 residues was established. The data were used for docking a structural model of 6S RNA to the high-resolution structure of the σ70-RNAP holoenzyme (Fig. 2).35,36 The docking results are consistent with nucleotides in or next to conserved bulges in the internal stem of 6S RNA being close to σ70 elements known to be involved in DNA binding and promoter melting. In this model, the sequence region around 6S RNA position U44 within the 5′-portion of the CB, known to be the start site for pRNA transcription, is in close vicinity to σ70 domain 3.2, also termed “σ-finger.”37 This σ70 element reaches deep into the active site of RNAP consistent with the fact that U44 must be close to the nucleotide addition center of the enzyme.18 In line with a 6S RNA truncation analysis,38 the model additionally suggests that roughly the CB-proximal half of the terminal (closing) stem of 6S RNA (roughly P1” and P2 in Fig. 1A), which was out of reach of the σ70-tethered FeBABE moieties, is located in the major RNAP cleft formed by the β and β' subunits known to take up the downstream regions of DNA promoters.
Figure 2. Three-dimensional model of 6S RNA bound to the E. coli σ70-RNAP holoenzyme (adapted from ref. 35). The RNAP α-subunits are depicted in cyan-gray shades, the β-subunit in anthracite, β’ in dark brown and σ70 in golden brown. The orange arrow indicates the site of pRNA initiation, with the pRNA template strand depicted in orange as well. Colored regions of 6S RNA indicate sites of RNA hydrolysis owing to FeBABE moieties conjugated to single cysteines of functional E. coli σ70-variants: FeBABE at position 581 (magenta), 496 (blue), 376 (green), 422 (yellow) or 517 (red) of σ70. For further details, see reference 36.
Still, the molecular basis for specific recognition of RNA instead of promoter DNA is unclear, as similarities to promoter consensus sequences are not identifiable in the 6S RNA core structure. Inspection of the FeBABE-induced 6S RNA hydrolysis sites mapped in the vicinity of functional σ70 domains (known to have DNA binding or strand separation activity) revealed prevalent cleavage at or near conserved 6S RNA bulges. Such bulges are known to interrupt the regular A-form helix, which widens the major groove and may facilitate the interaction with amino acid side chains.39 The observation that 6S RNA mutations converting these bulges to regular helical structures impaired RNAP binding is consistent with their role in RNAP recognition. In one case, the internal loop (between P5 and P6 in Fig. 1A) next to the apical loop was restored in one mutant, but with altered sequences on both sides. However, this failed to restore affinity for RNAP, suggesting that specific 6S RNA recognition by RNAP not only requires a defined pattern of helical elements, bulges and internal loops, but also depends on nucleotide identities or sequence patterns.36
Effects of E. coli 6S RNA on Transcription
The fact that 6S RNA regulates transcription is clearly undisputed; however, the molecular mechanism(s) leading to inhibition and how this inhibition is turned into a differential downregulation of cellular transcription units are open questions that are also matters of debate. Furthermore, it is not yet fully understood how regulation of transcription by 6S RNAs intertwines with other regulatory pathways that control transcription. For example, there is a potential functional link between 6S RNA and the co-transcribed ygfA gene, which is involved in folate pool regulation, purine synthesis, as well as formation of biofilms and multidrug-resistant persister cells.24-26 Moreover, a tight link exists between 6S RNA and the ppGpp metabolism of the cell, which affects growth rate regulation and adaptation of the translational capacity to starvation conditions (see below).12,40
In E. coli, 6S RNA is thought to sequester almost all RNAP holoenzymes under late stationary growth conditions, but no ubiquitous inhibition of σ70-dependent promoters is observed.10,13 This finding may suggest that 6S RNA:RNAP complex formation is not a static enzyme sequestration but a dynamic and selective process. The latter view is in line with the observation that pRNA synthesis in B. subtilis is also substantial during extended stationary phase5 (see below), maintaining conditions of constant binding competition between 6S RNA and DNA promoters. Initial experiments in E. coli indicated a strong preference for the negative regulation of σ70 promoters with an extended -10 recognition motif.13 This strict promoter specificity could not be substantiated in all cases in subsequent in vitro studies, which indicated that the outcome of inhibition by 6S RNA is not a simple and general discrimination of exponential vs. stationary phase-specific promoters.11 As in vitro studies are generally at risk of missing the cellular conditions for specificity, in vivo analyses were conducted which, however, also yielded some controversial results.10,12 Secondary effects of 6S RNA in the activation of stationary phase-specific (σ38-dependent) promoters were reported as well13 and, similar to the extended -10 motif, a weak -35 promoter element was identified to be a determinant for 6S RNA-sensitive promoters. Microarray analysis confirmed that two-thirds of the E. coli promoters predicted to be affected according to these features were indeed downregulated by 6S RNA.10 In an independent genome-wide transcriptome analysis, a large number of differentially expressed genes were identified when 6S RNA was depleted. Altered transcript levels were detected at early stationary phase but also during exponential growth. Interestingly, inhibition as well as activation was observed and many regulated genes were not under the control of σ70.12,41 As many regulators (e.g., FNR, regulator for anaerobic growth; TrpR, trp operon repressor; SlyA, antagonistic regulator of repression of the nucleoid-associated protein H-NS; IldR, dual regulator for transport and metabolism of L-lactate also involved in biofilm formation; FadR, fatty acid metabolism regulator)10,12 were affected in 6S RNA-depleted cells, not all of the observed changes in gene expression necessarily result from a direct effect of 6S RNA but might as well be indirect. Noteworthy, a significant increase in the basal level of the global regulatory compound ppGpp has been observed as a consequence of 6S RNA depletion. This could in part be attributed to derepressed synthesis of the ppGpp synthase RelA.40 However, the increase in ppGpp is also observed in a mutant strain lacking RelA and 6S RNA, indicating that the activity of the second ppGpp synthetase SpoT is also influenced by 6S RNA.12,41 In line with the known physiological role of ppGpp, a concerted reduction in ribosomal and other components of the translation machinery was detected at early stationary phase in 6S RNA-deficient cells, indicating that 6S RNA contributes to balancing the translational capacity of the cell.12,40,41
Function of E. coli 6S RNA in General Metabolism
Through the direct inhibition of a large fraction of σ70-dependent promoters at the end of exponential growth and the concomitant activating effect on transcription of many stationary phase-specific (σ38-dependent) genes, 6S RNA clearly contributes to the transcriptional adaptation of growing cells to stationary phase expression.10,12,13 Because the transition from exponential to stationary growth is induced by nutritional deprivation and/or the accumulation of environmental stress, a link between 6S RNA function and general stress adaptation is conceivable. In fact, 6S RNA-deficient cells have been shown to be at a disadvantage for survival under conditions of long-term stationary phase, while they can better cope with high pH.13,42 The latter effect is probably indirect and has been attributed to the 6S RNA-dependent inhibition of the regulator PspF, which acts in response to extracytoplasmic stress conditions and upon infection by filamentous phages.43 The observed effect of 6S RNA on the cellular concentration of the master regulator for stringent control and growth rate regulation, ppGpp, underlines the functional involvement of 6S RNA in stress adaptation. Independent studies indicate that both E. coli ppGpp synthetases, RelA, and SpoT, can account for the 6S RNA-dependent increase of the basal ppGpp pool.12,40,41 An interlinked network regulating σ factor competition and stress adaptation involving the regulators ppGpp, its synergistic cofactor DksA, the antisigma factor Rsd, and 6S RNA has recently been proposed.44 Genome-wide transcriptome analysis supports the involvement of 6S RNA as a component of several other stress regulatory networks. Respective examples showing that mRNA levels are altered in a 6S RNA-dependent way comprise the cold shock protein CspA, the general stress proteins UspG and UspF, or FNR, a global transcriptional regulator under anaerobic growth conditions.12 The study also demonstrated that 6S RNA, directly or indirectly, affects several key metabolic functions. Notable are altered mRNA levels of enzymes involved in purine metabolism. For example, transcript levels of the genes guaD and add, encoding guanine deaminase and adenosine deaminase, respectively, are significantly increased in a 6S RNA-deficient strain. Likewise, the gene encoding FolD, a bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/ cyclo-hydrolase, which is involved in C1 metabolism, is expressed at higher levels in 6S RNA-deficient cells. Interestingly, this enzyme is functionally related to the ygfA gene product whose mRNA is expressed as a bicistronic transcript with 6S RNA. These observations support the proposition that 6S RNA has a defined role in regulating central metabolic functions of the cell. Such a role is consistent with the proposal that 6S RNA fulfills an important task when resources become scarce or cells face other forms of stress. Hence, 6S RNA might act as resource sentinel, integrating stress adaptation with central metabolism in order to safeguard the economic use of scarce cellular components, energy, and nutrition.45,46
6S RNA Acts as a Template for RNAP—Findings in the E. coli System
The σ70-RNAP holoenzyme utilizes 6S RNA as a template for de novo synthesis of pRNAs,11,18 with transcription starting from a defined position (U44) within the 5′-strand of the CB (Fig. 1A). In the presence of elevated NTP concentrations, the majority of pRNAs reaches a length between 10 and approximately 20 nt, with the longer ones remaining stably attached to the 6S RNA template. A burst of pRNA synthesis occurs in vivo under outgrowth conditions when stationary growing cells face a nutritional upshift.18,22 The reaction is fast and the maximal pRNA concentration is reached 3 to 4 min after upshift. Transcription of pRNAs results in dissociation of the inhibitory RNAP:6S RNA complex, such that RNAP can resume transcription at DNA promoters. The molecular basis of complex dissociation has been explored by biochemical studies (see also below).5,21,22,38,47 Structural analyses of 6S RNAs, free and annealed to a pRNA 13- or 20-mer,21,47 have substantiated a defined conformational change of the 6S RNA secondary structure upon pRNA transcription and stable pRNA:6S RNA hybrid formation. As a consequence of base-pairing of pRNA with the RNA template sequence, the pre-existing internal 6S RNA helix P2 is disrupted, permitting formation of an extended 9-bp hairpin in the 3′-CB (Fig. 1A). Structural probing in living cells indicated that the same chain of events also occurs in vivo.47 The rearranged 6S RNA structure is obviously incompatible with the architecture of RNAP, which results in the release of σ70 followed by dissociation of core RNAP (for more details, see below). After leaving the RNAP complex, 6S RNA, still bound to pRNA, is thought to become vulnerable to attack by cellular nucleases leading to its rapid decay.22,47
Clearly, the immediate consequence of pRNA synthesis is the termination of the 6S RNA-mediated transcriptional inhibition via disruption of the 6S RNA:RNAP complex. Thus, the transcriptional regulator 6S RNA encodes its own cis-acting regulatory RNA.48 Whether or not this is the only function for pRNAs is still unsolved.
The synthesis of pRNAs has also been demonstrated for 6S RNAs from other bacteria, either by deep sequencing or biochemical methods.5,15,19,49 6S RNAs are also interchangeable to some extent. For example, several cyanobacterial 6S RNAs were shown to act as templates for E. coli RNAP, yielding pRNAs slightly longer than observed with the homologous E. coli 6S RNA.20
The pRNA-Induced Structural Rearrangement of 6S RNAs—Mechanistic Aspects
In addition to the E. coli system, the change in 6S RNA structure upon pRNA synthesis has also been shown for B. subtilis 6S-1 RNA.50 However, this 6S RNA rearranges its structure in a different manner to disrupt binding to RNAP. Here, unwinding of the endogenous helix P2 (Fig. 1B) upon pRNA synthesis makes nucleotides of the 3′-strand of P2 accessible to formation of a new base-pairing interaction with residues downstream of the pRNA initiation site in the 5′-portion of the central bulge (5′-CB). This new base-pairing interaction, demonstrated by enzymatic and chemical probing, was termed the “central bulge (CB) collapse helix” (Fig. 1B).50 A current question in the field concerns the mechanistic relevance of formation of this CB collapse helix: is this structural rearrangement mechanistically, thermodynamically and/or kinetically crucial in the RNAP release process, or might formation of the pRNA:6S-1 RNA hybrid and concomitant disruption of P2 be sufficient for the biological purpose, i.e., fast release of RNAP?
Interestingly, 6S RNA from the hyperthermophilic bacterium Aquifex aeolicus may combine both types of the pRNA-induced structural rearrangement, i.e., formation of a “CB collapse helix” as in B. subtilis 6S-1 RNA and formation of a hairpin structure in the 3′-portion of the CB as in E. coli 6S RNA (Fig. 1C). Experimental studies are needed to validate this structural rearrangement model proposed for A. aeolicus 6S RNA.50
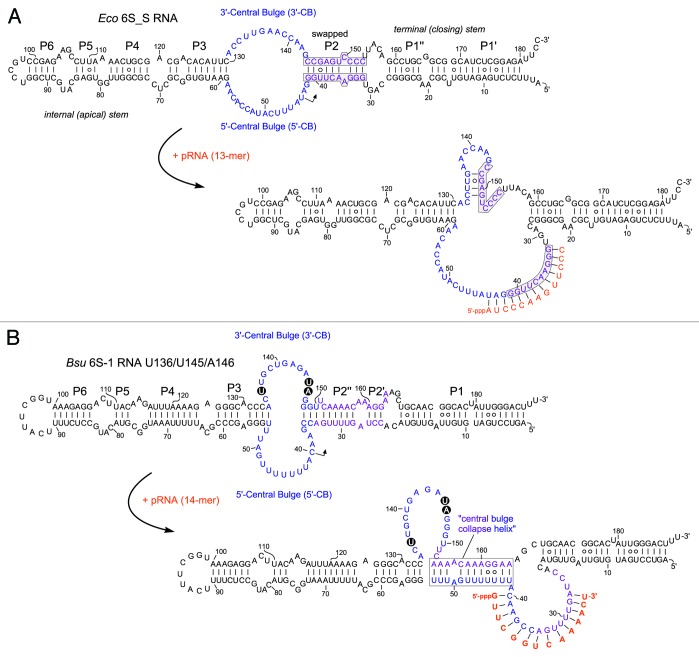
A kinetic study on the 6S RNA:RNAP interaction has recently been reported for the E. coli system.21 In addition to the σ70-RNAP:6S RNA ground state complex (termed state S2) with unchanged 6S RNA structure, the authors identified an intermediate RNAP:6S RNA state (S3), where σ70 has already left the complex and where associated pRNA 9-mers prevail. The pRNA 9-mers are then elongated by the core RNAP to pRNA 13-mers that persistently rearrange 6S RNA such that it dissociates from RNAP (state S4). The kinetic assays were performed at stoichiometric amounts of RNAP and heparin; in the absence of heparin, longer pRNAs were observed, suggesting that the energy barrier for RNAP:6S RNA dissociation is increased in the absence of heparin owing to non-specific RNA–protein interactions that stabilize the complex.21 Detection of the S3 state was made possible by slowing down pRNA transcription through omission of ATP in the presence of ApU for pRNA priming. In this setup, pRNAs were slowly extended to 13-mers.21 Furthermore, a 6S RNA mutant unable to form the extended hairpin in the 3′-CB was tested. This 6S RNA variant (termed 6S_S; Fig. 3A) slowed down 6S RNA release from RNAP (state S4), pRNAs in its S3 state were now predominantly 14- instead of 9-mers, and pRNAs in the S4 state were increased in length from 18 to 28 nt. It was concluded that formation of the extended hairpin in the 3′-CB is crucial for effectively destabilizing interactions between the 3′-CB of 6S RNA and σ70–RNAP that are initially present in the S2 ground state of the RNAP:6S RNA complex. All these findings are consistent with a scrunching mechanism (known from initial transcription at DNA promoters)51 involved in pRNA transcription: longer pRNAs in the S3 state (14 vs. 9 nt) with the 6S_S mutant RNA unable to form the extended hairpin can be attributed to the need to pull more ssRNA from both the top and bottom strands of the CB into the active core of RNAP during pRNA synthesis in order to build up the same tensile strain for repelling σ70 as in the presence of the hairpin. According to this model, the structural rearrangement of 6S RNA is already accomplished in the S3 state. Final dissociation of core RNAP and 6S RNA may be explained by the growing length of the pRNA:6S RNA hybrid helix which increases the stiffness of rearranged 6S RNA. Longer pRNAs in the S4 state (18–28 nt vs. 13 nt) with 6S_S RNA can as well be explained by reduced strain when the 3′-CB remains flexible as in 6S_S RNA.
Figure 3. Structure of (A) the E. coli 6S_S mutant RNA21 and (B) the B. subtilis U136/U145/A146 mutant 6S-1 RNA,50 including their putative pRNA-induced, rearranged structures. For comparison with the corresponding wild type 6S RNA structures, see Figure 1A and B. (A) In 6S_S RNA, the strands of helix P2 were swapped to preserve base pairing (marked by gray boxes); this structural change abolished the capacity to form an extended stable hairpin in the 3′-CB (RNAfold prediction). (B) Model of the B. subtilis mutant 6S-1 RNA structure before and after the pRNA-induced structural rearrangement (according to ref. 54). The three base exchanges are highlighted.
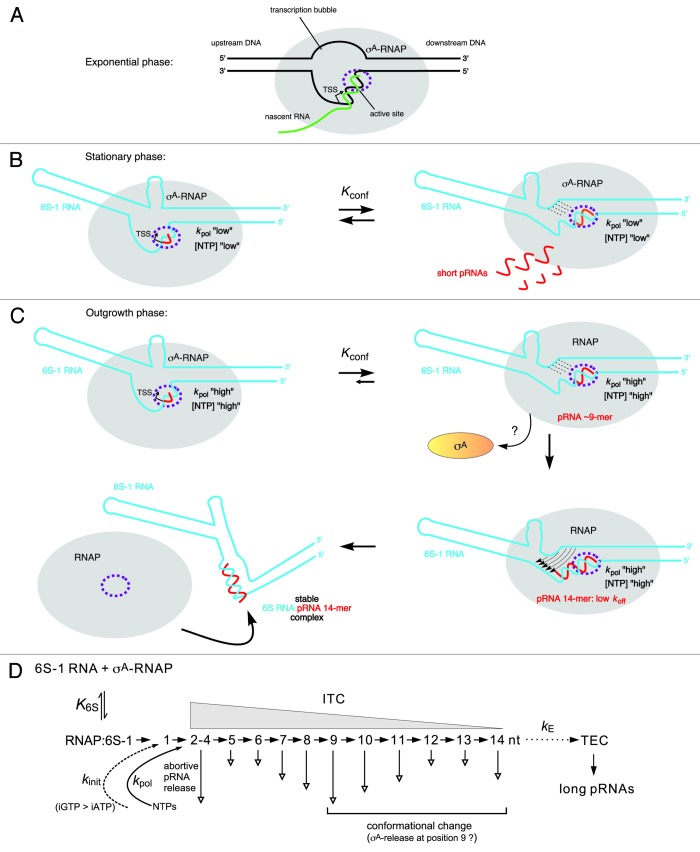
The first mechanistic model for pRNA synthesis and the structural 6S RNA rearrangement has been proposed for B. subtilis 6S-1 RNA.50 The details described in the following are illustrated in Figure 4, and the basic principles of the model (Fig. 4B and C) also pertain to the E. coli system. Two findings in a study combining deep sequencing (differential RNA sequencing = dRNA-seq) and biochemical analyses were the ignition sparks to put the model forward: dRNA-seq revealed very low 6S-1 pRNA levels in early exponential phase, substantial pRNA levels in stationary phase, and an apparent burst of pRNA synthesis under outgrowth conditions (3 min after late stationary cells had been diluted 1:5 in fresh LB medium). Moreover, under all conditions, shorter pRNAs (∼8 to 10-mers; length confirmed by dRNA-seq using poly(C) instead of poly(A) tailing; unpublished results) prevailed, but under outgrowth conditions, the fraction of longer pRNAs (∼14-mers) increased relative to stationary phase cells.5 These findings gave rise to the proposal that the B. subtilis housekeeping RNAP holoenzyme (σA-RNAP) synthesizes predominantly ≤ 9-mers on 6S-1 RNA in an idling cycle of abortive pRNA transcription (Fig. 4B), which does not lead to disruption of the 6S-1 RNA:RNAP complex. Only under outgrowth conditions when cells enter a new exponential growth phase and nutrients including NTPs are resupplied, longer 6S-1 pRNAs (∼14-mers) are increasingly synthesized to form stable pRNA:6S-1 RNA hybrids that persistently rearrange the 6S-1 RNA structure and induce dissociation of 6S-1:RNAP complexes (Fig. 4C).5,50 According to this model, several parameters play a role in this process: (1) kpol, the rate constant for pRNA elongation, which may have a specific value for every nucleotide addition step (nt +2, +3, +4 etc.); (2) koff, the rate constant for pRNA dissociation, which is high for short pRNAs and decreases with increasing pRNA length; (3) Kconf, (Fig. 4B and C) describing the conformational equilibrium of 6S-1 RNA between its ground state and the pRNA-induced rearranged structure triggering RNAP release. Based on the recent finding that the overall rate of pRNA synthesis by B. subtilis σA-RNAP (but not E. coli σ70-RNAP) is strongly influenced by the identity of the initiating nucleotide (iNTP), favorably GTP,49 a rate constant kinit for pRNA initiation may be invoked as well.52 It is proposed that an interplay of kinit, kpol, koff, and Kconfdecides whether RNAP synthesizes short pRNAs that dissociate from 6S-1 RNA before being able to stably rearrange the RNA’s structure (“the idling cycle of abortive pRNA transcription”) or whether the enzyme succeeds in synthesizing longer pRNAs (∼14-mers) that stably rearrange the 6S-1 RNA structure to terminate the idling cycle of abortive 6S-1 pRNA transcription by releasing the enzyme from the 6S-1 RNA-mediated block (Fig. 4D). The prevalence of 6S-1 RNA 8 to 10-mers and ∼14-mers seen in vivo by dRNA-seq5 and also in vitro5,50 may be related to a sequence idiosyncrasy of 6S-1 pRNAs which carry four A residues in a row from position 9 to 12 (5′-GUU CGG UCA AAA CU), where RNAP may stutter owing to transient depletion of the local ATP concentration. This leads to an interesting commonality between E. coli 6S RNA and B. subtilis 6S-1 RNA: the former encodes four consecutive G residues at positions 10–13 (5′-AUC GGC UCA GGG G) and the latter four A residues at positions 9–12, and both give rise to similar pRNA length species (9- and 13-mers in E. coli, 8 to 10-mers and 14-mers in B. subtilis). Future studies will address the assumed relevance of these homotetranucleotide stretches for processivity of pRNA synthesis. If one combines the findings obtained for B. subtilis 6S-1 RNA and E. coli 6S RNA and assumes that B. subtilis σA-RNAP also traverses an intermediate S3 state (Fig. 4C, top right), then the finding that 8 to 10-mers represent the most abundant 6S-1 pRNA length species in vivo5 may suggest that in the S3 state (after σA dissociation triggered by pRNA 8 to 10-mer synthesis), a large fraction of pRNA 8 to 10-mers dissociate from RNAP before their elongation by core RNAP, allowing σA to rebind to core RNAP to start a new pRNA synthesis cycle. If such a back-and-forth binding of σA indeed occurs needs to be demonstrated. If yes, the process of pRNA-induced RNAP release may be further influenced by the availability of σA in addition to NTP availability.
Figure 4. Model of the pRNA length-controlled structural rearrangement of B. subtilis 6S-1 RNA and its release from σA-RNAP, including mechanistic components inferred from the E. coli system. (A) In exponential phase, cellular 6S-1 RNA concentrations are low, only small amounts of pRNAs are synthesized and σA-RNAP is predominantly involved in gene transcription. The sketch illustrates a complex of RNAP and open promoter DNA; the latter is thought to be mimicked by 6S RNA.3 (B) 6S-1 RNA levels raise in late exponential and stationary phase and a larger fraction of σA-RNAP is sequestered in complexes with 6S-1 RNA. In late stationary phase, nutrients including NTPs are scarce and abortive transcripts/short pRNAs dissociate instead of being elongated to longer pRNAs because the rate constant for nucleotide addition is relatively slow (kpol “low”). Here, 6S-1 RNAs begin to transiently rearrange their structure (right panel), but fall back to the ground state (left panel) owing to pRNA dissociation; it is predicted that 6S-1 RNA molecules oscillate between their ground state and the transiently rearranged structure; if this also involves σA release and rebinding to RNAP (depicted in panel C) as suggested for the E. coli system upon pRNA 9-mer synthesis, remains to be seen. (C) In a newly initiated exponential growth phase (outgrowth), nutrients are resupplied and NTP levels increase. As a consequence, kpol increases (kpol “high”) and the fraction of longer pRNAs rises (12 to 14-mers), which stably rearrange 6S-1 RNA structure and trigger release of RNAP. An increase in the fraction of longer pRNA ~14-mers during outgrowth in vivo has been detected by dRNA-seq.5 Since the sigma factor dissociates from RNAP upon synthesis of a pRNA 9-mer in the E. coli system, we have tentatively included this mechanistic element also for the B. subtilis system. An open question is if the 6S-1 RNA rearrangement is already complete after synthesis of a pRNA 9-mer (as in the E. coli system) or incomplete until a stable RNAP:pRNA ~14-mer complex is formed. In the first three sketches, the hairpin in the 3′-CB is shown with a curved stem to indicate its equilibrium with an open structure. The two helical arms are shown as slightly bent to consider that formation of the hairpin constricts this side of the central bulge. In the top sketch on the right, stippled lines depict the progressive disruption of helix P2; thin arrows in the bottom sketch on the right illustrate formation of the central bulge collapse helix (see Fig. 1 B). The fully rearranged 6S-1 RNA structure is depicted in the bottom sketch on the left; the orientation of the helical elements is not known and thus arbitrary. (D) Kinetic scheme for pRNA synthesis in B. subtilis, adapted from abortive transcription initiation at DNA promoters.50,73 Ground state binding of 6S-1 RNA to σA-RNAP is governed by the equilibrium constant K6S. Initiation of pRNA synthesis depends on the rate constant for initiation, kinit, which has been shown to be faster with GTP than ATP in B. subtilis, but not in E. coli.49 With pRNA polymerization, the RNAP:6S-1 RNA complex acts as an initial transcribing complex (ITC) releasing abortive transcripts of various length (here mainly 2–14 nt) to different extents (indicated by the length of the vertical arrow; inferred from dRNA-seq data using poly(A)5 and poly(C) tailing (unpublished data) of cellular RNAs. Only pRNAs of ≥ 12 nt stably rearrange the 6S-1 RNA structure owing to their sufficiently low koff. The rate constant kE describes the transition from an ITC to a productive transcription elongation complex (TEC) and may be of limited relevance to pRNA transcription in vivo.
Whereas much is known about 6S RNA rearrangements on the level of secondary structure, essentially nothing is known about the changes on the level of tertiary structure including the molecular details of how RNAP is released from 6S RNA sequestration. For B. subtilis one may speculate that 6S-1 RNA:pRNA hybrids form a rigid “V-like” tertiary structure after the central bulge collapse, thereby disrupting the interactions to RNAP. As E. coli 6S RNA:pRNA hybrids form an extended hairpin in the 3′-CB but no CB collapse helix, their tertiary structure as well as the molecular details of RNAP release may to some extent differ in comparison with B. subtilis 6S-1 RNA. As a commonality, both 6S RNA systems are likely to lose their malleable rod shape upon rearrangement to adopt a more rigid overall structure incompatible with RNAP binding. Unravelling the three-dimensional nature of 6S RNA rearrangements will contribute to a more profound understanding of 6S RNA release processes.
A B. subtilis 6S-1 RNA mutant with a destabilized hairpin in the 3′-CB (Fig. 3B) showed evidence of increased conformational flexibility, had a two- to 3-fold lower affinity for σA-RNAP and gave rise to a pRNA length pattern shifted to longer transcripts relative to its wild type counterpart.50 The latter finding resembles what has been observed with the aforementioned E. coli 6S_S RNA mutant unable to form a stable extended hairpin in the 3′-CB (Fig. 3A). Evidently, when rearranged 6S RNA structures are made less rigid and distinct by mutations, longer pRNAs and more runoff transcripts are synthesized and required to displace the more malleable mutant 6S RNAs from RNAP.
B. subtilis 6S-2 RNA—The Paralog of 6S-1 RNA
Unlike E. coli and the majority of bacteria, a group of Firmicutes including B. subtilis express two 6S RNAs, termed 6S-1 and 6S-2 RNA (genes bsrA and bsrB) in B. subtilis.7,17,53 6S-2 RNA was observed to reach its highest levels between early and mid-exponential growth phase, with expression levels decreasing toward extended stationary phase; in contrast, 6S-1 RNA reaches its highest level in the late exponential and stationary phase.3,5,17 Somewhat deviating from the just mentioned 6S-2 RNA expression profile, 6S-2 RNA levels were also reported to be rather constant at all growth stages.7,19 We recently observed strain-specific differences in 6S-2 RNA expression profiles (unpublished data), which could explain the reported inconsistencies.
On the basis of similarities in function and expression profile, B. subtilis 6S-1 RNA is considered to be the functional homolog of E. coli 6S RNA. A 6S-1 RNA gene knockout in the B. subtilis strain 168 background was found to cause a delayed outgrowth phenotype (retardation of a new exponential phase after diluting stationary phase cells in fresh medium)49 and led to a faster exhaustion of nutrients, such that cells began to sporulate earlier.45 The latter phenotype indicates that 6S-1 RNA contributes to an economical utilization of nutrients and metabolites, similar to what has been found for E. coli 6S RNA (see above).
First evidence prompting the classification of 6S-2 RNA as a genuine 6S RNA came from the RNA’s co-immunoprecipitation with the σA housekeeping RNAP.7 Using 6S-2 RNAs from other Firmicutes, a consensus secondary structure was predicted that fulfills the criteria of bona fide 6S RNAs, i.e., a central, largely single-stranded internal loop region flanked by imperfect helical arms.7,52 σA-RNAP initiates pRNA synthesis on 6S-2 RNA in vitro in the 5′-CB as expected for a canonical 6S RNA.3,5,52 However, pRNA synthesis on B. subtilis 6S-2 RNA is initiated with ATP instead of GTP as in the case of 6S-1 RNA.5,52 In comparison to 6S-1 RNA, in vitro pRNA synthesis from 6S-2 RNA results in a broader pRNA length spectrum (13 to 16-mers vs. the 6S-1 pRNA 14-mer) and is less effective at lower NTP concentrations (e.g., 20 or 50 µM of each NTP).49,52 It was recently shown that B. subtilis σA-RNAP favors the synthesis of pRNAs that are initiated with GTP, explaining why pRNA synthesis from 6S-1 RNA is usually more effective than that from 6S-2 RNA starting with ATP.49 Consistently, mutating the first pRNA template nucleotide in the chromosomal gene of 6S-1 RNA from C to U, giving rise to 6S-1 pRNAs initiated with A instead of G, reduced the levels of 6S-1 pRNA synthesis in vivo.5 A recent in vitro study compared the functional properties of B. subtilis 6S-1 and 6S-2 RNAs in much detail. This revealed that (1) both 6S RNAs bind with similar affinity to RNAP and (2) inhibit in vitro transcription from DNA promoters with comparable efficacy, and (3), as with 6S-1 RNA, pRNA synthesis on 6S-2 RNA rearranges the RNA’s structure to induce dissociation of RNAP:6S-2 RNA complexes.52 Despite these commonalities, differences between 6S-1 and 6S-2 RNA are evident as well. Beyond the unequal identity of the iNTP, the major pRNA species derived from the two 6S RNAs differ in their G,C-content (6S-2 pRNA 13 to 16-mers: 3 × G,C; 6S-1 pRNA 14-mer: 6 × G,C). This could explain why a 6S-2 pRNA with a minimum length of 20 nt was required to form 6S-2 RNA:pRNA hybrid structures that underwent only negligible dissociation within the experimental time frame. A comparable stability of 6S-1 pRNA complexes was already achieved with a pRNA 14-mer.52 This difference in 6S RNA:pRNA hybrid stability might have the effect that nascent 6S-2 pRNAs more frequently dissociate from the 6S-2 RNA template before being elongated to longer pRNAs, which may impede the escape of RNAP from the sequestration by 6S-2 RNA. Furthermore, also after dissociation from RNAP, 6S-2 pRNAs may more rapidly dissociate from 6S-2 RNA than pRNAs of equal length dissociate from 6S-1 RNA, thus accelerating pRNA decay or favoring rebinding of 6S-2 RNA to a new RNAP molecule.
Up to now, evidence for 6S-2 RNA-derived pRNA synthesis in vivo is lacking. Whereas 6S-1 pRNAs could be detected by dRNA-seq and northern blot experiments in B. subtilis cells grown under standard conditions, none of the techniques so far detected significant amounts of pRNAs derived from 6S-2 RNA.5,19 The in vivo role of 6S-2 RNA was addressed by analyzing the growth phenotypes of B. subtilis 6S RNA-knockout strains.49 Whereas the above mentioned 6S-1-knockout strain showed a delayed outgrowth phenotype, mutant strains with a 6S-2-knockout or a 6S-1/6S-2 double knockout displayed normal growth phenotypes. A wild-type-like growth phenotype of the Δ6S-1 strain could be restored by expressing 6S-1 RNA from a complementation plasmid or by further deleting the 6S-2 RNA gene.49 A possible interpretation of these findings is that 6S-2 RNA inhibits RNAP and 6S-1 RNA somehow helps overcome or prevents the block of RNAP by 6S-2 RNA.52 These findings, that is the absence of evidence for 6S-2 pRNA synthesis in vivo5,19 and the lower efficiency of 6S-2 vs. 6S-1 pRNA synthesis in vitro, particularly at lower NTP concentrations and owing to a less favorable iATP, prompted the Wassarman group to posit that pRNA synthesis from 6S-2 RNA may not occur in vivo at all.49 If this turned out to be correct, then one could hypothesize that the 6S-2 RNA paralog lost its original biological function only very recently on evolutionary time scales, as the RNA still displays the basic mechanistic capabilities of 6S RNAs in vitro.52 This has raised the question how the 6S-2 RNA-mediated blockade of RNAP may be lifted in vivo, if not by pRNA synthesis in cis. A simple displacement of 6S-2 RNA by excess amounts of 6S-1 RNA in the absence of transcription could be experimentally excluded, as could be the possibility that σA-RNAP:6S-2 RNA complexes able to synthesize very short 6S-2 pRNA (5 to 6-mers) may provide sufficient dynamics to replace 6S-2 RNA with 6S-1 RNA.52
At present, we advocate that the issue if pRNA synthesis from 6S-2 RNA may occur in vivo at some stage and to an extent that allows RNAP molecules to escape from the 6S-2 RNA block should be treated as unsettled. Although several lines of evidence (the phenotypes of the just mentioned 6S RNA knockout strains; no detection of 6S-2 pRNAs by northern blots and RNA-seq hitherto; an unfavorable iATP) argue against 6S-2 pRNA synthesis in vivo, there is still a possibility that 6S-2 pRNAs escaped in vivo detection owing to (1) their rapid decay, (2) use of northern probes of insufficient specificity (note that a 6S-2 pRNA 15-mer, 5′-AAA GGU UAA AAC UUA, carries stretches of three and four A residues), or (3) unknown technical reasons that have prevented their inclusion in RNA-seq libraries. Also, low efficiency of 6S-2 pRNA synthesis at 20–50 μM each NTP in vitro is not too informative as long as the intracellular NTP availability is not firmly defined. In an early study, the ATP concentration was determined to be 3 to 4-fold higher than that of the other three NTPs.54 Combined with a recent study reporting an ATP concentration of ~60 μM in exponentially growing B. subtilis cells,55 one may infer a GTP concentration of 15–20 μM. In contrast, 1–3 mM GTP were determined for B. subtilis in another analysis.56 In the E. coli system, ~4–10 mM ATP and ~1–5 mM GTP were measured in exponentially growing cells.57 In view of these conflicting reports, the intracellular concentration of NTPs available for transcription may well be in the lower mM rather than in the μM range.
Noticeable Cases of 6S RNAs in Other Bacteria
The 6S RNA of the hyperthermophilic bacterium A.aeolicus (growth temperature: optimum 85 °C, maximum 95 °C) was identified in an experimental RNomics study as the most abundant ncRNA in this bacterium apart from rRNAs and tRNAs.58 This ~160-nt long 6S RNA variant is at the lower length limit among 6S RNAs that usually have sizes close to 200 nt. A. aeolicus 6S RNA is predicted to have one of the most stable structures among known 6S RNAs, with a ΔG of -96 kcal/mol predicted by RNAfold under standard conditions for its minimal free energy (MFE), rod-shaped structure. For comparison, MFE structures of -80 kcal/mol are predicted for E. coli 6S RNA (184 nt) and -63 kcal/mol for B. subtilis 6S-1 RNA (190 nt). Synthesis of pRNAs has been demonstrated (unpublished results), indicating that the RNA functions as a genuine 6S RNA in the hyperthermophilic host. Owing to its rigid structure rich in G-C base pairs, a shortened version of the RNA was subjected to crystallization screens. A 12-bp fragment of its terminal stem region, derived from RNA hydrolysis within crystallization droplets, yielded resolvable crystals diffracting at 2.6 Å. The X-ray structure revealed a regular A-form duplex containing three G°U wobble-type base pairs, one of which was involved in intermolecular contacts through a ribose-zipper motif at the crystal-packing interface.59 The case of A. aeolicus 6S RNA is remarkable, as it demonstrates that this regulatory process is able to operate under extreme temperature conditions. It is also astounding that A. aeolicus has maintained a 6S RNA riboregulator despite the extreme reduction of its genome to 1.6 Mbp, in turn, emphasizing the important role of 6S RNA in regulating the dynamics of bacterial transcriptomes.
Another notable 6S RNA variant is expressed in the human pathogen Helicobacter pylori, which also possesses a small genome (~1.7 Mbp) and which has a particularly low genomic G,C-content of ~33%. This 6S RNA variant has a length of 181 nt with a ΔG of merely -52 kcal/mol for the predicted MFE structure.15 dRNA-seq identified two types of 6S RNA-templated pRNA transcripts, 12 to 13-meric canonical pRNAs initiated in the 5′-CB, and pRNA* 17-mers initiated in the 3′-CB.15 This finding indicated that H. pylori 6S RNA can bind to the cognate RNAP in reverse orientations, giving rise to the two types of pRNA transcripts. Interestingly, duplex formation of both pRNA types with 6S RNA favors formation of the same type of extended hairpin in the 3′-CB50 as seen for E. coli 6S RNA (see above).21,47 It remains to be seen whether the miscellaneous RNAP binding mode of H. pylori 6S RNA reflects a functional diversification or an impreciseness without functional consequences. A possibility is that 6S RNA:pRNA complexes enter a decay pathway different from that of 6S RNA:pRNA* complexes.
Other Potential Interaction Partners of 6S RNA and RNAP
Interesting future issues will include the question as to how the 6S RNA:RNAP regulatory system is affected or modulated by other interaction partners and how such interactions are intertwined with global RNA metabolism and other pathways regulating RNAP activity. Additional binding partners of 6S RNA as well as RNAP have indeed been identified in E. coli. The group of Renée Schroeder isolated and identified proteins binding to 6S RNA combining affinity chromatography of aptamer-tagged 6S RNA and mass spectrometry.60 Beyond the β-subunit of RNAP, the study identified Hfq, ribosomal protein S1, EF-Tu, polynucleotide phosphorylase (PNPase), poly(A) polymerase I (PAP I), RNase III, and the DNA-binding protein HU-α (HU-2) as potential interaction partners of 6S RNA. Co-purification of protein S1 and EF-Tu likely occurred indirectly via protein–protein interactions involving Hfq and/or RNAP.60 Remarkably, 6S RNA bound to Hfq with affinity in the 20 nM range, with evidence for more than one 6S RNA molecule binding to an Hfq hexamer. Interaction of 6S RNA with Hfq, PAP I, PNPase, and RNase III may be relevant for 6S RNA decay, as all four proteins are part of an RNA degradation and processing network in E. coli. The components of this network are organized into supramolecular structures, which appear as extended, cytoplasmic membrane-associated assemblies that coil around the periphery of the cell.61,62 6S RNA interaction with Hfq might also play a role in mediating or modulating the intracellular access of RNAP to 6S RNA, as in stationary phase E. coli cells Hfq was not only detected in close proximity to the inner membrane, but also in the cytoplasm and at the nucleoid.61,63 It is also conceivable that RNAPs transcribing 6S RNA genes are able to directly bind their transcription products, either with or without the help of Hfq.
Regarding RNAP binding to other RNAs, E. coli RNAP was demonstrated to 3′-extend RyhB RNA and an RNA consisting of the 5′-terminal 50 nt of MicF RNA. Full-length MicF RNA bound to RNAP with high affinity, and appeared to be partially degraded by E. coli RNAP, likely via the enzyme’s described endonucleolytic RNA cleavage activity.64 Finally, five RNAs from an E. coli genomic library co-immunoprecipitated with RNAP.60 As RyhB and MicF are stress-induced sRNAs, their interaction with RNAP may reflect a transcriptional control pathway associated with the respective stress responses.
A View Beyond Bacteria
Another interesting (non-bacterial) riboregulator is the ~180-nt B2 RNA, which inhibits mammalian RNA polymerase II (Pol II) in response to heat shock and other cellular stresses.65,66 A specific region of B2 RNA aligns the RNA’s own 3′-end for self-templated addition of 18 nt by Pol II, a process that triggers dissociation of inactive ternary Pol II:DNA promoter:B2 RNA complexes, further requiring a so-far-unknown cellular factor. Despite the similarity to the mode of 6S RNA action, a major mechanistic difference of B2 RNA relative to 6S RNAs is the alteration of B2 RNA’s primary structure instead of synthesis of de novo transcripts. In conclusion, regulations of RNAPs exploiting their RdRP acitivities may be more widespread than as yet evident.
Concluding Remarks
In E. coli, 6S RNA knockouts combined with transcriptome analyses have provided evidence that the RNA plays a role in long-term cell survival, is interdigitated with other regulatory pathways that control transcription in response to stresses, and takes part in cellular networks regulating metabolic functions. For B. subtilis it has been shown that a 6S-1 RNA knockout leads to faster turnover of nutrients, such that cells begin to sporulate earlier than those of the parental strain under conditions of limited nutrients.45 Beyond these phenotypes averaged over the entire bacterial population, an interesting aspect of future studies will be the question if 6S RNAs help to increase transcriptome adaptivity, in the sense that bacterial subpopulations can better adapt to new conditions, for example, to access new locations of nutrient sources. Here, the intensely studied field of bacterial motility comes into play. Motility includes swimming, swarming, and twitching of bacteria, involving a variety of genes that encode and regulate the different motility machineries.67,68 It has been observed for E. coli69 as well as for B. subtilis70 that a cell population can often be divided into two subpopulations, (1) sessile cells that essentially remain at one location (“waiting for better times”) and (2) a subpopulation of swimming cells that explore new habitats for better conditions (“being at risk to encounter even worse conditions”). Simultaneous implementation of both survival strategies increases the chance that at least some cells survive and serve as germ cells for a successor population. With respect to the finding that 6S RNA functions in economizing on nutrients, one may speculate that cellular motility programs may also be regulated by 6S RNA, considering the importance of such programs for population survival. However, it should be noted that laboratory strains, such as B. subtilis 168 and PY79, are defective in flagellum and biosurfactant formation and thus swarming, necessitating the use of wild-type strains (e.g., strain 3610) for corresponding investigations.70-72 Notably, deletion of σ38 (gene rpoS), the sigma factor that accumulates in E. coli cells upon nutrient deprivation, stress, or entry into stationary phase, results in an increase in the subpopulation of cells with increased motility.69
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft, SPP 1258 and IRTG 1384.
References
- 1.Brownlee GG. Sequence of 6S RNA of E. coli. Nat New Biol. 1971;229:147–9. doi: 10.1038/newbio229147a0. [DOI] [PubMed] [Google Scholar]
- 2.Hindley J. Fractionation of 32P-labelled ribonucleic acids on polyacrylamide gels and their characterization by fingerprinting. J Mol Biol. 1967;30:125–36. doi: 10.1016/0022-2836(67)90248-3. [DOI] [PubMed] [Google Scholar]
- 3.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–84. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axmann IM, Holtzendorff J, Voss B, Kensche P, Hess WR. Two distinct types of 6S RNA in Prochlorococcus. Gene. 2007;406:69–78. doi: 10.1016/j.gene.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Beckmann BM, Burenina OY, Hoch PG, Kubareva EA, Sharma CM, Hartmann RK. In vivo and in vitro analysis of 6S RNA-templated short transcripts in Bacillus subtilis. RNA Biol. 2011;8:839–49. doi: 10.4161/rna.8.5.16151. [DOI] [PubMed] [Google Scholar]
- 6.Weissenmayer BA, Prendergast JG, Lohan AJ, Loftus BJ. Sequencing illustrates the transcriptional response of Legionella pneumophila during infection and identifies seventy novel small non-coding RNAs. PLoS One. 2011;6:e17570. doi: 10.1371/journal.pone.0017570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trotochaud AE, Wassarman KM. A highly conserved 6S RNA structure is required for regulation of transcription. Nat Struct Mol Biol. 2005;12:313–9. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- 8.Lee SY, Bailey SC, Apirion D. Small stable RNAs from Escherichia coli: evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J Bacteriol. 1978;133:1015–23. doi: 10.1128/jb.133.2.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–23. doi: 10.1016/S0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 10.Cavanagh AT, Klocko AD, Liu X, Wassarman KM. Promoter specificity for 6S RNA regulation of transcription is determined by core promoter sequences and competition for region 4.2 of sigma70. Mol Microbiol. 2008;67:1242–56. doi: 10.1111/j.1365-2958.2008.06117.x. [DOI] [PubMed] [Google Scholar]
- 11.Gildehaus N, Neusser T, Wurm R, Wagner R. Studies on the function of the riboregulator 6S RNA from E. coli: RNA polymerase binding, inhibition of in vitro transcription and synthesis of RNA-directed de novo transcripts. Nucleic Acids Res. 2007;35:1885–96. doi: 10.1093/nar/gkm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neusser T, Polen T, Geissen R, Wagner R. Depletion of the non-coding regulatory 6S RNA in E. coli causes a surprising reduction in the expression of the translation machinery. BMC Genomics. 2010;11:165. doi: 10.1186/1471-2164-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J Bacteriol. 2004;186:4978–85. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faucher SP, Friedlander G, Livny J, Margalit H, Shuman HA. Legionella pneumophila 6S RNA optimizes intracellular multiplication. Proc Natl Acad Sci U S A. 2010;107:7533–8. doi: 10.1073/pnas.0911764107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe T, Sugiura M, Sugita M. A novel small stable RNA, 6Sa RNA, from the cyanobacterium Synechococcus sp. strain PCC6301. FEBS Lett. 1997;416:302–6. doi: 10.1016/S0014-5793(97)01237-4. [DOI] [PubMed] [Google Scholar]
- 17.Ando Y, Asari S, Suzuma S, Yamane K, Nakamura K. Expression of a small RNA, BS203 RNA, from the yocI-yocJ intergenic region of Bacillus subtilis genome. FEMS Microbiol Lett. 2002;207:29–33. doi: 10.1111/j.1574-6968.2002.tb11023.x. [DOI] [PubMed] [Google Scholar]
- 18.Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–3. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- 19.Cavanagh AT, Sperger JM, Wassarman KM. Regulation of 6S RNA by pRNA synthesis is required for efficient recovery from stationary phase in E. coli and B. subtilis. Nucleic Acids Res. 2012;40:2234–46. doi: 10.1093/nar/gkr1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rediger A, Geissen R, Steuten B, Heilmann B, Wagner R, Axmann IM. 6S RNA - an old issue became blue-green. Microbiology. 2012;158:2480–91. doi: 10.1099/mic.0.058958-0. [DOI] [PubMed] [Google Scholar]
- 21.Panchapakesan SS, Unrau PJE. E. coli 6S RNA release from RNA polymerase requires σ70 ejection by scrunching and is orchestrated by a conserved RNA hairpin. RNA. 2012;18:2251–9. doi: 10.1261/rna.034785.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wurm R, Neusser T, Wagner R. 6S RNA-dependent inhibition of RNA polymerase is released by RNA-dependent synthesis of small de novo products. Biol Chem. 2010;391:187–96. doi: 10.1515/bc.2010.018. [DOI] [PubMed] [Google Scholar]
- 23.Hsu LM, Zagorski J, Wang Z, Fournier MJ. Escherichia coli 6S RNA gene is part of a dual-function transcription unit. J Bacteriol. 1985;161:1162–70. doi: 10.1128/jb.161.3.1162-1170.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanguenin L, Lara-Núñez A, Pribat A, Mageroy MH, Gregory JF, 3rd, Rice KC, de Crécy-Lagard V, Hanson AD. Moonlighting glutamate formiminotransferases can functionally replace 5-formyltetrahydrofolate cycloligase. J Biol Chem. 2010;285:41557–66. doi: 10.1074/jbc.M110.190504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen S, Lewis K, Vulić M. Role of global regulators and nucleotide metabolism in antibiotic tolerance in Escherichia coli. Antimicrob Agents Chemother. 2008;52:2718–26. doi: 10.1128/AAC.00144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64:515–24. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 27.Kim KS, Lee Y. Regulation of 6S RNA biogenesis by switching utilization of both sigma factors and endoribonucleases. Nucleic Acids Res. 2004;32:6057–68. doi: 10.1093/nar/gkh939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Willkomm DK, Hartmann RK. 6S RNA - an ancient regulator of bacterial RNA polymerase rediscovered. Biol Chem. 2005;386:1273–7. doi: 10.1515/BC.2005.144. [DOI] [PubMed] [Google Scholar]
- 29.Lee JY, Park H, Bak G, Kim KS, Lee Y. Regulation of transcription from two ssrS promoters in 6S RNA biogenesis. Mol Cells. 2013;36:227–34. doi: 10.1007/s10059-013-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neusser T, Gildehaus N, Wurm R, Wagner R. Studies on the expression of 6S RNA from E. coli: involvement of regulators important for stress and growth adaptation. Biol Chem. 2008;389:285–97. doi: 10.1515/BC.2008.023. [DOI] [PubMed] [Google Scholar]
- 31.Chae H, Han K, Kim KS, Park H, Lee J, Lee Y. Rho-dependent termination of ssrS (6S RNA) transcription in Escherichia coli: implication for 3′ processing of 6S RNA and expression of downstream ygfA (putative 5-formyl-tetrahydrofolate cyclo-ligase) J Biol Chem. 2011;286:114–22. doi: 10.1074/jbc.M110.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klocko AD, Wassarman KM. 6S RNA binding to Esigma(70) requires a positively charged surface of sigma(70) region 4.2. Mol Microbiol. 2009;73:152–64. doi: 10.1111/j.1365-2958.2009.06758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meares CF, Datwyler SA, Schmidt BD, Owens J, Ishihama A. Principles and methods of affinity cleavage in studying transcription. Methods Enzymol. 2003;371:82–106. doi: 10.1016/S0076-6879(03)71006-4. [DOI] [PubMed] [Google Scholar]
- 34.Owens JT, Chmura AJ, Murakami K, Fujita N, Ishihama A, Meares CF. Mapping the promoter DNA sites proximal to conserved regions of sigma 70 in an Escherichia coli RNA polymerase-lacUV5 open promoter complex. Biochemistry. 1998;37:7670–5. doi: 10.1021/bi980188n. [DOI] [PubMed] [Google Scholar]
- 35.Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase σ70 holoenzyme. J Biol Chem. 2013;288:9126–34. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steuten B, Setny P, Zacharias M, Wagner R. Mapping the spatial neighborhood of the regulatory 6S RNA bound to Escherichia coli RNA polymerase holoenzyme. J Mol Biol. 2013;425:3649–61. doi: 10.1016/j.jmb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–80. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shephard L, Dobson N, Unrau PJ. Binding and release of the 6S transcriptional control RNA. RNA. 2010;16:885–92. doi: 10.1261/rna.2036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weeks KM, Crothers DM. Major groove accessibility of RNA. Science. 1993;261:1574–7. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- 40.Cavanagh AT, Chandrangsu P, Wassarman KM. 6S RNA regulation of relA alters ppGpp levels in early stationary phase. Microbiology. 2010;156:3791–800. doi: 10.1099/mic.0.043992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geissen R, Steuten B, Polen T, Wagner R. E. coli 6S RNA: a universal transcriptional regulator within the centre of growth adaptation. RNA Biol. 2010;7:564–8. doi: 10.4161/rna.7.5.12969. [DOI] [PubMed] [Google Scholar]
- 42.Trotochaud AE, Wassarman KM. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J Bacteriol. 2006;188:3936–43. doi: 10.1128/JB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–61. doi: 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 44.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–57. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 45.Cavanagh AT, Wassarman KM. 6S-1 RNA function leads to a delay in sporulation in Bacillus subtilis. J Bacteriol. 2013;195:2079–86. doi: 10.1128/JB.00050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steuten B, Schneider S, Wagner R. 6S RNA: recent answers - future questions. Mol Microbiol. 2014;91:641–8. doi: 10.1111/mmi.12484. [DOI] [PubMed] [Google Scholar]
- 47.Steuten B, Wagner R. A conformational switch is responsible for the reversal of the 6S RNA-dependent RNA polymerase inhibition in Escherichia coli. Biol Chem. 2012;393:1513–22. doi: 10.1515/hsz-2012-0237. [DOI] [PubMed] [Google Scholar]
- 48.Kugel JF, Goodrich JA. An RNA transcriptional regulator templates its own regulatory RNA. Nat Chem Biol. 2007;3:89–90. doi: 10.1038/nchembio0207-89. [DOI] [PubMed] [Google Scholar]
- 49.Cabrera-Ostertag IJ, Cavanagh AT, Wassarman KM. Initiating nucleotide identity determines efficiency of RNA synthesis from 6S RNA templates in Bacillus subtilis but not Escherichia coli. Nucleic Acids Res. 2013;41:7501–11. doi: 10.1093/nar/gkt517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beckmann BM, Hoch PG, Marz M, Willkomm DK, Salas M, Hartmann RK. A pRNA-induced structural rearrangement triggers 6S-1 RNA release from RNA polymerase in Bacillus subtilis. EMBO J. 2012;31:1727–38. doi: 10.1038/emboj.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burenina OY, Hoch PG, Damm K, Salas M, Zatsepin TS, Lechner M, Oretskaya TS, Kubareva EA, Hartmann RK. Mechanistic comparison of Bacillus subtilis 6S-1 and 6S-2 RNAs--commonalities and differences. RNA. 2014;20:348–59. doi: 10.1261/rna.042077.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuma S, Asari S, Bunai K, Yoshino K, Ando Y, Kakeshita H, Fujita M, Nakamura K, Yamane K. Identification and characterization of novel small RNAs in the aspS-yrvM intergenic region of the Bacillus subtilis genome. Microbiology. 2002;148:2591–8. doi: 10.1099/00221287-148-8-2591. [DOI] [PubMed] [Google Scholar]
- 54.Lopez JM, Marks CL, Freese E. The decrease of guanine nucleotides initiates sporulation of Bacillus subtilis. Biochim Biophys Acta. 1979;587:238–52. doi: 10.1016/0304-4165(79)90357-X. [DOI] [PubMed] [Google Scholar]
- 55.Meyer FM, Jules M, Mehne FM, Le Coq D, Landmann JJ, Görke B, Aymerich S, Stülke J. Malate-mediated carbon catabolite repression in Bacillus subtilis involves the HPrK/CcpA pathway. J Bacteriol. 2011;193:6939–49. doi: 10.1128/JB.06197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–9. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willkomm DK, Minnerup J, Hüttenhofer A, Hartmann RK. Experimental RNomics in Aquifex aeolicus: identification of small non-coding RNAs and the putative 6S RNA homolog. Nucleic Acids Res. 2005;33:1949–60. doi: 10.1093/nar/gki334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo J, Dock-Bregeon AC, Willkomm DK, Hartmann RK, Westhof E. Structure of an A-form RNA duplex obtained by degradation of 6S RNA in a crystallization droplet. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:634–9. doi: 10.1107/S1744309113013018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Windbichler N, von Pelchrzim F, Mayer O, Csaszar E, Schroeder R. Isolation of small RNA-binding proteins from E. coli: evidence for frequent interaction of RNAs with RNA polymerase. RNA Biol. 2008;5:30–40. doi: 10.4161/rna.5.1.5694. [DOI] [PubMed] [Google Scholar]
- 61.Hoch PG, Hartmann RK. Supramolecular membrane-associated assemblies of RNA metabolic proteins in Escherichia coli. Biochem J. 2014;458:e1–3. doi: 10.1042/BJ20131676. [DOI] [PubMed] [Google Scholar]
- 62.Taghbalout A, Yang Q, Arluison V. The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem J. 2014;458:11–22. doi: 10.1042/BJ20131287. [DOI] [PubMed] [Google Scholar]
- 63.Diestra E, Cayrol B, Arluison V, Risco C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS One. 2009;4:e8301. doi: 10.1371/journal.pone.0008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surratt CK, Milan SC, Chamberlin MJ. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991;88:7983–7. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–9. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 66.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, Goodrich JA. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013;32:781–90. doi: 10.1038/emboj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–76. doi: 10.1038/nrmicro1900. [DOI] [PubMed] [Google Scholar]
- 68.Kearns DB. A field guide to bacterial swarming motility. Nat Rev Microbiol. 2010;8:634–44. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ojima Y, Hakamada K, Nishinoue Y, Nguyen MH, Miyake J, Taya M. Motility behavior of rpoS-deficient Escherichia coli analyzed by individual cell tracking. J Biosci Bioeng. 2012;114:652–6. doi: 10.1016/j.jbiosc.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 70.Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–94. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ghelardi E, Salvetti S, Ceragioli M, Gueye SA, Celandroni F, Senesi S. Contribution of surfactin and SwrA to flagellin expression, swimming, and surface motility in Bacillus subtilis. Appl Environ Microbiol. 2012;78:6540–4. doi: 10.1128/AEM.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol. 2004;52:357–69. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 73.Hsu LM. Monitoring abortive initiation. Methods. 2009;47:25–36. doi: 10.1016/j.ymeth.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]