Abstract
Cyanobacteria constitute a heterogeneous phylum of oxygen-producing, photosynthetic prokaryotes. They are susceptible to various stress conditions like heat, salt, or light stress, all inducing the cyanobacterial heat shock response (HSR). Cyanobacterial small heat shock proteins (sHsps) are known to preserve thylakoid membrane integrity under stress conditions, thereby protecting the photosynthesis machinery. In Synechocystis sp PCC 6803, synthesis of the sHsp Hsp17 is regulated by an RNA thermometer (RNAT) in the 5′-untranslated region (5′-UTR) of the hsp17 mRNA. RNATs are direct temperature sensors that control expression of many bacterial heat shock and virulence genes. They hinder translation at low temperatures by base pairing, thus blocking ribosome access to the mRNA.
To explore the temperature range in which RNATs act, we studied various RNAT candidates upstream of sHsp genes from mesophilic and thermophilic cyanobacteria. The mesophilic cyanobacteria Anabaena variabilis and Nostoc sp chromosomally encode two sHsps each. Reporter gene studies suggested RNAT-mediated post-transcriptional regulation of shsp expression in both organisms. Detailed structural analysis of the two A. variabilis candidates revealed two novel RNAT types. The first, avashort, regulates translation primarily by masking of the AUG translational start codon. The second, featuring an extended initial hairpin, thus named avalong, presumably makes use of complex tertiary interaction. The 5′-UTR of the small heat shock gene hspA in the thermophile Thermosynechococcus elongatus is predicted to adopt an extended secondary structure. Structure probing revealed that the ribosome binding site was blocked at temperatures below 55 °C. The results of this study demonstrate that cyanobacteria commonly use RNATs to control expression of their small heat shock genes.
Keywords: post-transcriptional control, heat shock response, small heat shock protein, RNA thermometer, cyanobacteria
Introduction
Cyanobacteria are present on earth for ~3.5 billion years1 and N2-fixing cyanobacteria are considered to be the progenitors of chloroplasts in higher plants.2 These oxygen-producing, photosynthetic microorganisms are responsible for half of the global CO2 fixation.3 They constitute a heterogeneous group of five orders (Chroococcales, Pleurocapsales, Oscillatoriales, Nostocales, and Stigonematales), including unicellular and filamentous species. Although most cyanobacteria prefer moderate growth temperatures, they are also found in extreme habitats.4 In their natural environment, cyanobacteria are frequently exposed to stress conditions, including temperature changes, variations in osmolarity, nutrient supply, or light intensity. Global analyses in the unicellular, mesophilic freshwater-cyanobacterium Synechocystis sp PCC 6803 (henceforth called Synechocystis) revealed the induction of heat shock gene expression under heat,5 hyperosmotic,6 salt,7 H2O2-,8 high light stress,9 and at increased pH.10 Heat shock genes encode chaperones, proteases, and small heat shock proteins (sHsps). Chaperones and proteases mediate refolding or degradation, respectively, of misfolded proteins typically in an ATP-dependent process. The third party in the heat shock response (HSR), the sHsps, are ATP-independent molecular chaperones that bind to misfolded proteins and maintain them in a folding-competent state.11 Deletion of sHsp genes tends to have a much more severe effect in cyanobacteria than in other microorganisms.12 In Synechocystis, deletion of hsp17 resulted in reduced oxygen production.13 Further, the content of chlorophyll a was low suggesting substantial defects in the photosynthetic apparatus.14
Photosystem II (PS II) is highly heat sensitive15 and chloroplast small heat shock proteins (cp-sHsps) are able to protect the cp-PS II from heat.16 Cyanobacterial sHsps can likewise prevent heat inactivation of PS II.17 Localization studies revealed the association of cyanobacterial sHsps with the thylakoid membranes and carboxysomes.18,19 Subsequent to heat stress, small heat shock proteins are distributed in the cytosol, but re-directed to the thylakoid membranes under prolonged heat stress.18 Following a heat shock, cyanobacterial phycobilisomes (PBS), thylakoid membrane-associated light-harvesting complexes, first dissociate and subsequently aggregate.20 Small heat shock proteins are capable of preventing PBS aggregation.20,21 Further, they can inhibit heat-induced photobleaching by direct interaction with phycocyanins.20
Heat shock gene expression is usually controlled by a combination of transcriptional and post-transcriptional mechanisms.22 In diverse α- and γ-proteobacteria, synthesis of sHsps is transcriptionally regulated by an alternative heat shock sigma factor and post-transcriptionally by RNA thermometers (RNATs), typically located in the 5′-untranslated region (5′-UTR) of the mRNA.23 RNATs operate by temperature-dependent alterations of their secondary structure. The Shine-Dalgarno (SD) sequence and occasionally the AUG start codon are masked by intramolecular base pairing at low temperatures. The secondary structure is relieved upon a temperature increase, thus enabling formation of the translation initiation complex.24
Dual control of heat shock gene expression by alternative sigma factors and an RNAT has recently been reported for Synechocystis. The sigma factors SigB and SigC are needed for heat shock gene transcription.25,26 The first cyanobacterial RNAT was reported in the 5′-UTR of the hsp17 mRNA.14 In contrast to the two widely distributed RNAT classes, the ROSE elements (repression of heat shock gene expression)23,27 and the fourU elements,28-31 the hsp17 RNAT constitutes an own class. The SD sequence is sequestered by nearly perfect canonical base pairing at low temperatures. An internal bulge is critical for melting of the RNA structure at heat shock temperatures.14 A Synechocystis strain with a chromosomally integrated stabilized, permanently “closed” hsp17 RNAT exhibited the same heat and high-light stress susceptibility as an hsp17 deletion strain. Interestingly, this was also partially true for an “open” hsp17 RNAT variant with enhanced Hsp17 synthesis when combined heat and light stress was applied.14 Apparently, the amount of sHsps must be tightly regulated according to the cellular demand under stress conditions.
In the present study, we investigated the occurrence of RNATs in other cyanobacterial species, namely the filamentous, mesophilic cyanobacteria Nostoc sp PCC 7120 (also known as Anabaena sp), and Anabaena variabilis ATCC 29413, and in the thermophilic unicellular cyanobacterium Thermosynechococcus elongatus BP-1. For all three organisms, we provide evidence for RNAT-mediated regulation of sHsp synthesis.
Results
Abundance of small heat shock genes in cyanobacteria
RNA thermometers are often associated with small heat shock genes and this is also true for Synechocystis.14 In order to identify new cyanobacterial RNAT candidates, we searched for sHsp genes in cyanobacterial genome sequences and found that several species feature up to six copies (Table 1). The thermophile T. elongatus and the terrestrial cyanobacterium Gloeobacter violaceus PCC 7421 encode one sHsp like the unicellular, mesophilic aquatic cyanobacteria Synechocystis and Synechococcus elongatus PCC 7942. Nostoc azollae 0708 is the only filamentous cyanobacterium with just one sHsp gene. The cyanobacteria with two sHsps constitute a heterogeneous group without any obvious common feature. Cyanobacteria with three or more sHsps are all filamentous except for Acaryochloris marina.32 As Nostoc punctiforme is found in diverse habitats, the large number of six encoded sHsps might reflect a special need for stress adaptation.
Table 1. Annotated small heat shock genes in selected cyanobacteria from various habitats.
| Species | Order | Habitat | Morphology | Number of small heat shock genes |
|---|---|---|---|---|
| Synechocystis sp PCC 6803 | Chroococcales | Fresh water, mesophilic |
unicellular | 1 (hsp17) |
| Thermosynechococcus elongatus BP-1 | Chroococcales | Fresh water, thermophilic |
unicellular | 1 (hspA) |
|
Synechococcus elongatus PCC 7942 |
Chroococcales | Marine, mesophilic | unicellular | 1 (Synpcc7942_2401) |
| Nostoc azollae 0708 | Nostocales | Plant symbiont, mesophilic |
filamentous | 1 (Aazo_2049) |
|
Gloeobacter violaceus PCC 7421 |
Gloeobacterales | Terrestrial, mesophilic | unicellular | 1 (glr2703) |
| Nostoc sp PCC 7120 | Nostocales | Aquatic, mesophilic | filamentous | 2 (alr0286, alr1809) |
| Cyanothece sp. PCC 7424 | Chroococcales | Terrestrial, mesophilic | unicellular | 2 (PCC7424_3788, PCC7424_4819) |
|
Microcystis aeruginosa NIES-843 |
Chroococcales | Fresh water, mesophilic | Colonial or unicellular | 2 (MAE_11440, MAE_45710) |
|
Synechococcus sp JA-3–3Ab |
Chroococcales | marine, thermophilic | unicellular | 2 (CYA_1442, CYA_1888) |
|
Anabaena variabilis ATCC 29413 |
Nostocales | Aquatic, mesophilic | filamentous | 3 (ava3076, ava4812, avaC0143) |
| Trichodesmium erythraeum IMS101 | Oscillatoriales | Marine, mesophilic | filamentous | 4 (Tery_3061, Tery_3062,Tery_3063, Tery_3064) |
| Acaryochloris marina MBIC11017 | Chroococcales | Marine, coral symbiont mesophilic | unicellular | 4 (AM1_1464, AM1_2993, AM1_D0093, AM1_D0147) |
|
Nostoc punctiforme PCC 73102 |
Nostocales | Diverse, mesophilic | filamentous | 6 (Npun_F0198, Npun_F0784, Npun_F1627, Npun_R1930, Npun_F2984, Npun_F3131) |
The two mesophilic species further inspected in this study are Nostoc sp PCC 7120 (Anabaena sp) and Anabaena variabilis ATCC 29413. They are closely related filamentous cyanobacteria. Nostoc sp chromosomally encodes the two sHsps Alr0286 and Alr1809.33 Alr0286 was shown to be a more effective chaperone than Alr1809 in E. coli.34 A. variabilis codes for three small heat shock proteins. Two are chromosomally (Ava3076 and Ava4812) and the third one, AvaC0143, is encoded on plasmid C.35 AvaC0143 exhibits 98% identity to sHsp Npun_F2984 of N. punctiforme and was probably acquired via horizontal gene transfer.
New RNAT candidates in Nostoc sp and Anabaena variabilis
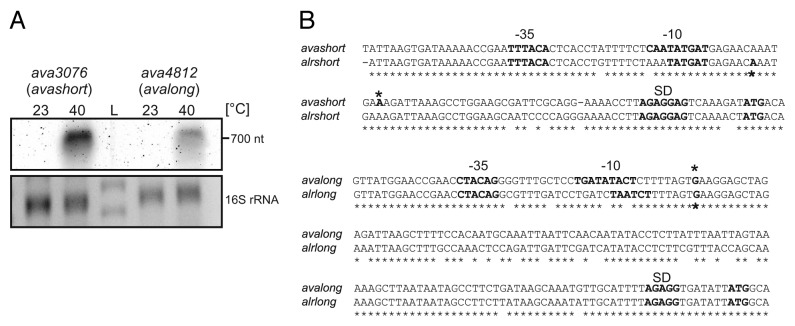
Secondary structure predictions with sequences upstream of the annotated start codons suggested RNAT-like structures for the chromosomally encoded sHsps Alr0286/Alr1809 in Nostoc sp and Ava3076/Ava4812 in A. variabilis (see below). To map the transcriptional start sites (TSS) and to investigate temperature-dependent expression of ava3076 and ava4812 in vivo, total RNA was isolated from A. variabilis kept at 23 °C or heat shocked to sub-lethal 40 °C for 1 h. Northern blot analysis showed heat-induced transcription of both small heat shock genes (Fig. 1A). 5′RACE (rapid amplification of cDNA ends) combined with in silico promoter prediction was applied to determine the TSS of these transcripts. For ava3076, the TSS was mapped at position -52 with regard to the adenine of the AUG codon, whereas the 5′UTR of ava4812 was much longer with transcription initiating at -125 Nt (Fig. 1B).
Figure 1. Transcriptional analysis of avashort und avalong genes from A. variabilis. (A) Northern analysis of RNA isolated from A. variabilis kept at 23 °C or heat shocked to 40 °C for 1 h. Avashort and avalong mRNA was detected with specific digoxygenine-labeled RNA probes. (B) Sequence alignment of avashort/alrshort and avalong/alrlong. Sequence alignment of the small heat shock gene promoter regions and 5′UTRs from A. variabilis ATCC 29413 (avashort, avalong) and the homologous genes (alrshort, alrlong) from Nostoc PCC 7120 performed using ClustalW2.84,85 Similarity is indicated by asterisks. For avashort and avalong, promoter sequences were computed with BPROM from the softberry webserver, transcriptional start sites (*) were determined by 5′RACE. For Nostoc, start sites were published before.3 Both sequence alignments reveal high similarity. Promoter sequences (-35, -10), transcriptional start sites (*), Shine-Dalgarno sequence (SD), and ATG start codon are depicted.
Transcription of the equivalent Nostoc sp shsp genes was heat-inducible like the A. variabilis homologs (data not shown). Their TSS have previously been determined by differential RNA-seq.3 The corresponding short and long UTRs of the shsp genes in Nostoc sp and A. variabilis are very similar (Fig. 1B) and will henceforth be called alrlong/avalong and alrshort/avashort.
To test for putative RNAT function, the 5′-UTRs were cloned into the well-established pBAD2-bgaB reporter gene system29 to obtain translational fusions to bgaB coding for a thermo-stable β-galactosidase (Fig. 2A). A fusion to the Synechocystis hsp17-5′UTR14 served as control. All fusions showed temperature-dependent induction in E. coli when a heat shock from 30–42 °C was applied (Fig. 2B). Due to the high similarity of the Nostoc and A. variabilis RNAT candidates, we focused on avalong and avashort in further studies.
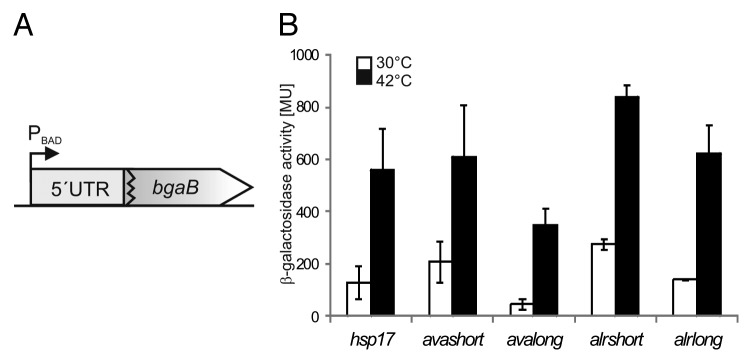
Figure 2. Temperature-dependent translational regulation of reporter activity. (A) Schematic drawing of translational fusion of RNAT of interest to bgaB (thermostable β-galactosidase) in pBAD2-bgaB.29 (B) Reporter gene assay of translational fusions of cyanobacterial RNAT candidates to bgaB tested in E. coli at 30 °C (white columns) or 42 °C (black columns). The well-studied hsp17-RNAT from Synechocystis14 served as positive control. Both the fusion from Anabaena (avashort/avalong) as well as those from Nostoc (alrshort/alrlong) exhibited temperature-induced reporter activity after heat shock from 30–42 °C. The presented results are mean values of a double measurement; mean standard deviation is indicated by error bars.
As the predicted avalong structure with a ΔG of -25.1 kcal mol−1 is more stable than avashort (ΔG -11.4 kcal mol−1), it should melt at higher temperatures. To validate this assumption, comparative β-galactosidase measurements were performed within a temperature range from 30–42 °C (Fig. 3). The control RNAT of the Synechocystis hsp17 gene (ΔG -5.5 kcal mol−1) exhibited a gradual increase of reporter activity resulting in a 3-fold higher expression at 42 °C. The temperature-dependent increase in reporter activity of the translational fusion to avashort was similar. The avalong-bgaB fusion exhibited a more pronounced response to rising temperature featuring a 6-fold increase in reporter activity at 42 °C compared with 30 °C.
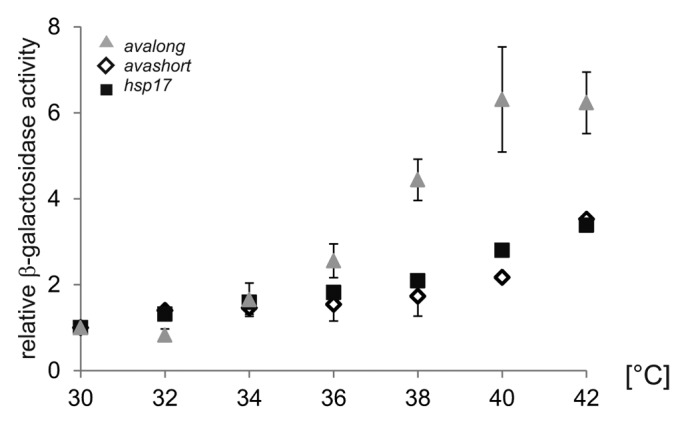
Figure 3. Reporter activity over temperature range. To elucidate the temperature-dependent RNAT activity of the avashort and the avalong-5′UTRs, reporter activity of both RNAT-bgaB fusions as well as the control hsp17-bgaB were analyzed in E. coli over a temperature range from 30–42 °C in 2 degree steps. The presented results are mean values of a double measurement; mean standard deviation is indicated by error bars. For normalization, the respective 30 °C value was set to 1.
Accessibility of the start codon is controlled in avashort
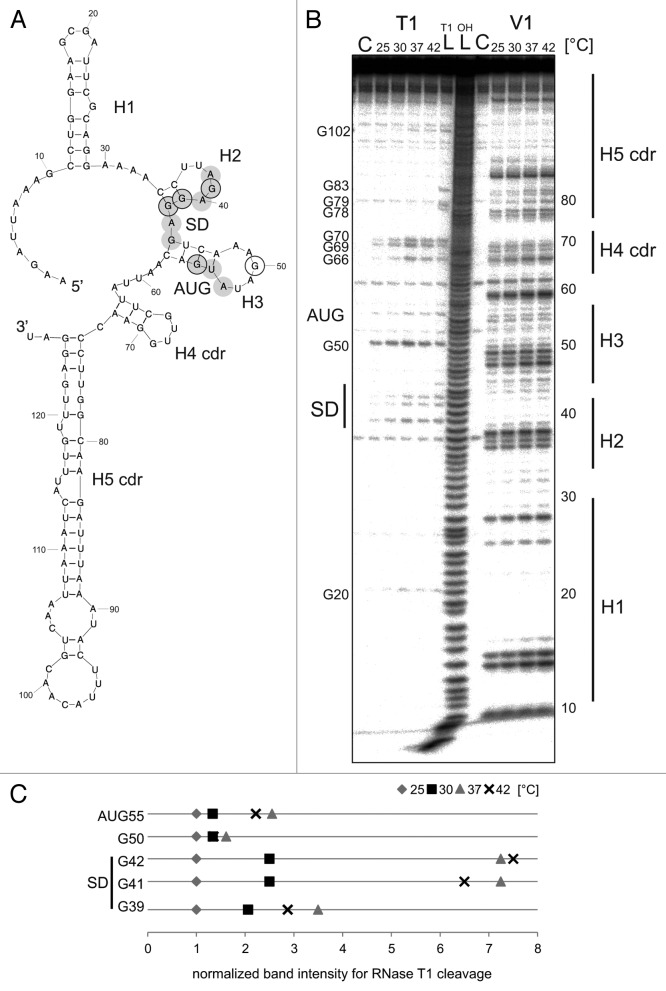
To address the molecular details of the different temperature response of avashort and avalong, we examined the structures of their 5′UTRs bioinformatically and experimentally. The 5′UTR of avashort is predicted to form a three hairpin structure (Fig. 4A) exhibiting a proximal hairpin (H1), a small hairpin (H2) involving the SD sequence, and a third hairpin (H3) with the AUG start codon. The first 72 Nt of the coding region are predicted to fold into a short fourth hairpin (H4 cdr) as well as an extended fifth hairpin (H5 cdr).
Figure 4. Temperature-dependent structural alterations of avashort-RNAT. (A) Secondary structure prediction of the avashort-5′UTR computed with mfold,86 including 72 Nt coding region (cdr) as well as the first three Nt of the blunt end restriction site EcoRV. SD and AUG are highlighted by gray circles. Black encircled Nt are referred to in the quantification in (C). The 5′ structure forms three hairpins (H1-H3), the SD sequence is partly base paired in H2, and the AUG is embedded in H3. The cdr forms H4 and H5. (B) In vitro transcribed avashort RNA (avashort-5′UTR and 72 Nt) was 5′ labeled and partially digested with RNase T1 (0.001U) and RNase V1 (0.0025U) at the indicated temperatures. For orientation, an alkaline ladder (LOH) was loaded; for the control (C) enzyme was replaced by A. dest. (C) Quantification of altered RNase T1 cleavage intensity between 25–42 °C for selected guanines. Band intensities were quantified from (B) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 30 °C set as 1.
To map the RNA structure, in vitro-transcribed avashort RNA was 5′labeled and digested with RNase T1 (prefers single-stranded guanines) and RNase V1 (double-strand specific) at 25 °C, 30 °C, 37 °C, or 42 °C, and separated on a denaturing 8% polyacrylamide gel (Fig. 4B, quantification of relevant T1 cuts in Fig. 4C). The absence of RNase T1 cleavage at G10, G14, G15, G18, G25, G28, and G29, as well as moderate cleavage of G20, located in the top loop of H1, confirmed formation of H1. G39, G41, and G42 of the SD sequence showed enhanced susceptibility to T1 with increasing temperature suggesting formation of H2 at lower temperatures. G44 of the SD was protected up to 42 °C reflecting its position in the more stable hairpin H3. As expected, G50 in the loop region of H3 was prone to cleavage at all temperatures. T1 cleavage at G55 of the AUG codon increased with increasing temperature supporting the prediction that the start codon is masked at low temperatures and accessible at heat shock temperature.
The overall V1 cleavage pattern supports the formation of several hairpins (Fig. 4B). The predicted binding of the SD sequence by C34 and C35 is not evident. Instead, prominent V1 cuts at Nt 36–39 suggest base-pairing of the uridines U36 and U37. V1 cleavage of Nt 47–49 indicates that the top loop of H3 is not as open as predicted probably involving an additional base pair between A48 and U52.
To analyze the in vivo importance of the predicted short anti-SD sequence formed by C34 and C35, the reporter plasmid pBAD2-avashort was subjected to site-directed mutagenesis. C34 and C35 were substituted by various other nucleotides, all intended to destabilize H2. All substitutions resulted in a wild-type-like temperature response in E. coli at 30 °C and at 42 °C (Fig. 5A). Thus, masking of the SD sequence by base pairing with C34 and C35 is not crucial for functionality of the avashort RNAT. Substitutions by adenines of U36 and U37, which might be base-paired according to the structure-probing experiment, likewise had no influence on reporter gene expression (data not shown). In contrast, exchange of C46 to adenine predicted to destabilize interaction with the AUG start codon led to a clear derepression at 30 °C (Fig. 5B) indicating that the avashort UTR primarily operates by masking the start codon.
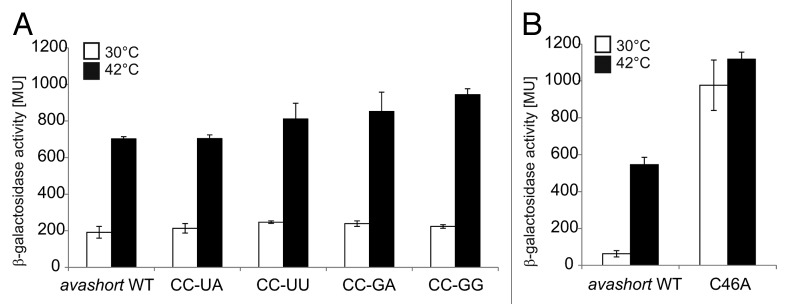
Figure 5. Reporter assay of avashort-bgaB WT and variants. (A) The putative anti-SD forming cytosines C34 and C35 were replaced by UA, UU, GA, and GG, respectively using randomized site-directed mutagenesis. The avashort WT exhibited a 3.5-fold induction of reporter activity after shift from 30–42 °C. All four variants exhibited WT-like activity. (B) Binding of AUG was loosened by changing the G55-C46 pair into a G55-A46 mismatch. Analyzed in the reporter assay, replacement of the cytosine by adenine resulted in complete destabilization of the avashort-RNAT structure visible in almost equal galactosidase activity levels at 30–42 °C. Both the results shown in (A and B) are mean values of a double measurement; mean standard deviation is given by error bars. WT, wild-type.
Structural alterations liberate the SD sequence of avalong at heat shock temperatures
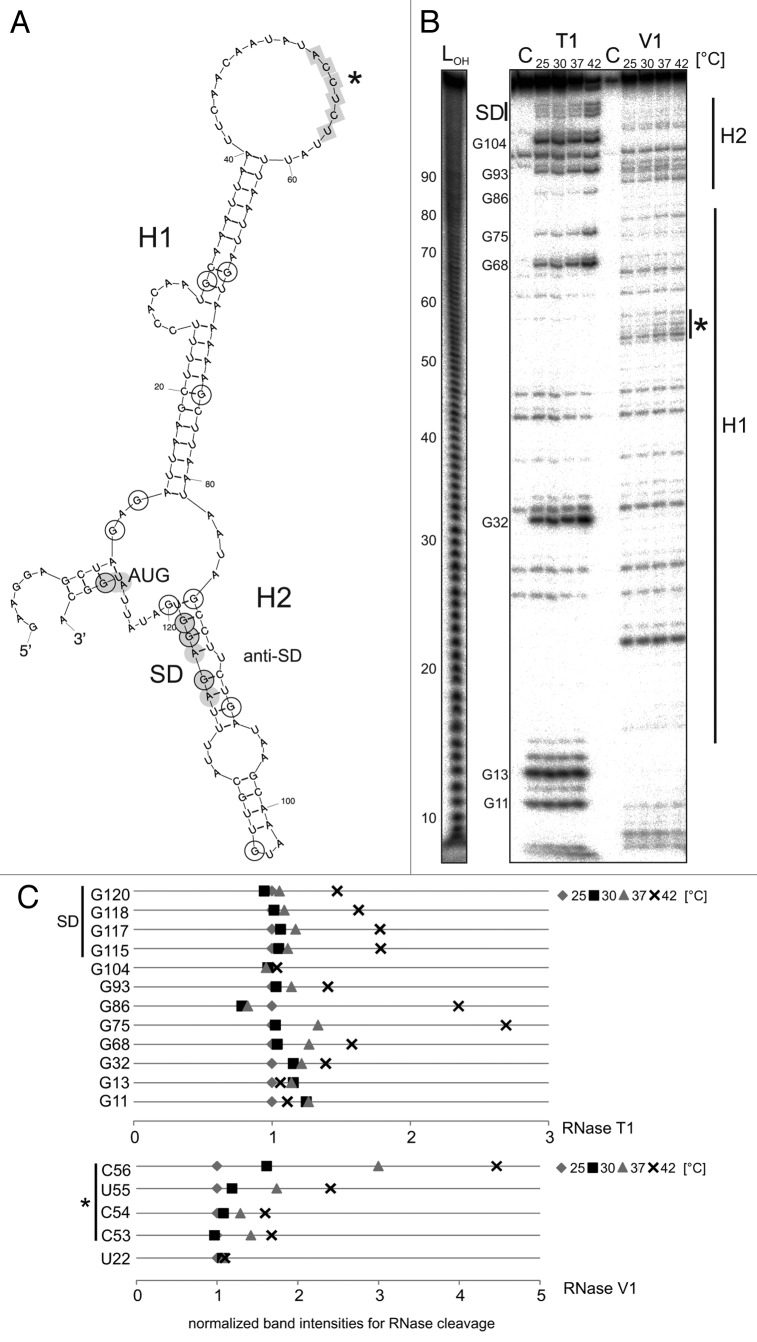
The avalong UTR is predicted to form an extended first hairpin (H1) with a large top loop and a shorter second hairpin (H2) containing the SD sequence (Fig. 6A). The SD sequence is paired by a ROSE-like anti-SD sequence. Instead of a bulged G residue in the conserved U(U/C)GCU sequence,23,27 avalong contains a bulged U residue. Another remarkable feature is a 6-Nt stretch complementary to the SD sequence in the top loop of H1 possibly acting as alternative anti-SD sequence (marked by an asterisk in Fig. 6A). Structure probing was conducted to validate the predicted structure and to analyze temperature-dependent alterations. In vitro transcribed avalong RNA was digested with RNase T1 and RNase V1 at various temperatures (Fig. 6B, quantification of selected residues in Fig. 6C). Digestion with RNase T1 revealed high susceptibility of guanines G11, G13, and G32 in H1 consistent with their exposed location (G11 and G13) or their position next to a large internal loop (G32). Higher temperatures led to increased T1 susceptibility of G68 and G75, suggesting melting of the stem structure. In H2, RNase T1 susceptibility of G86 in the anti-SD as well as G120, G118, G117, and G115 in the SD sequence increased at 42 °C, indicating liberation of the ribosome binding region at heat shock temperature. G104 is positioned in the single-stranded top loop of H2, and thus, accessible at any temperature. Cleavage at several positions in the water control (Fig. 6B) and few RNase V1 cuts in H1 might reflect its high AU content and suggest an overall lower structural stability of the mesophilic UTR compared with the thermophilic hspA UTR of similar shape (see below). The prominent RNase V1 cut at Nt 22 together with the absence of any T1 cuts, however, support base pairing of that region, and thus, H1 formation. V1 cuts between Nt 90–100 and 110–113 provide evidence for formation of H2. The 6-Nt putative alternative anti-SD sequence in the loop of the H1 (Nt 52–57) was susceptible to RNase V1 suggesting a double-stranded conformation not compatible with the predicted large loop. Tertiary interactions might be responsible for this effect (see below).
Figure 6. Temperature-dependent structural alterations of avalong-RNAT. (A) Secondary structure prediction of the avalong-5′UTR as well as the first three Nt of the blunt end restriction site EcoRV computed with mfold.86 The structure exhibits two RNA hairpins (H1 and H2), the first possesses a large terminal loop containing a putative alternative anti-SD sequence (marked in gray, asterisk). The second hairpin is short, containing the SD sequence base paired with a ROSE-like motif23,27 exhibiting a bulged uridine (U90). SD and AUG are highlighted in gray circles, the putative anti-SD sequence is marked by squares. Black encircled/framed Nt are referred to in the quantification in (C). (B) Enzymatic structure probing of avalong-RNAT. In vitro transcribed, 5′labeled avalong RNA was partially digested with RNase T1 (0.002U) and RNase V1 (0.01U) at the indicated temperatures. For the control (C) A. dest was added instead of enzyme. An alkaline ladder (LOH) was loaded for orientation. The asterisk marks the cleavages for the alternative anti-SD (C). Quantification of altered RNase T1 (upper panel) and V1 (lower panel) cleavage intensity between 25–42 °C for selected nucleotides. Band intensities were quantified from (B) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 25 °C set as 1.
The avalong-UTR is a ROSE-like thermosensor and might involve tertiary interactions
To examine the in vivo importance of the H1 top loop as well as the ROSE-like anti-SD sequence, site-directed mutagenesis was performed on pBAD2-avalong. The first variant was constructed by deleting Nt 31–70 constituting the upper stem-loop region of H1 (Δloop1). In the second variant, the predicted bulged U90 residue opposite the SD sequence was deleted (ΔU90). The third variant was constructed to evaluate whether the intramolecular base pairing around the SD sequence is important to prevent translation at lower temperatures. The structure was loosened by two mismatches introduced by exchanging the C residues at positions 88 and 91 against adenines (CC88/91AA). All three variants were tested in reporter assays in E. coli at 30 °C or 42 °C (Fig. 7A). The wild-type fusion showed the expected heat-inducible reporter activity (compare with Fig. 2B). Deletion of the loop region of H1 resulted in high β-galactosidase activity already at 30 °C, suggesting destabilization of the overall structure and better accessibility of the SD sequence, thus providing evidence for long-range tertiary interactions. This seems to involve more than just the putative anti-SD sequence in loop 1 as exchange of C53 and C54 (avalong*2A) or of the entire CCUC motif (Nt 53–56, avalong*4A) against adenines did not affect reporter activity at low temperatures and stimulated heat induction (Fig. 7B). Deletion of the bulged U90 residue opposite the SD sequence abrogated reporter activity even at heat shock temperature. In contrast, the CC88/91AA exchange led to full de-repression at 30 °C as shown for equivalent mutations in ROSE-like RNATs.27,36-38
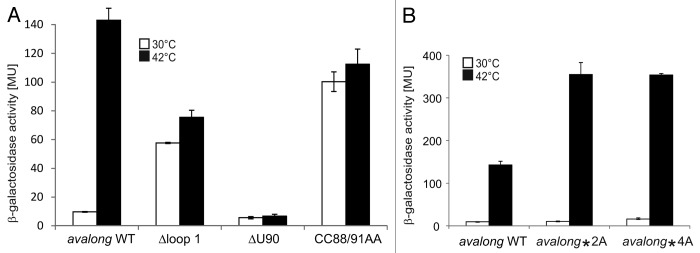
Figure 7. Reporter gene assay with avalong-RNAT WT and variants. (A) Using site-directed mutagenesis, the terminal H1-loop (Nt 31–70), the bulged uridine 90 in the ROSE-like anti-SD was deleted (ΔU90) and two flanking GC pairs were opened by exchanging the cytosines at positions 88 and 91 against adenines (CC88/91AA). Compared with the WT exhibiting a 10-fold increase after heat shock from 30–42 °C, deletion of the top-loop resulted in high β-galactosidase activity at 30 °C and decreased heat-induction after the shift to 42 °C. In contrast, deletion of the predicted bulged U90 provoked low levels of reporter activity both under heat shock and non-heat shock conditions. A loosened structure around the SD led to a high reporter gene expression at 30 °C and 42 °C. The results reflect mean values of a double-measurement; mean standard deviation is given by error bars. (B) Measurement of β-galactosidase activity at 30 °C and 42 °C of avalong-variants harboring a mutated alternative anti-SD sequence. Neither the substitution of C53, C54 by adenines (avalong*2A), nor of the CCUC motif (Nt 53–56, avalong*4A) led to a derepression at low temperatures compared with the wild-type.
A putative RNA thermometer acting at high temperatures
HspA is the only Hsp17 homolog in the thermophilic cyanobacterium Thermosynechococcus elongatus (Table 1). Chaperone activity of HspA was demonstrated in the close relative Thermosynechococcus vulcanus.39 Like transcription of T. elongatus hspA,40 transcription of T. vulcanus hspA is heat-induced and the transcriptional start site (TSS) was mapped -120 nucleotides (Nt) relative to the adenine of the AUG start codon.41 Constitutive transcription was observed when transcriptional lacZ fusions of T. vulcanus hspA fragments were expressed in Escherichia coli.42 Binding of a protein from crude T. elongatus extract to a DNA inverted repeat in the hspA-5′-UTR under non-heat stress, suggested the involvement of a repressor protein in transcriptional control.43 Translational fusions of the T. vulcanus hspA-5′UTR to lacZ exhibited only low β-galactosidase activity, which led to the proposal of a regulatory element inhibiting translation,42 potentially acting like an RNAT.44 Interestingly, the T. elongatus and T. vulcanus hspA-5′UTRs display some similarity to the Synechocystis hsp17-5′UTR around their SD sequences, suggesting the presence of RNATs in these thermophilic organisms.14
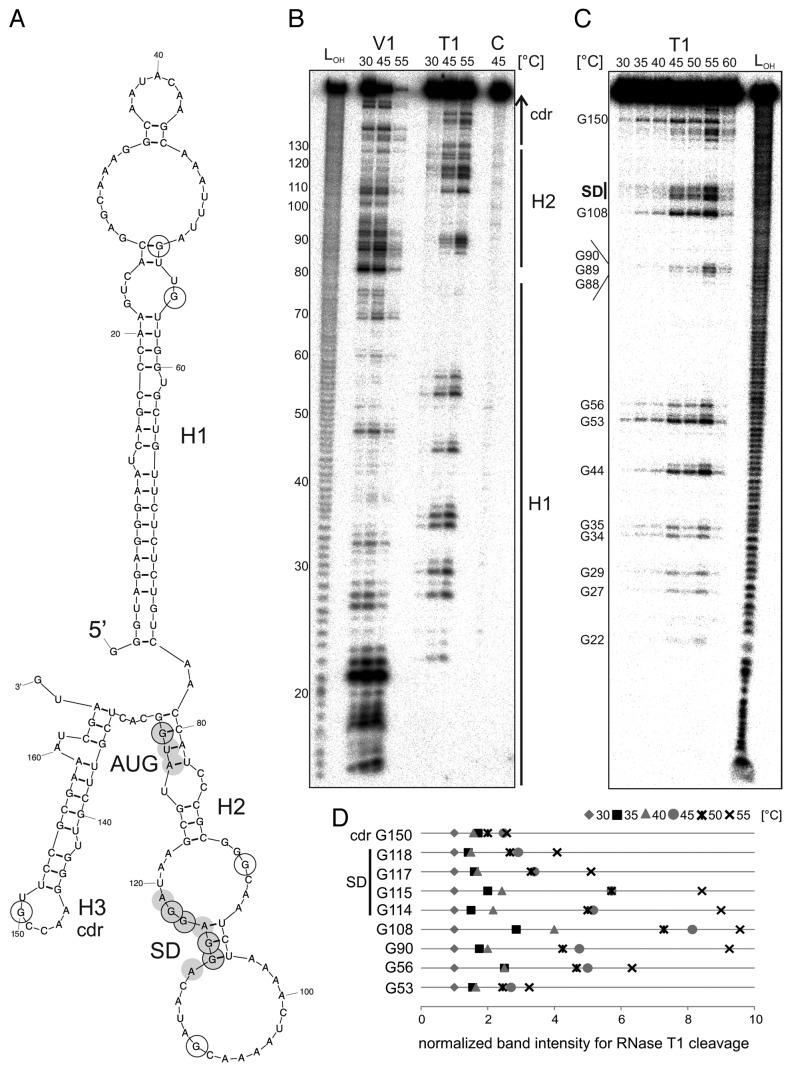
The predicted structure of the hspA-5′UTR resembles the avalong 5′UTR, as a long needle-like structure precedes the hairpin that blocks the translation initiation region (Fig. 8A). A TSS at 126 nucleotides Nt upstream of the start codon was assumed according to the previously mapped T. vulcanus hspA TSS (Fig. S1). Since the established reporter gene systems for studying RNAT29 are restricted to mesophilic hosts like E. coli, and thus, unsuitable for in vivo analysis of thermophilic RNAT candidates, we used in vitro methods to investigate whether the T. elongatus hspA-5′-UTR acts as an RNAT. For structural analysis, an extended hspA-RNA fragment was used, containing the hspA-5′UTR and 40 Nt of the coding region predicted to form a third hairpin (H3). In vitro transcribed hspA RNA was 5′-labeled and subjected to partial digestion with RNase V1 and RNase T1 at 30 °C, 45 °C, or 55 °C, followed by separation on an 8% denaturing polyacrylamide gel (Fig. 8B).
Figure 8. Temperature-dependent structural alterations of the hspA-5′UTR. (A) Model of the T. elongatus hspA-5′UTR and 40 Nt coding region as well as the first two Nt of the blunt end restriction site HpyCH4v according to a secondary structure prediction computed with mfold.86 The structure exhibits two RNA hairpins H1 and H2 in the 5′UTR, the second containing SD region and AUG start codon, as well as a third H3 in the coding region. SD sequence und AUG start codon are highlighted by gray circles. Black encircled Nt are referred to in the quantification in (D). (B) Enzymatic structure probing of the hspA-5′UTR with 40 Nt coding region. In vitro transcribed RNA was 5′labeled81 and subjected to partial digestion82 with RNase V1 (0.001U) and T1 (0.002U) at the indicated temperatures. For the control, (C) A. dest was added instead of enzyme. An alkaline ladder (LOH) was loaded for orientation. Samples were separated on an 8% denaturing polyacrylamide gel. (C) Temperature stability of the hspA RNA structure. Labeled RNA was digested by RNase T1 (0.002U) within a temperature range from 30–60 °C in 5 degree steps. The probing reveals slight accessibility of the SD region for RNase T1 at 45 °C and high susceptibility at 55 °C. Please note that RNase T1 was inactive at 60 °C. (D) Quantification of altered RNase T1 cleavage intensity between 30–55 °C for selected guanines. Band intensities were quantified from (C) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 30 °C set as 1. cdr, coding region.
Formation of hairpin H1 composed of a GC-rich stem from Nt 2–21 and Nt 57–76 with a large top loop was corroborated by efficient cleavage of the double-strand-specific RNase V1 at Nt 14–23 at all tested temperatures (Fig. 8B), although the complementary region (Nt 57–76) showed weaker V1 cuts. Conversely and consistent with stem formation, RNase T1 did not cleave in these two regions at all (Fig. 8B and C). Probing with RNase A (preferentially cleaving single-stranded RNA at U or C residues) confirmed stability of the H1 stem up to 65 °C in vitro (data not shown). Multiple V1 cuts between Nt 80 and 95 indicate a stable H2 structure at 30 °C and 45 °C. A decrease in V1 cuts at 55 °C, despite efficient cleavage in the H1 stem region from Nt 14–23, suggests temperature-dependent melting of the H1 top loop and H2. This is consistent with the reverse RNase T1 pattern for the H2 guanines G88–90, being only accessible at 55 °C, as well as of the SD guanines G114, G115, G117, and G118 at 55 °C.
To further analyze the temperature response of H2, hspA RNA was digested with RNase T1 in a temperature range from 30–60 °C in steps of 5 degrees (Fig. 8C; note that the RNase was almost completely inactive at 60 °C). Quantification of temperature-dependent changes in RNase T1 cleavage intensity is given in Figure 8D for selected guanines. Residues G88–90 were best accessible to RNase T1 at 55 °C. Likewise, the SD guanines 114, 115, 117, and 118 were barely attacked at low temperature but prone to cleavage at 55 °C, indicating liberation of the SD sequence. The combined V1 and T1 probing results suggested heat-stable formation of the H1 stem and a heat-labile H2 structure. Further, the temperature responsiveness of the predicted single-stranded positions G88, G89, G90, and G108, suggests that tertiary interactions might be involved in masking these residues at low temperature.
Formation of H3 in the coding region can be inferred from V1 cuts around Nt 130–140 and around 155–160 (Fig. 8B). Accessibility of G150 by RNaseT1 already at 30 °C supports its predicted location in the H3 loop, whereas increased susceptibility of G143–145 by RNaseT1 indicate H3 melting at heat shock temperature (Fig. 8C).
Discussion
Transcriptional and translation control of the cyanobacterial heat shock response
Upon heat shock, cyanobacteria express a multitude of genes like many other bacteria including E. coli.45-47 In contrast to this model organism, cyanobacteria encode additional DnaK and GroEL homologs, suggesting a higher requirement for heat shock proteins and/or their functional diversification in photosynthetic microorganisms. DnaK2 and DnaJ2 from Synechococcus sp PCC 7942 were shown to interact with RNase E,48 whereas DnaK3 can associate with thylakoid membranes,49 probably stabilizing them like sHsps. Further, GroEL was shown to form stable complexes with ribulose-1,5 bisphosphate carboxylase/oxygenase (RubisCO) in pea chloroplasts in vitro.50 Deletion of the chaperone gene htpG resulted in a yellow-greenish appearance of Synechococcus instead of the wild-type blue-green coloring. Thus, HtpG might play a role in phycobiliprotein synthesis or degradation.51 Furthermore, HtpG is associated with regulation of tetrapyrrole biosynthesis.52 The FtsH protease is associated with PS II in Synechocystis53 and degrades UV-damaged D1 and D2 protein.54
The obvious impact of heat shock proteins on photosynthesis necessitates a tight regulation of the HSR in cyanobacteria. The most obvious difference compared with E. coli is the lack of a σ32 homolog, although some heat-related alternative sigma factors are present.25,26,55 Expression of the cyanobacterial groESL1 operon is regulated by the HrcA/CIRCE system,12 a regulatory system well studied in Bacillus subtilis56 and common in diverse microorganisms.57 The HrcA repressor protein binds to the inverted repeat motif CIRCE (controlling inverted repeat of chaperone expression). Depending on the species, expression of groEL2 can be regulated via HrcA/CIRCE as e.g., in Synechocystis12 or by some unknown mechanisms as in Thermosynechococcus.40 Besides alternative sigma factors or repressor proteins, the sensor kinase Hik34 was identified as a negative regulator, leading to transcriptional repression of a number of heat shock genes e.g., htpG, groESL1, and hsp17 expression under non-heat stress conditions in Synechocystis.58
As cyanobacteria are capable of performing oxygenic photosynthesis, light intensity is an additional important growth parameter. Accordingly, light triggers the heat-dependent induction of htpG, groESL1, groEL2, and hsp17 in Synechocystis.59 Except for hsp17, expression was lower in the dark. Application of DCMU (3-[3,4-dichlorophenyl]-1,1-dimethylurea), an inhibitor of the photosynthetic electron transport chain, led to suppression of light-regulated groE expression in Synechocystis. Subsequent analysis revealed light-responsive regulation of Synechocystis groESL1 and groEL2 genes by regulatory K- and N-boxes on the DNA level.60 Further, transcription as well as translation of HSR components are affected by both temperature and thylakoid membrane physical order, defining the thylakoid membranes as the primary stress sensor determining the set point of HSR induction.19,52 A simple RNA thermometer comprised of a single hairpin was found to be responsible for temperature-dependent control of translation in Synechocystis.14
Anabaena features two RNATs applying different modes of action
In this study, we describe two novel RNAT in mesophilic cyanobacteria. Avashort and avalong differ not only in length but also in sequence, structure, and mode of action. In contrast to previously studied RNATs, avashort acts by modulating the accessibility of the AUG start codon in response to temperature. Although some RNAT include the start codon in masking of the SD sequence,27,61 a strict requirement for AUG-mediated regulation is unique. Interestingly, an RNA structure in the 5′UTR of the chloroplast psbD mRNA in the eukaryotic green alga Chlamydomonas reinhardtii was shown to mask the translation start codon.62 It is tempting to speculate that this structure responds to temperature changes, thereby modulating expression of D2 protein.
The avalong-5′UTR folds into two RNA hairpins with a heat-stable, needle-like hairpin 1. The top loop of H1 contains a 6-Nt stretch complementary to the SD sequence, possibly acting as alternative anti-SD sequence. Base-pairing of that region was supported by structure probing experiments. It is conceivable that in a dynamic temperature-modulated structure, the ROSE-like anti-SD in H2 and the alternative anti-SD in H1 compete for binding to the SD sequence. Another possibility is interaction of the alternative anti-SD in H1 with some other part of the avalong-5′UTR. For example, the 6-Nt stretch in H1 could bind to the most proximal 5′end of avalong, as the sequence from Nt 1–14 is highly AG-rich. It has been reported that 5′end located AG-rich sequences, resembling ribosome binding sites, stabilize the respective transcript.63 The sequence similarity of the 5′end of the avalong-5′UTR to the conserved 5′end of the Yfr2 family of cyanobacterial non-coding RNAs starting with 5′GUGAGGA-3′ or a similar sequence64 raises the intriguing possibility that the exposed anti-SD sequence in the H1 top loop could act in trans to regulate expression of unknown target genes.
Evidence for a thermophilic RNA thermometer
Many RNAT control the expression of heat shock and virulence genes in the mesophilic temperature range.24 Here, we asked whether this mechanism can also operate in thermophiles. Base pairing of the SD sequence the hspA-5′UTR from T. elongatus up to 55 °C supports the existence of a zipper-like RNAT. Possibly, the GC-rich H1 of hspA, which is stable up to 65 °C, functions as a stabilizing element. Most likely, translation inhibition involves complex tertiary interactions with the SD-containing second hairpin. A detailed molecular analysis of regulatory elements acting in the thermophilic temperature range is presently hampered by the lack of suitable reporter systems. Therefore, the exact mode of action of the hspA UTR remains to be elucidated and the involvement of additional factors contributing to the temperature response cannot be excluded. The exposed positioning of an inverted repeat in H1 (Fig. S1) might facilitate interaction with RNA-binding proteins as it is common in cyanobacterial cold stress regulation.65,66 This region has previously been implicated in transcriptional control by binding of a yet unknown repressor protein on DNA level.41-43 A global transcriptional regulator of heat stress genes in Synechocystis is Sll1130 featuring a DNA-binding PemK domain. Absence of this protein resulted in overexpression of heat-related genes e.g., htpG and hsp17 or the heat- and iron-regulated genes isiA and isiB.67 In T. elongatus, Tlr0462 is an ortholog of Sll1130 with 36% sequence identity (CyanoBase ortholog table for Sll1130). Tlr0462 is not a PemK-like protein but is assigned to the bacterial BAX inhibitor (BI)-1 superfamily. Whether Tlr0462 has similar functions in T. elongatus as Sll1130 in Synechocystis and whether it is the unidentified DNA-binding protein proposed by Kojima, et al. remains to be elucidated.43
Importance of RNA-based regulation in photosynthetic bacteria
In the last decade, the enormous importance and frequency of RNA-based regulation in cyanobacteria has been unraveled. Presumably, 8–10% of the Synechocystis genome is directly or indirectly regulated by antisense RNAs (asRNAs).68 Comparative genome analyses identified potential cyanobacterial riboswitches selective for thiamine pyrophosphate, cobalamin,69 and glutamine.70 Extensive RNA-based regulation was revealed for photosynthesis components. Two cis-encoded asRNAs are present in the 5′UTRs of psbA2 and psbA3 coding for D1 protein of PS II in Synechocystis.71 Furthermore, an AU-box motif probably involved in transcript stability was identified in the 5′UTRs of psbA genes from different pro- and eukaryotic photosynthetic organisms.72 A potential asRNA was identified in the intergenic region of the psaAB operon in Synechocystis, encoding photosystem I components.73 The prevalence of RNA-mediated regulation has recently been recognized in other photosynthetic bacteria. The trans-acting sRNA PcrZ negatively affects photosynthesis gene expression in Rhodobacter sphaeroides.74 Moreover, a large repertoire of sRNAs is involved in environmental control of gene expression in Rhodobacter,75 Roseobacter,76 and Prochlorococcus.77
The frequency of regulatory RNAs in photosynthetic microorganisms emphasizes their importance in environmental adaptation processes. RNATs add a simple, temperature-responsive layer of control to the diverse regulatory mechanisms for cyanobacterial heat shock gene expression. As RNATs are an integral part of the mRNA, signal transduction is rapid, allowing an immediate response to heat stress. This might be the reason for their universal use in controlling the synthesis of sHsps, which play a fundamental role in the bacterial chaperone network, in particular in cyanobacteria.
Experimental Procedures
Bacterial growth conditions
Anabaena variabilis ATCC 29413 and Nostoc (Anabaena) sp PCC 7120 were grown in BG-11 medium at 23 °C with continuous shaking and illumination with 100 μEm−2s−1.
Thermosynechococcus elongatus BP-1 was grown as described.78
E. coli was grown in LB medium at the indicated temperatures. In case, the following additives were applied to the given final concentrations: Ampicillin (150 µg ml−1); Kanamycin (50 µg ml−1); 5-bromo-4-chloro-indolyl-β-D-galactopyranoside (X-gal) (40 µg ml−1). Addition of 0.01% (w/v) L-arabinose was applied to induce transcription from pBAD2-constructs.
Plasmid construction
Construction and handling of plasmids was performed according to standard protocols.79 Enzymes were obtained from Thermo Scientific. All plasmids generated in this study are listed in Table S1.
5′UTRs of interest were PCR amplified from chromosomal DNA using the corresponding primer pairs listed in Table S1, cloned blunt end into the SmaI site of pUC18,80 and subjected to automated sequencing (eurofins). Translational fusions to bgaB were obtained by NheI/EcoRI cloning into pBAD2-bgaB.29 To generate RNAT-bgaB variants, plasmids pBAD2-avahort and pBAD2-avalong served as templates for site-directed mutagenesis using the primers listed in Table S1.
Runoff-plasmids for in vitro transcription were constructed as follows: The RNAT of interest (and in case additional coding region) was PCR amplified, adding a T7-promoter sequence (GAAATTAATA CGACTCACTA TAGGG) to the 5′end and a blunt end digestion side to the 3′end. The used blunt end site was EcoRV in the case of avashort and avalong and HpyCH4v for hspA-RNAT. In the applied hspA-construct, an internal HpyCH4v site was used, giving rise to an RNA template containing the RNAT and 40 Nt coding region. After linearization of the plasmid, 4 μg were used as template for in vitro transcription using T7 RNA polymerase (Thermo Fisher Scientific).
Enzymatic structure probing
RNA was 5′end labeled as described before81 and subjected to partial digestion with RNases.82 The applied ribonucleases T1 and V1 were purchased from Ambion. The RNases were used in the following concentrations: hspA 0.002U T1, 0.001U V1; avalong 0.002U T1, 0.01U V1; avashort 0.001U T1, 0.0025U V1. As control served samples where A. dest was added instead of enzyme. For orientation, an alkaline hydrolysis ladder was set up.81 For identification of the appropriate guanines, a T1 ladder was made in digesting avashort-RNA at 55 °C. Quantification of band intensities was performed using Alpha Ease software (Alpha Innotech, Biozym).
RNA isolation
Anabaena was grown to late stationary phase (oD730 18.6) at 23 °C before the culture was split and half was subjected to a 1 h heat shock at 40 °C. After harvesting, pellets were washed with 1 ml ice-cold TE buffer (40 mM Tris, 1 mM EDTA, pH 8) and stored at -80 °C until further treatment. After thawing on ice for 3 h, pellets were solved in 1 ml Trizol with 500 μl mixed 0.5 mm and 0.1 mm zirconia/silica beads (BioSpec). After 5 min incubation at room temperature, cells were disrupted by vortexing for 2 × 30 s per sample. The supernatant was transferred into a new tube, 1 vol ethanol was added, and samples were homogenized by vortexing. Seven hundred μl were used for RNA isolation with Direct-zol RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s manual.
Northern analysis
Northern blot hybridization was performed as described.29 RNA probes were synthesized by in vitro transcription with T7 RNA polymerase (Thermo Fisher Scientific) using DIG RNA labeling mix (Roche) according to the manufacturer’s manual. Signals were examined via chemiluminescence detection (FluorChem SP, Alpha Innotech, Biozym).
5′RACE (rapid amplification of cDNA ends)
A. variabilis was grown at 23 °C under continuous shaking and illumination till stationary phase previous to splitting of the culture and subsequent heat shock of one-half to 42 °C for 10 min prior to RNA isolation. 5′RACE for determination of 5′ends was performed as described83 with minor modifications.36 Applied adaptor oligonucleotides, gene-specific primers 529 (avashort), and 461 (avalong) are listed in Table S1.
β-galactosidase assay
E. coli harboring pBAD2-constructs were grown in 25 ml LB at 30 °C to oD600 0.5. Addition of 0.01% (w/v) L-arabinose induced transcription from pBAD promoter prior to splitting of the culture and transfer of 10 ml to pre-warmed flasks at 42 °C for 30 min. For measuring β-galactosidase activity within a temperature range from 30–42 °C, cells were heat shocked from 30 °C to the respective temperature. The enzymatic assay was conducted as described.36
Bioinformatic tools
Genome sequence data was assessed from the NCBI microbial genome database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/microbial_taxtree.html).
Potential Sll1130 orthologs were retrieved from CyanoBase (http://genome.microbedb.jp/cyanobase/Synechocystis/genes/sll1130).
Formatting was done using a sequence editor program (http://www.fr33.net/seqedit.php). ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/)84,85 and the LALIGN server (http://www.ch.embnet.org/software/LALIGN_form.html) were used to perform sequence alignments. Plasmid sequencing data was analyzed with Chromas lite Version 2.01 (Technelysium Pty Ltd). BPROM from the SoftBerry webserver was used to predict promoter sequences (http://linux1.softberry.com/berry.phtml). RNA secondary structure prediction was computed using the mfold Web Server86 (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Iris Maldener (Tübingen) for the gift of Anabaena variabilis ATCC 29413 and Nostoc sp PCC 7120 and to Matthias Rögner and Ulrich Kück for help with the cultivation of cyanobacteria. We acknowledge Mats Leifels for initial experiments on avashort and avalong pBAD2-variants. We thank all members of the RNA group and Lisa-Marie Bittner for critical reading of the manuscript and helpful discussions. Funding was provided by the German Research Foundation (DFG priority program SPP 1258: Sensory and regulatory RNAs in Prokaryotes) to FN, by grants from the “Ruth und Gert Massenberg Stiftung” and the “Ruhr University Research School” to Cimdins A and by a fellowship of the “Studienstiftung des Deutschen Volkes” to Kortmann J.
References
- 1.Schopf JW. Microfossils of the Early Archean Apex chert: new evidence of the antiquity of life. Science. 1993;260:640–6. doi: 10.1126/science.260.5108.640. [DOI] [PubMed] [Google Scholar]
- 2.Falcón LI, Magallón S, Castillo A. Dating the cyanobacterial ancestor of the chloroplast. ISME J. 2010;4:777–83. doi: 10.1038/ismej.2010.2. [DOI] [PubMed] [Google Scholar]
- 3.Mitschke J, Vioque A, Haas F, Hess WR, Muro-Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci U S A. 2011;108:20130–5. doi: 10.1073/pnas.1112724108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castenholz RW. Phylum BX. Cyanobacteria. In: Boone DR, Castenholz RW, eds. Bergey’s Manual of Systematic Bacteriology, The Archaea and the Deeply Branching and Phototrophic Bacteria. New York: Springer, 2001:473-599. [Google Scholar]
- 5.Suzuki I, Simon WJ, Slabas AR. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J Exp Bot. 2006;57:1573–8. doi: 10.1093/jxb/erj148. [DOI] [PubMed] [Google Scholar]
- 6.Paithoonrangsarid K, Shoumskaya MA, Kanesaki Y, Satoh S, Tabata S, Los DA, Zinchenko VV, Hayashi H, Tanticharoen M, Suzuki I, et al. Five histidine kinases perceive osmotic stress and regulate distinct sets of genes in Synechocystis. J Biol Chem. 2004;279:53078–86. doi: 10.1074/jbc.M410162200. [DOI] [PubMed] [Google Scholar]
- 7.Marin K, Kanesaki Y, Los DA, Murata N, Suzuki I, Hagemann M. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 2004;136:3290–300. doi: 10.1104/pp.104.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandenbroucke K, Robbens S, Vandepoele K, Inzé D, Van de Peer Y, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–16. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 9.Mary I, Tu CJ, Grossman A, Vaulot D. Effects of high light on transcripts of stress-associated genes for the cyanobacteria Synechocystis sp. PCC 6803 and Prochlorococcus MED4 and MIT9313. Microbiology. 2004;150:1271–81. doi: 10.1099/mic.0.27014-0. [DOI] [PubMed] [Google Scholar]
- 10.Summerfield TC, Sherman LA. Global transcriptional response of the alkali-tolerant cyanobacterium Synechocystis sp. strain PCC 6803 to a pH 10 environment. Appl Environ Microbiol. 2008;74:5276–84. doi: 10.1128/AEM.00883-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narberhaus F. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamoto H, Suzuki M, Kojima K. Targeted inactivation of the hrcA repressor gene in cyanobacteria. FEBS Lett. 2003;549:57–62. doi: 10.1016/S0014-5793(03)00768-3. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Prochaska DJ, Fang F, Barnum SR., Sr. A 16.6-kilodalton protein in the cyanobacterium Synechocystis sp. PCC 6803 plays a role in the heat shock response. Curr Microbiol. 1998;37:403–7. doi: 10.1007/s002849900400. [DOI] [PubMed] [Google Scholar]
- 14.Kortmann J, Sczodrok S, Rinnenthal J, Schwalbe H, Narberhaus F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 2011;39:2855–68. doi: 10.1093/nar/gkq1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Havaux M. Stress tolerance of photosystem II in vivo: antagonistic effects of water, heat, and photoinhibition stresses. Plant Physiol. 1992;100:424–32. doi: 10.1104/pp.100.1.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heckathorn SA, Downs CA, Sharkey TD, Coleman JS. The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol. 1998;116:439–44. doi: 10.1104/pp.116.1.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakamoto H, Suzuki N, Roy SK. Constitutive expression of a small heat-shock protein confers cellular thermotolerance and thermal protection to the photosynthetic apparatus in cyanobacteria. FEBS Lett. 2000;483:169–74. doi: 10.1016/S0014-5793(00)02097-4. [DOI] [PubMed] [Google Scholar]
- 18.Nitta K, Suzuki N, Honma D, Kaneko Y, Nakamoto H. Ultrastructural stability under high temperature or intensive light stress conferred by a small heat shock protein in cyanobacteria. FEBS Lett. 2005;579:1235–42. doi: 10.1016/j.febslet.2004.12.095. [DOI] [PubMed] [Google Scholar]
- 19.Horváth I, Glatz A, Varvasovszki V, Török Z, Páli T, Balogh G, Kovács E, Nádasdi L, Benkö S, Joó F, et al. Membrane physical state controls the signaling mechanism of the heat shock response in Synechocystis PCC 6803: identification of hsp17 as a “fluidity gene”. Proc Natl Acad Sci U S A. 1998;95:3513–8. doi: 10.1073/pnas.95.7.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamoto H, Honma D. Interaction of a small heat shock protein with light-harvesting cyanobacterial phycocyanins under stress conditions. FEBS Lett. 2006;580:3029–34. doi: 10.1016/j.febslet.2006.04.047. [DOI] [PubMed] [Google Scholar]
- 21.Sakthivel K, Watanabe T, Nakamoto H. A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in cyanobacteria under oxidative stress. Arch Microbiol. 2009;191:319–28. doi: 10.1007/s00203-009-0457-z. [DOI] [PubMed] [Google Scholar]
- 22.Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72:545–54. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. RNA thermometers are common in alpha- and gamma-proteobacteria. Biol Chem. 2005;386:1279–86. doi: 10.1515/BC.2005.145. [DOI] [PubMed] [Google Scholar]
- 24.Kortmann J, Narberhaus F. Bacterial RNA thermometers: molecular zippers and switches. Nat Rev Microbiol. 2012;10:255–65. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 25.Tuominen I, Pollari M, Aguirre von Wobeser E, Tyystjärvi E, Ibelings BW, Matthijs HC, Tyystjärvi T. Sigma factor SigC is required for heat acclimation of the cyanobacterium Synechocystis sp. strain PCC 6803. FEBS Lett. 2008;582:346–50. doi: 10.1016/j.febslet.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 26.Tuominen I, Pollari M, Tyystjärvi E, Tyystjärvi T. The SigB sigma factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 2006;580:319–23. doi: 10.1016/j.febslet.2005.11.082. [DOI] [PubMed] [Google Scholar]
- 27.Nocker A, Hausherr T, Balsiger S, Krstulovic NP, Hennecke H, Narberhaus F. A mRNA-based thermosensor controls expression of rhizobial heat shock genes. Nucleic Acids Res. 2001;29:4800–7. doi: 10.1093/nar/29.23.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol Microbiol. 2007;65:413–24. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 29.Klinkert B, Cimdins A, Gaubig LC, Roßmanith J, Aschke-Sonnenborn U, Narberhaus F. Thermogenetic tools to monitor temperature-dependent gene expression in bacteria. J Biotechnol. 2012;160:55–63. doi: 10.1016/j.jbiotec.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Kouse AB, Righetti F, Kortmann J, Narberhaus F, Murphy ER. RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS One. 2013;8:e63781. doi: 10.1371/journal.pone.0063781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Böhme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog. 2012;8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rynearson TA, Palenik B. Learning to read the oceans: genomics of marine phytoplankton. In: Lesser M, ed. Advances in Marine Biology: Academic Press, 2011:1-39. [DOI] [PubMed] [Google Scholar]
- 33.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, et al. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:205, 13, 27, 53. doi: 10.1093/dnares/8.5.205. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Huang W, Li M, Wu Q. Purification and characterization of two small heat shock proteins from Anabaena sp. PCC 7120. IUBMB Life. 2005;57:449–54. doi: 10.1080/15216540500138402. [DOI] [PubMed] [Google Scholar]
- 35.Copeland A, Lucas S, Lapidus A, Barry K, Detter JC, Glavina T, Hammon N, Israni S, Pitluck S, Saunders EH, et al. Complete sequence of Anabaena variabilis ATCC 29413. US DOE Joint Genome Institute, 2005. [Google Scholar]
- 36.Gaubig LC, Waldminghaus T, Narberhaus F. Multiple layers of control govern expression of the Escherichia coli ibpAB heat-shock operon. Microbiology. 2011;157:66–76. doi: 10.1099/mic.0.043802-0. [DOI] [PubMed] [Google Scholar]
- 37.Waldminghaus T, Gaubig LC, Klinkert B, Narberhaus F. The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 2009;6:455–63. doi: 10.4161/rna.6.4.9014. [DOI] [PubMed] [Google Scholar]
- 38.Chowdhury S, Ragaz C, Kreuger E, Narberhaus F. Temperature-controlled structural alterations of an RNA thermometer. J Biol Chem. 2003;278:47915–21. doi: 10.1074/jbc.M306874200. [DOI] [PubMed] [Google Scholar]
- 39.Roy SK, Hiyama T, Nakamoto H. Purification and characterization of the 16-kDa heat-shock-responsive protein from the thermophilic cyanobacterium Synechococcus vulcanus, which is an alpha-crystallin-related, small heat shock protein. Eur J Biochem. 1999;262:406–16. doi: 10.1046/j.1432-1327.1999.00380.x. [DOI] [PubMed] [Google Scholar]
- 40.Sato S, Ikeuchi M, Nakamoto H. Expression and function of a groEL paralog in the thermophilic cyanobacterium Thermosynechococcus elongatus under heat and cold stress. FEBS Lett. 2008;582:3389–95. doi: 10.1016/j.febslet.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 41.Roy SK, Nakamoto H. Cloning, characterization, and transcriptional analysis of a gene encoding an alpha-crystallin-related, small heat shock protein from the thermophilic cyanobacterium Synechococcus vulcanus. J Bacteriol. 1998;180:3997–4001. doi: 10.1128/jb.180.15.3997-4001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kojima K, Nakamoto H. Post-transcriptional control of the cyanobacterial hspA heat-shock induction. Biochem Biophys Res Commun. 2005;331:583–8. doi: 10.1016/j.bbrc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Kojima K, Nakamoto H. Specific binding of a protein to a novel DNA element in the cyanobacterial small heat-shock protein gene. Biochem Biophys Res Commun. 2002;297:616–24. doi: 10.1016/S0006-291X(02)02256-8. [DOI] [PubMed] [Google Scholar]
- 44.Nakamoto H. Molecular chaperones and stress tolerance in cyanobacteria. In: Srivastava AK, Rai AN, Neilan BA, eds. Stress Biology of Cyanobacteria Molecular Mechanisms to Cellular Responses: CRC Press, 2013:114-38. [Google Scholar]
- 45.Borbély G, Surányi G, Korcz A, Pálfi Z. Effect of heat shock on protein synthesis in the cyanobacterium Synechococcus sp. strain PCC 6301. J Bacteriol. 1985;161:1125–30. doi: 10.1128/jb.161.3.1125-1130.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webb R, Sherman LA. The cyanobacterial heat-shock response and the molecular chaperones. In: Bryant DA, ed. The Molecular Biology of Cyanobacteria Advances in Photosynthesis and Respiration. Dordrecht: Springer, 1994:751-67. [Google Scholar]
- 47.Rajaram H, Chaurasia AK, Apte SK. Cyanobacterial Heat Shock Response: Role and Regulation of Molecular Chaperones. Microbiology. 2014 doi: 10.1099/mic.0.073478-0. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe S, Sato M, Nimura-Matsune K, Chibazakura T, Yoshikawa H. Protection of psbAII transcript from ribonuclease degradation in vitro by DnaK2 and DnaJ2 chaperones of the cyanobacterium Synechococcus elongatus PCC 7942. Biosci Biotechnol Biochem. 2007;71:279–82. doi: 10.1271/bbb.60647. [DOI] [PubMed] [Google Scholar]
- 49.Nimura K, Yoshikawa H, Takahashi H. DnaK3, one of the three DnaK proteins of cyanobacterium Synechococcus sp. PCC7942, is quantitatively detected in the thylakoid membrane. Biochem Biophys Res Commun. 1996;229:334–40. doi: 10.1006/bbrc.1996.1802. [DOI] [PubMed] [Google Scholar]
- 50.Lubben TH, Donaldson GK, Viitanen PV, Gatenby AA. Several proteins imported into chloroplasts form stable complexes with the GroEL-related chloroplast molecular chaperone. Plant Cell. 1989;1:1223–30. doi: 10.1105/tpc.1.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanaka N, Nakamoto H. HtpG is essential for the thermal stress management in cyanobacteria. FEBS Lett. 1999;458:117–23. doi: 10.1016/S0014-5793(99)01134-5. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe S, Kobayashi T, Saito M, Sato M, Nimura-Matsune K, Chibazakura T, Taketani S, Nakamoto H, Yoshikawa H. Studies on the role of HtpG in the tetrapyrrole biosynthesis pathway of the cyanobacterium Synechococcus elongatus PCC 7942. Biochem Biophys Res Commun. 2007;352:36–41. doi: 10.1016/j.bbrc.2006.10.144. [DOI] [PubMed] [Google Scholar]
- 53.Kashino Y, Lauber WM, Carroll JA, Wang Q, Whitmarsh J, Satoh K, Pakrasi HB. Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry. 2002;41:8004–12. doi: 10.1021/bi026012+. [DOI] [PubMed] [Google Scholar]
- 54.Cheregi O, Sicora C, Kós PB, Barker M, Nixon PJ, Vass I. The role of the FtsH and Deg proteases in the repair of UV-B radiation-damaged Photosystem II in the cyanobacterium Synechocystis PCC 6803. Biochim Biophys Acta. 2007;1767:820–8. doi: 10.1016/j.bbabio.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 55.Singh AK, Summerfield TC, Li H, Sherman LA. The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch Microbiol. 2006;186:273–86. doi: 10.1007/s00203-006-0138-0. [DOI] [PubMed] [Google Scholar]
- 56.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–93. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. doi: 10.1046/j.1365-2958.1999.01166.x. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki I, Kanesaki Y, Hayashi H, Hall JJ, Simon WJ, Slabas AR, Murata N. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 2005;138:1409–21. doi: 10.1104/pp.104.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asadulghani SY, Suzuki Y, Nakamoto H. Light plays a key role in the modulation of heat shock response in the cyanobacterium Synechocystis sp PCC 6803. Biochem Biophys Res Commun. 2003;306:872–9. doi: 10.1016/S0006-291X(03)01085-4. [DOI] [PubMed] [Google Scholar]
- 60.Kojima K, Nakamoto H. A novel light- and heat-responsive regulation of the groE transcription in the absence of HrcA or CIRCE in cyanobacteria. FEBS Lett. 2007;581:1871–80. doi: 10.1016/j.febslet.2007.03.084. [DOI] [PubMed] [Google Scholar]
- 61.Balsiger S, Ragaz C, Baron C, Narberhaus F. Replicon-specific regulation of small heat shock genes in Agrobacterium tumefaciens. J Bacteriol. 2004;186:6824–9. doi: 10.1128/JB.186.20.6824-6829.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klinkert B, Elles I, Nickelsen J. Translation of chloroplast psbD mRNA in Chlamydomonas is controlled by a secondary RNA structure blocking the AUG start codon. Nucleic Acids Res. 2006;34:386–94. doi: 10.1093/nar/gkj433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agaisse H, Lereclus D. STAB-SD: a Shine-Dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol Microbiol. 1996;20:633–43. doi: 10.1046/j.1365-2958.1996.5401046.x. [DOI] [PubMed] [Google Scholar]
- 64.Gierga G, Voss B, Hess WR. The Yfr2 ncRNA family, a group of abundant RNA molecules widely conserved in cyanobacteria. RNA Biol. 2009;6:222–7. doi: 10.4161/rna.6.3.8921. [DOI] [PubMed] [Google Scholar]
- 65.Ehira S, Hamano T, Hayashida T, Kojima K, Nakamoto H, Hiyama T, Ohmori M, Shivaji S, Sato N. Conserved temperature-dependent expression of RNA-binding proteins in cyanobacteria with different temperature optima. FEMS Microbiol Lett. 2003;225:137–42. doi: 10.1016/S0378-1097(03)00503-2. [DOI] [PubMed] [Google Scholar]
- 66.Ehira S, Ohmori M, Sato N. Role of the 5′-UTR in accumulation of the rbpA1 transcript at low temperature in the cyanobacterium Anabaena variabilis M3. FEMS Microbiol Lett. 2005;251:91–8. doi: 10.1016/j.femsle.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 67.Krishna PS, Rani BR, Mohan MK, Suzuki I, Shivaji S, Prakash JS. A novel transcriptional regulator, Sll1130, negatively regulates heat-responsive genes in Synechocystis sp. PCC6803. Biochem J. 2013;449:751–60. doi: 10.1042/BJ20120928. [DOI] [PubMed] [Google Scholar]
- 68.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ames TD, Breaker RR. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011;8:82–9. doi: 10.4161/rna.8.1.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakurai I, Stazic D, Eisenhut M, Vuorio E, Steglich C, Hess WR, Aro EM. Positive regulation of psbA gene expression by cis-encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012;160:1000–10. doi: 10.1104/pp.112.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agrawal GK, Kato H, Asayama M, Shirai M. An AU-box motif upstream of the SD sequence of light-dependent psbA transcripts confers mRNA instability in darkness in cyanobacteria. Nucleic Acids Res. 2001;29:1835–43. doi: 10.1093/nar/29.9.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mitschke J, Georg J, Scholz I, Sharma CM, Dienst D, Bantscheff J, Voss B, Steglich C, Wilde A, Vogel J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A. 2011;108:2124–9. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci U S A. 2012;109:16306–11. doi: 10.1073/pnas.1207067109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol. 2009;74:1497–512. doi: 10.1111/j.1365-2958.2009.06949.x. [DOI] [PubMed] [Google Scholar]
- 76.Berghoff BA, Glaeser J, Nuss AM, Zobawa M, Lottspeich F, Klug G. Anoxygenic photosynthesis and photooxidative stress: a particular challenge for Roseobacter. Environ Microbiol. 2011;13:775–91. doi: 10.1111/j.1462-2920.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 77.Steglich C, Futschik ME, Lindell D, Voss B, Chisholm SW, Hess WR. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuhl H, Kruip J, Seidler A, Krieger-Liszkay A, Bunker M, Bald D, Scheidig AJ, Rögner M. Towards structural determination of the water-splitting enzyme. Purification, crystallization, and preliminary crystallographic studies of photosystem II from a thermophilic cyanobacterium. J Biol Chem. 2000;275:20652–9. doi: 10.1074/jbc.M001321200. [DOI] [PubMed] [Google Scholar]
- 79.Sambrook JE, Russel D. Molecular cloning: a laboratory manual, 3ed. New York: Cold Spring Harbor Laboratory Press, 2001. [Google Scholar]
- 80.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–6. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 81.Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 1994;13:3599–607. doi: 10.1002/j.1460-2075.1994.tb06667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol Chem. 2008;389:1319–26. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- 83.Willkomm DK, Minnerup J, Hüttenhofer A, Hartmann RK. Experimental RNomics in Aquifex aeolicus: identification of small non-coding RNAs and the putative 6S RNA homolog. Nucleic Acids Res. 2005;33:1949–60. doi: 10.1093/nar/gki334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 85.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695, 9. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.