SUMMARY
Hippo signaling limits organ growth by inhibiting the transcriptional coactivator Yorkie. Despite the key role of Yorkie in both normal and oncogenic growth, the mechanism by which it activates transcription has not been defined. We report that Yorkie binding to chromatin correlates with histone H3K4 methylation, and is sufficient to locally increase it. We show that Yorkie can recruit a histone methyltransferase complex, through binding between WW domains of Yorkie and PPxY sequence motifs of NcoA6, a subunit of the Trithorax-related (Trr) methyltransferase complex. Cell culture and in vivo assays establish that this recruitment of NcoA6 contributes to Yorkie’s ability to activate transcription. Mammalian NcoA6, a subunit of Trr-homologous methyltransferase complexes, can similarly interact with Yorkie’s mammalian homologue YAP. Our results implicate direct recruitment of a histone methyltransferase complex as central to transcriptional activation by Yorkie, linking the control of cell proliferation by Hippo signaling to chromatin modification.
INTRODUCTION
The key output of most signal transduction pathways is an alteration of the cellular transcription program, through activation or inhibition of transcriptional activators and repressors. The Hippo signaling pathway plays a crucial role in limiting organ growth, which it accomplishes by down-regulating the transcriptional coactivator protein Yorkie (Yki) (Oh and Irvine, 2010; Yu and Guan, 2013). Components of the Hippo pathway were first identified by the tumorous overgrowths caused by mutations in Drosophila Hippo pathway genes (Reddy and Irvine, 2008). Subsequent studies have identified many additional pathway components, described additional biological functions, and established that Hippo signaling and its role in controlling organ growth are highly conserved amongst metazoa (Harvey et al., 2013; Yu and Guan, 2013). However, the molecular mechanism by which Yorkie, or its mammalian homologues YAP and TAZ, actually activate transcription has remained elusive.
The core of the Hippo pathway comprises two kinases, Hippo and Warts, which are regulated through several upstream regulatory branches that collectively cause Hippo signaling to be affected by diverse inputs, including cell junctional complexes linked to cell polarity and cell contact, the actomyosin cytoskeleton, and growth factor and G protein signaling pathways (Staley and Irvine, 2012; Yu and Guan, 2013). Hippo and Warts act in sequence to inhibit Yki by phosphorylating it and promoting its cytoplasmic localization. As a transcriptional coactivator, Yki needs to interact both with DNA-binding proteins, and with proteins that effect transcriptional activation, and while several partners of Yorkie have been identified (Hong and Guan, 2012; Oh and Irvine, 2010; Oh et al., 2013), our understanding of transcriptional activation by Yki remains incomplete. For example, structure-function studies of Yki have identified a pair of conserved protein-protein interaction motifs, the two WW domains of Yki, as playing an absolutely essential, but ill-defined, role in transcriptional activation (Oh and Irvine, 2009; Zhang et al., 2009; Zhang et al., 2011; Zhao et al., 2009). A protein that can interact with these WW domains, Wbp2, was identified, but its contribution to the requirement for the WW domains in transcription remains unclear, and its down-regulation only modestly impaired Yki activity in vivo (Zhang et al., 2011).
The transcription of eukaryotic genes correlates with changes in chromatin structure (Kharchenko et al., 2010; Li et al., 2007). Chromatin of silenced genes is typically associated with methylation of histone H3 lysine 27 (H3K27), whereas chromatin of active genes is associated with methylation of H3K4. Different regions of a gene tend to have distinct methylation profiles, for example, H3K4 monomethylation (H3K4me1) around enhancers, H3K4me2 over the gene body, and H3K4me3 around promoters. H3K4 methylation is accomplished by conserved, multi-subunit complexes. Biochemical and genetic studies have defined three Drosophila H3K4 histone methyltransferase (HMT) complexes and revealed that they each have distinct essential functions (Eissenberg and Shilatifard, 2010; Shilatifard, 2012). Set1 acts as a global H3K4 HMT, as reduction of Set1 results in a general decrease in H3K4me3 levels (Ardehali et al., 2011; Hallson et al., 2012; Mohan et al., 2011). In contrast, Trithorax (Trx) and Trithorax-related (Trr) have more specialized functions. Trx has been implicated in homeotic gene regulation, whereas Trr has been linked steroid hormone signaling (Sedkov et al., 2003; Shilatifard, 2012). More recent studies have also identified a broad requirement for Trr in H3K4 mono-methylation (me1) (Herz et al., 2012; Kanda et al., 2013).
Although H3K4 methylation is correlated with transcriptional activation, the molecular mechanism by which this methylation is established and its causal relationship to transcription are complex (Ruthenburg et al., 2007; Shilatifard, 2012; Smith and Shilatifard, 2010). In yeast, and to some degree in metazoa, H3K4 methylation occurs as a consequence of transcription rather than a cause of transcription, and the major H3K4me3, Set1/COMPASS, can be recruited to actively transcribed genes through RNA polymerase associated factor. However, in animals there is evidence that H3K4 methylation also contributes to transcriptional activation. Components of the Trr and Trx HMTs, or their mammalian homologues MLL1-4, are required for the normal expression of some eukaryotic genes, presumably because H3K4 methylation, which is recognized by a structural motif (PHD) found in several proteins associated with histone modification and transcription, recruits proteins that promote transcriptional activation. Moreover, recent biochemical studies established that H3K4 methylation could promote transcription in in vitro assays (Jiang et al., 2013).
Here, we establish a direct link between Yki’s ability to activate transcription and its ability to recruit the Trr HMT complex. We show that Yki promotes H3K4 methylation, and that this occurs via direct physical association between Yki and NcoA6, a subunit of the Trr HMT complex. Moreover, our results indicate that recruitment of NcoA6 contributes to transcriptional activation by Yki. Our results thus identify a key molecular mechanism by which Yki activates transcription, and provide an illustration of the mechanism and central importance of H3K4 methylation to transcriptional activation by a major signal transduction pathway.
RESULTS
Yki induces H3K4 methylation
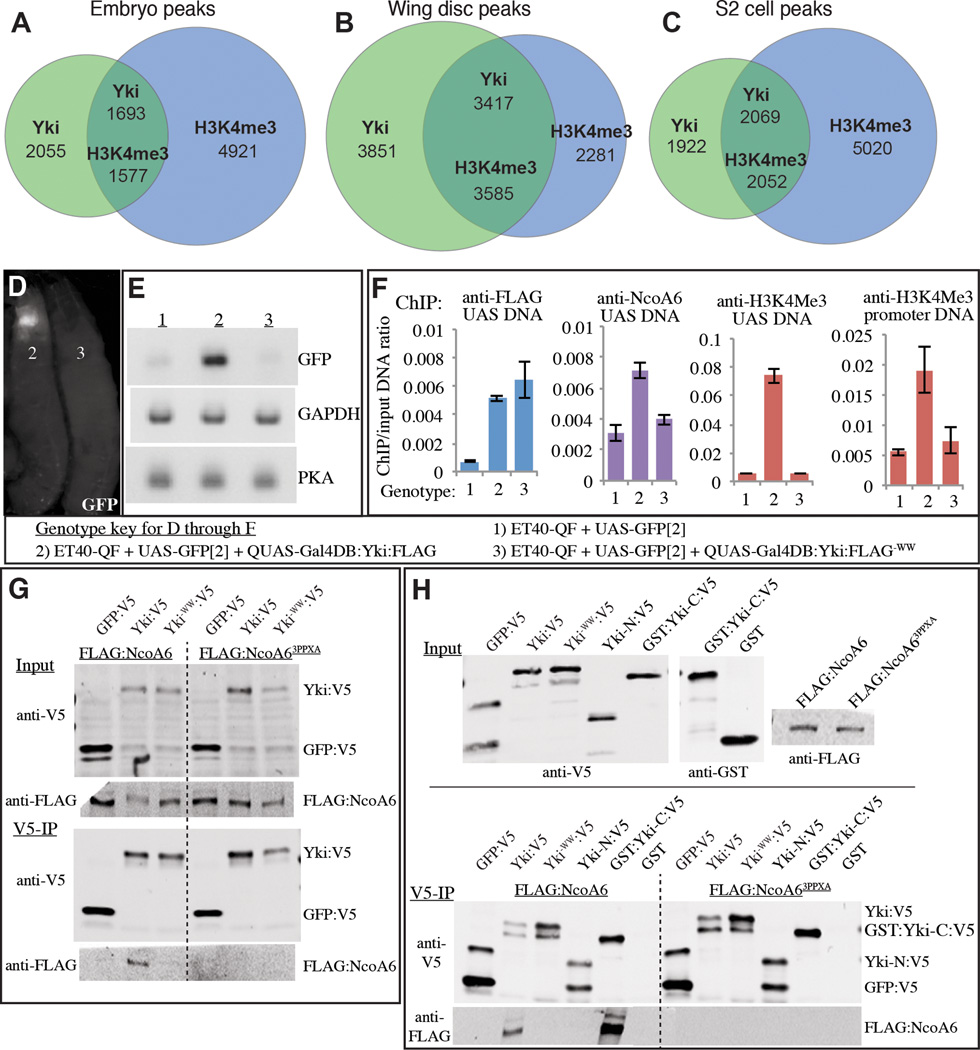
To investigate the transcriptional program initiated by Yki, we performed chromatin immunoprecipitation (ChIP). These experiments identified thousands of sites of Yki association with chromatin in Drosophila embryos (Oh et al., 2013), wing imaginal discs (Oh et al., 2013), and cultured S2 cells (this study). Analysis of the Yki-binding profile revealed substantial overlap between peaks of Yki association and peaks of H3K4me3 modification, as in each of these samples approximately 45–50% of Yki peaks overlap H3K4me3 peaks (Fig. 1A–C). Although it could be that Yki is preferentially targeted to H3K4me3 modified chromatin, or that Yki binding and H3K4me3 modification are regulated independently, this correlation raised the possibility that H3K4me3 modification is induced by Yki.
Figure 1. Induction of H3K4me3 by Yki.
(A–C) Venn diagrams showing overlap between Yki binding and H3K4 trimethylation (H3K4Me3) as revealed by genome-wide ChIP-seq analyses in embryos (A), wing discs (B) and S2 cells (C). Numbers of peaks are indicated; numbers in the overlap differ because a peak for one protein can overlap two peaks of the other. (D–F) Transcriptional activation and induction of H3K4Me3 by Yki. Gal4DB:Yki:FLAG and Gal4DB:Yki−WW:FLAG were expressed from QUAS transgenes using a ET40-QF driver, which is expressed in the imaginal discs and brain, genotypes are indicated by the key. (D,E) Shows expression of UAS-GFP in the 3rd instar larval discs and brain by GFP fluorescence (D) and reverse-transcription-PCR (E). (F) Yki and NcoA6 binding, and H3K4Me3 status around the UAS and transcription start sites, histograms show the results of ChIP-qPCR (average ChIP/Input ratio from triplicate qPCR, error bars indicate standard deviation). (G) Western blots showing coimmunoprecipitation of Yki:V5 or Yki−WW:V5 with Flag:NcoA6 or Flag: NcoA6−3PPxA from S2 cell extracts (GFP is a negative control). Two upper panels (Input) show blots on lysates, and two lower panels (V5-IP) show blots (anti-V5 and anti-Flag) on material precipitated by anti-V5 beads. (H) Western blot showing results of coimmunoprecipitation using bacterially expressed Yki:V5 and its mutated or fragmented forms, immobilized on V5-agarose beads, and incubated with S2 cell nuclear extracts from cells expressing Flag:NcoA6 or Flag:NcoA6−3PPxA. GFP:V5 and GST are negative controls. Upper panels show input material, lower panels show blots on material precipitated by anti-V5 beads. See also Fig. S1, Table S1.
To examine whether localization of Yki could be sufficient to promote H3K4me3, we combined the UAS-Gal4 and Q systems (Potter et al., 2010) to create a novel site of Yki localization on chromosomes. A fusion protein combining full-length Yki with the DNA binding domain of Gal4 (Gal4DB:Yki) was constructed, and then expressed in Drosophila imaginal tissues from a QUAS transgene under ET40-QF control. Gal4DB:Yki could activate transcription from a UAS-GFP transgene in vivo (Fig. 1D,E). Measurement of H3K4me3 levels by ChIP-PCR revealed a significant increase surrounding the UAS and promoter regions of UAS-GFP when Gal4DB:Yki was expressed (Fig. 1F). Thus, targeting Yki to a novel chromosomal locus is sufficient to locally increase H3K4me3.
Structure-function studies of Yki have implicated a pair of conserved protein-protein interaction motifs, the two WW domains of Yki, as playing an essential role in transcriptional activation (Oh and Irvine, 2009; Zhang et al., 2009; Zhang et al., 2011; Zhao et al., 2009). To investigate the mechanism by which Yki induces H3K4me3, we created and characterized a mutant isoform, Gal4DB:Yki−WW, which contains point mutations in these WW domains. As expected, Gal4DB:Yki−WW failed to activate transcription of UAS-GFP (Fig. 1D,E). Moreover, quantitative ChIP-PCR confirmed that Gal4DB:Yki and Gal4DB:Yki−WW bound similarly to UAS-GFP, but Gal4DB:Yki−WW failed to enhance H3K4me3 modification (Fig. 1F). Thus, the WW domains of Yki are required for its ability to induce H3K4me3.
Yki binds NcoA6 through WW domain – PPxY interactions
In principle the requirement for WW domains in promoting H3K4me3 could either be a cause, or a consequence, of the requirement for the WW domains in Yki-promoted transcription. WW domains interact with a short sequence motif, PPxY, hence the requirement for WW domains implies that Yki must partner with a protein containing PPxY motifs to activate transcription. To investigate the possibility that Yki recruits histone-methylating complexes through its WW domains, we searched for PPxY sequence motifs within all of the subunits of the three Drosophila H3K4 HMT complexes (Mohan et al., 2011; Shilatifard, 2012). Only one protein, NcoA6, a component of the Trr HMT complex, contains multiple PPxY motifs (Fig. S1), which in our experience is usually required for strong binding to proteins with WW domains.
To assay for potential binding between Yki and NcoA6, epitope-tagged isoforms were co-expressed in S2 cells. FLAG:NcoA6 was specifically precipitated by Yki:V5, and not a control protein, GFP:V5 (Fig. 1G). Direct physical interaction between Yki and NcoA6 was confirmed by co-precipitation using bacterially-expressed Yki, and this binding activity mapped to the C-terminal half of Yki, which contains the WW domains (Fig. 1H). Moreover, mutation of either the WW domains of Yki, or the PPxY motifs of NcoA6, abolished detectable binding between them, both in S2 cells and using bacterial lysates, confirming that Yki and NcoA6 proteins interact through their WW and PPxY motifs (Fig. 1G,H). Moreover, ChIP revealed that wild-type, but not WW mutant, Gal4DB:Yki could recruit NcoA6 to UAS-GFP chromatin in vivo (Fig. 1F).
NcoA6 and Trr contribute to transcriptional activation by Yki
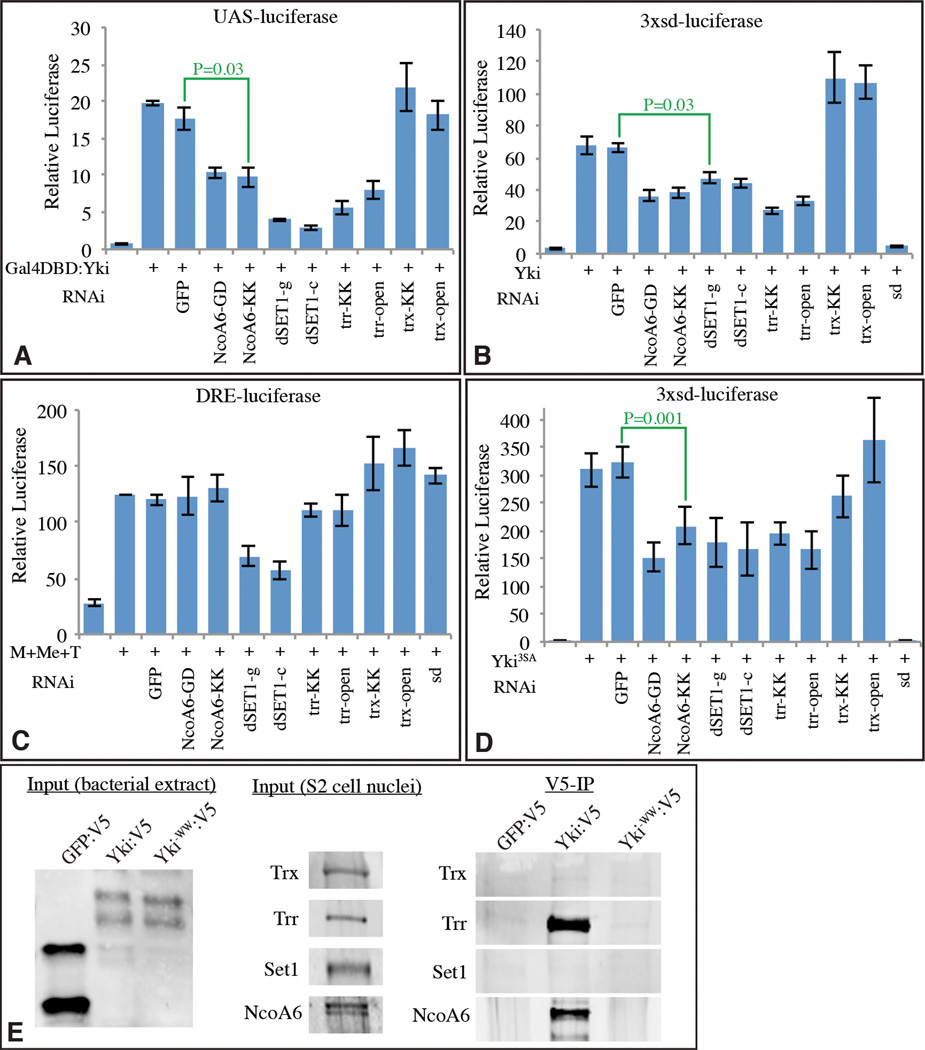
To evaluate the potential functional requirement for NcoA6 in Yki-mediated transcription, we employed transcriptional reporter assays in S2 cells. Expression of Gal4DB:Yki increases transcription of a UAS-luciferase reporter (Fig. 2A)(Huang et al., 2005; Oh and Irvine, 2009). RNAi-mediated downregulation of NcoA6, using either of two distinct dsRNAs, decreased this UAS-luciferase expression (Fig. 2A, effectiveness of RNAi in Supplementary Fig. S2). Moreover, activation of a distinct reporter to which Yki is recruited by its DNA-binding partner Scalloped (Sd), 3xSd-luciferase (Zhang et al., 2008), is also reduced by NcoA6 RNAi (Fig. 2B). Conversely, a Yki-independent reporter, the Ubx-DRE (Oh and Irvine, 2011), was not affected by NcoA6 RNAi (Fig. 2C). To rule out the possibility that NcoA6 influences Yki through an effect on Hippo signaling, we repeated assays with the 3xSd luciferase reporter using an activated form of Yki (Yki3SA), which contains mutations in all three Warts phosphorylations sites (Oh and Irvine, 2009). Yki3SA-induced transcription was similarly reduced by knockdown of NcoA6 (Fig. 2D), implying that NcoA6 acts downstream of Hippo pathway regulation. This is consistent with previous studies of the requirement for WW domains, as mutation of the WW domains eliminates the transcriptional activity of both wild-type and activated forms of Yki, without preventing its nuclear localization (Oh and Irvine, 2009).
Figure 2. NcoA6 and Trr are required for transcriptional activation by Yki.
Histograms showing results of luciferase assays (depicted as average firefly/renilla ratio from triplicate experiments, where error bars indicate SD) using (A) UAS-luciferase, (B, D) 3XSd-luciferase, or (C) DRE-luciferase reporters in S2 cells transfected to express Gal4DB:Yki, Yki, TkvQ235D (T), Mad (M), or Medea (Me) as indicated. Double-stranded RNAs for RNAi against the specific genes were also added as indicated; the abbreviations after the gene symbol identify distinct the dsRNAs used for each gene. P values for statistical significance obtained from pairwise t tests for sample comparisons are indicated in green. E) Physical interaction between HMT complexes and Yki. Yki:V5, Yki−WW:V5, or, as a control, GFP:V5 were purified from bacterial lysates on anti-V5 beads and mixed with nuclear extracts from S2 cells. Interaction of endogenous NcoA6 and Trr with Yki was detected, but no interaction with Set1 or Trx was observed. See also Fig. S2
NcoA6 is specifically associated with Trr HMT complexes (Mohan et al., 2011). In S2 cells, knockdown of Trr reduced the transcription of Yki-activated reporters similarly to NcoA6 knockdown (Fig. 2A–D), consistent with the expectation that the requirement for NcoA6 reflects its participation within Trr HMT complexes. For comparison, we also examined the consequences of reduction of Trx and Set1. Trx RNAi had no affect on Yki-dependent reporters (Fig. 2A–D). Knock-down of Set1 lowered the transcription of Yki-dependent reporters, but also reduced a Yki-independent reporter (Fig. 2A – D), which could reflect its global affects on H3K4me3. These functional studies were complemented by binding experiments, which assayed the ability of bacterially expressed Yki to co-precipitate components of each of the H3K4 HMT complexes from S2 cell nuclear extracts. Both NcoA6 and Trr, but not SET1 or Trx, could be specifically co-precipitated by Yki, and this co-precipitation required Yki’s WW domains (Fig. 2E).
Recruitment of NcoA6 is sufficient to account for transcriptional activation by Yki
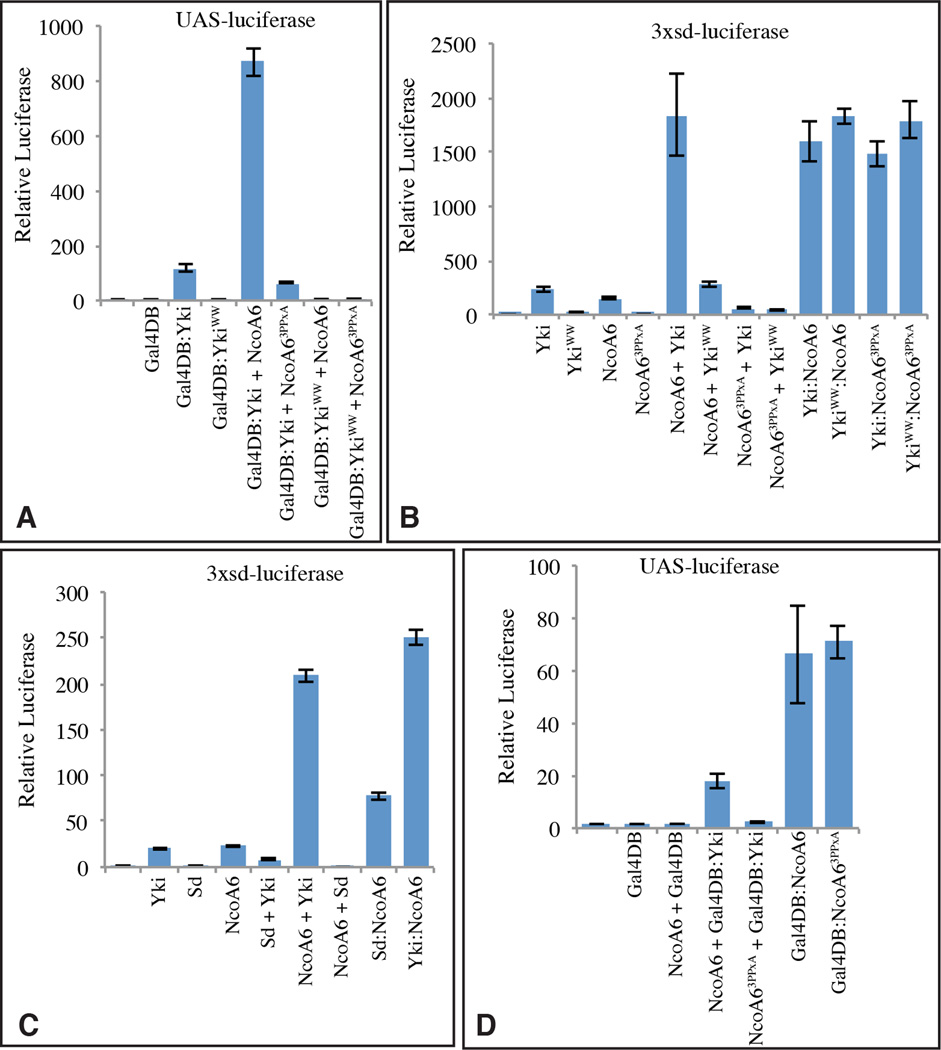
A role for NcoA6 in transcriptional activation by Yki was further supported by the observation that over-expression of NcoA6 could stimulate transcription of Yki reporters in S2 cells, including a several fold-increase in expression of UAS-luciferase by Gal4DB:Yki, and a similar increase in expression of 3xSd-luciferase by Yki (Fig. 3A,B). This cooperative increase in transcription associated with Yki and NcoA6 co-transfection was dependent upon both the WW domains of Yki, and the PPxY motifs of NcoA6 (Fig. 3A,B), implying that it depends upon direct physical interaction between them.
Figure 3. Recruitment of NcoA6 is sufficient for transcriptional activation by Yki.
Histograms showing results of luciferase assays (depicted as average firefly/renilla ratio from triplicate experiments, where error bars indicate SD) using (A,D) UAS-luciferase, (B, C) 3XSd-luciferase reporters in S2 cells transfected to express the indicated proteins. See also Fig. S3
The central role of NcoA6 recruitment in Yki’s ability to activate transcription was confirmed by characterization of a series of Yki:NcoA6 fusion proteins. Expression of a Yki:NcoA6 fusion protein strongly activated expression of 3xSd-luciferase, comparable to the expression induced by co-expression of Yki and NcoA6 (Fig. 3B). Strikingly, transcriptional activation induced by Yki:NcoA6 fusion proteins no longer depends upon either the WW domains of Yki, or the PPxY motifs of NcoA6, as these motifs could be mutated within the fusion protein without impairing transcriptional activation (Fig. 3B). Thus, direct interaction with NcoA6 is both necessary and sufficient to account for the absolute requirement for the WW domains of Yki in transcriptional activation.
The observations described above imply that recruitment of NcoA6 is essential to transcriptional activation by Yki. We next investigated the sufficiency of NcoA6 recruitment to Yki’s transcriptional activation function. If Yki’s key role in transcription is to act as an adapter protein, linking the Trr HMT complex to DNA binding proteins, then direct fusion of NcoA6 to a DNA binding partner might bypass the requirement for Yki. This was tested by creating a Sd:NcoA6 fusion protein. Indeed, even though over-expression of Sd alone normally represses transcription, presumably due to interaction with co-repressors (Koontz et al., 2013), Sd:NcoA6 robustly activated transcription from the 3xSd-luciferase reporter even without addition of Yki (Fig. 3C). Thus, as long as NcoA6 is recruited, Yki was not required for transcription from this Yki-dependent reporter gene.
The PPxY motifs of NcoA6 are required for transcriptional activation on Yki-dependent reporters (Fig. 3A,B). However, they were no longer required when NcoA6 was directly fused to Yki (Fig. 3B). To confirm that these PPxY motifs are only required for recruitment of NcoA6 to DNA via Yki, and not for its ability to activate transcription, we created Gal4DB:NcoA6 fusion proteins. Indeed, both wild-type and PPxA mutant isoforms of Gal4DB:NcoA6 were potent activators of transcription of a UAS-luciferase reporter (Fig. 3D).
Co-localization of Yki and Trr on chromosomes
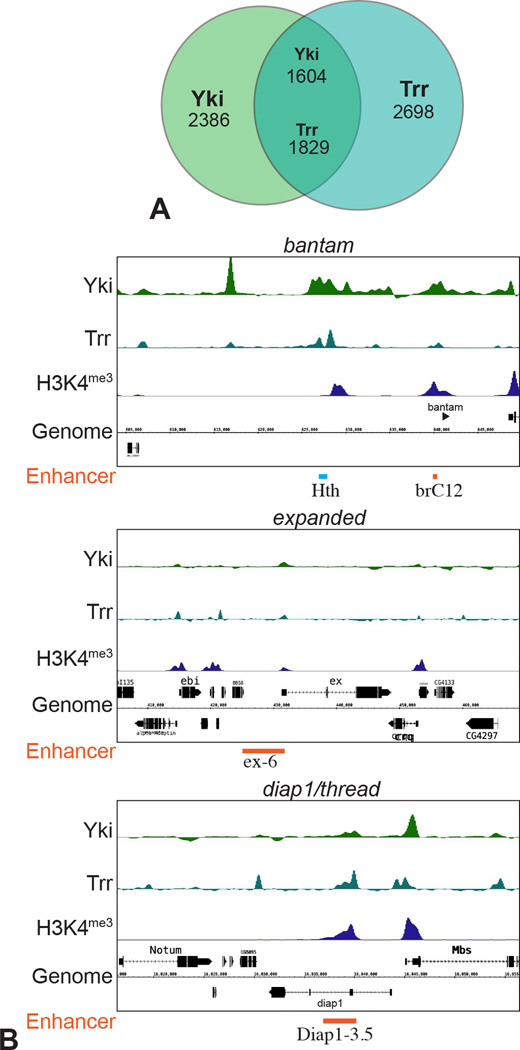
As NcoA6 is a conserved component of the Trr HMT complex, Yki-NcoA6 interaction should recruit Trr in vivo. To investigate this, we compared available ChIP-seq data describing the chromosomal localization of Trr in S2 cells (Herz et al., 2012) to ChIP-seq data we generated describing the chromosomal localization of Yki in S2 cells. A substantial overlap between these binding peaks was observed, as roughly 40% of all peaks across the genome overlap (Fig. 4A). Moreover, overlap between Yki and Trr peaks at specific known Yki target genes, including bantam, expanded, and Diap1 (thread), was identified, and these Yki/Trr peaks also overlap both functionally characterized Yki-regulated enhancers, and H3K4me3 peaks (Fig. 4B). Thus, analysis of the chromosomal localization of Yki and Trr provides further support for a functional connection between them. As Trr has previously been found to interact functionally and physically with the Drosophila ecdysone receptor (Johnston et al., 2011; Sedkov et al., 2003), and mammalian homologues of Trr interact with multiple mammalian steroid hormone receptors (Goo et al., 2003; Lee et al., 2006; Vicent et al., 2011), Trr binding that does not overlap Yki likely reflects association of Trr with other transcription factors.
Figure 4. Co-localization of Yki and Trr on chromatin.
(A) Overlap between Yki- and Trr-binding sites, where numbers indicate the numbers of peaks. Numbers in the overlap differ because one peak for one protein can overlap two peaks of the other. (B) Plot of ChIP peaks at three loci (bantam, expanded and diap1/thread) regulated by Yki. Yki-responsive enhancers that have been identified at these loci (Oh and Irvine, 2011; Oh et al., 2013; Zhang et al., 2008) are indicated in orange at the bottom, and a previously identified Hth- and Yki-binding region (Peng et al., 2009) is indicated in light blue. See also Fig. S4
In vivo requirement for NcoA6 in Yki-dependent growth and transcription
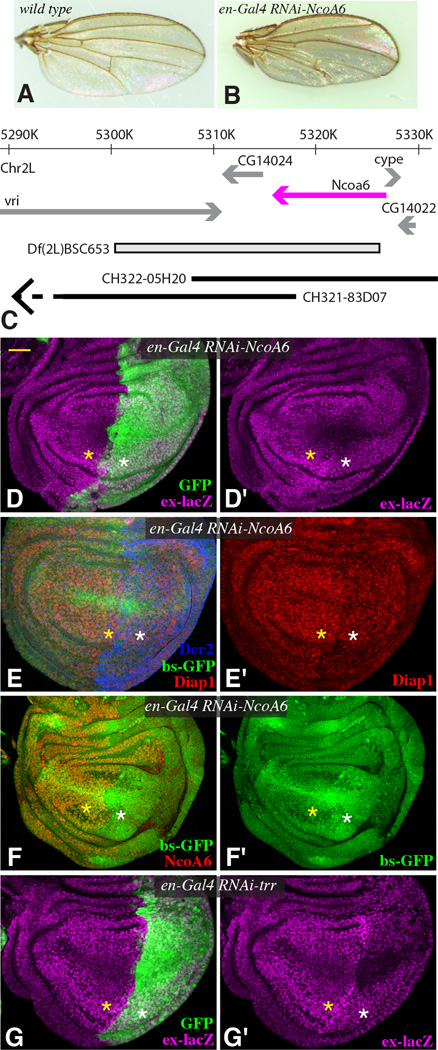
If NcoA6 is required for Yki’s transcriptional activity in vivo, then like Yki it should be required for cell proliferation and cell survival. Indeed, RNAi-mediated down-regulation of NcoA6 in developing wings or eyes reduced wing or eye size, and induced apoptosis, similar to loss of yki (Figs 5A,B, S3, S4). Moreover, eye overgrowth induced by activation of Yki was suppressed by reduction of NcoA6 (Fig S3D). Thus, NcoA6 is required for Yki-promoted growth.
Figure 5. in vivo requirement for NcoA6.
(A,B) Adult female wings from (A) en-Gal4 (wild type control) (B) en-Gal4; UAS-RNAi-NcoA6 [TRiP.HMS00664]. C) Map of the genomic region surrounding NcoA6 (magenta), indicating DNA deleted by Df(2L)BSC653 and included within BAC clones CH322-05H20 and CH321-83D07 (the left end of this clones extends beyond the region shown). (D–G) Influence of Trr HMT complex on Yki target genes in wing discs. Projections through five confocal sections of wing discs; panels marked by prime symbols show separated channels. Yellow asterisks identify regions with normal gene expression, and white asterisks identify regions with expression of RNAi lines and altered target gene expression. Scale bar (top left, yellow) = 32 µm. (D,G) en-Gal4 UAS-GFP ex-lacZ UAS-Dcr2, with (D) UAS-RNAi-NcoA6 [vdrc36480] (G) UAS-RNAi-trr[TRiP.JF03242] showing expression of ex-lacZ (magenta) and with posterior cells marked by GFP (green). (E,F) en-Gal4 bs-GFP; UAS-Dcr2 and with UAS-RNAi-NcoA6 [TRiP.HMS00664] incubated on 25°C (E) and 29°C (F), showing expression of Diap1 (red) and bs-GFP (green), and with posterior cells marked by Dcr2 (blue, in E) or lack of NcoA6 (red, in F). See also Fig. S5
As we were not able to obtain null alleles of NcoA6, we combined Bac clones with a chromosomal deficiency to confirm the genetic requirement for NcoA6 and its ability to interact with Yki. Df(2L)BSC653 removes NcoA6 and two neighboring genes (Fig. 5C). The BAC clones CH322-05H20 and CH321-83D07 together encompass all of the DNA deleted by Df(2L)BSC653, with CH322-05H20 including NcoA6 and CH321-83D07 including neighboring genes (Fig. 5C). Using recombineering (Venken et al., 2009), HA-tagged wild-type and PPxA mutant versions of NcoA6 were created within CH322-05H20 and transformed into Drosophila. Df(2L)BSC653 is embryonic lethal, and this lethality was fully rescued by the combination of CH322-05H20 and CH321-83D07. However, the absence of CH322-05H20, which corresponds genetically to specific deletion of NcoA6, was embryonic lethal, confirming that NcoA6 is an essential gene. The CH322-05H20 clone expressing the PPxA mutant version of NcoA6 also failed to rescue NcoA6 deletion, and these animals died at or before the first larval instar. Since the activity of NcoA6 on transcriptional reporters only required PPxY motifs for interaction with Yki (Fig. 4), this observation implies that NcoA6 interaction with Yki is essential in vivo. The PPxA mutant also exhibited a dominant-negative effect, as wild-type flies homozygous for CH322-05H20 encoding PPxA mutant NcoA6, but not wild-type CH322-05H20, have increased apoptosis in the wing disc, which is consistent with impaired Yki activity (Fig. S4I,J).
To further assess requirements for NcoA6 and Trr in Yki activity in vivo, we used en-Gal4 (drives expression in posterior cells) or AyGal4 (drives expression in clones) to down-regulate NcoA6 or Trr by expression of UAS-RNAi constructs. Three different Yki target genes were examined: bantam (a micro-RNA gene whose expression is revealed by an inverse sensor, bs-GFP), thread (revealed by antibody staining for its protein product, Diap1), and expanded (revealed by an ex-lacZ reporter). Expression of each of these Yki targets was reduced by RNAi of NcoA6 or Trr, using either of two independent UAS-RNAi transgenes (Figs 5,S3). Reduction of NcoA6 or Trr could also suppress the effects of Yki activation on these transgenes (Fig S5).
Conversely, the other two Drosophila HMT complexes were not detectably required for Yki activity in vivo, as neither Trx nor Set1 RNAi suppressed activated-Yki phenotypes (Fig S5). Antibody staining confirmed that these RNAi lines were effective, as Set1 RNAi reduced total nuclear levels of H3K4me3 (Fig. S3)(Mohan et al., 2011), and Trx RNAi reduced anti-Trx staining (Fig S4A,B). Thus, we infer that Yki-promoted transcription is specifically dependent upon the Trr HMT complex. It has recently been reported that mutation or knockdown of Trr could increase growth in some circumstances, while decreasing growth or inducing apoptosis in others (Kanda et al., 2013). We observed decreases in growth and induction of apoptosis when Yki, NcoA6, or Trr were knocked down by RNAi in wing discs (Fig. S4), but our results do not exclude the possibility that in some contexts Trr could be recruited by other transcription factors that inhibit growth.
We could also detect the reported general requirement for Trr in H3K4me1 in wing discs (Herz et al., 2012; Kanda et al., 2013), but could not detect an effect of NcoA6 or Yki knockdown on total H3K4me1 levels, even though their levels were reduced by RNAi (Fig. S4 and data not shown). A substantial fraction of Yki-bound loci are also associated with elevated H3K4me1, especially at loci lacking elevated H3K4me3 (Table S1). Trr has been implicated in induction of both H3K4me1 and H3K4me3, and we infer that Yki-mediated recruitment of NcoA6 has the potential ability to locally increase H3K4 methylation through recruitment of a Trr complex, which depending on the context may result preferentially in H3K4me1 or H3K4me3.
Interaction of YAP with NCOA6 in human cells
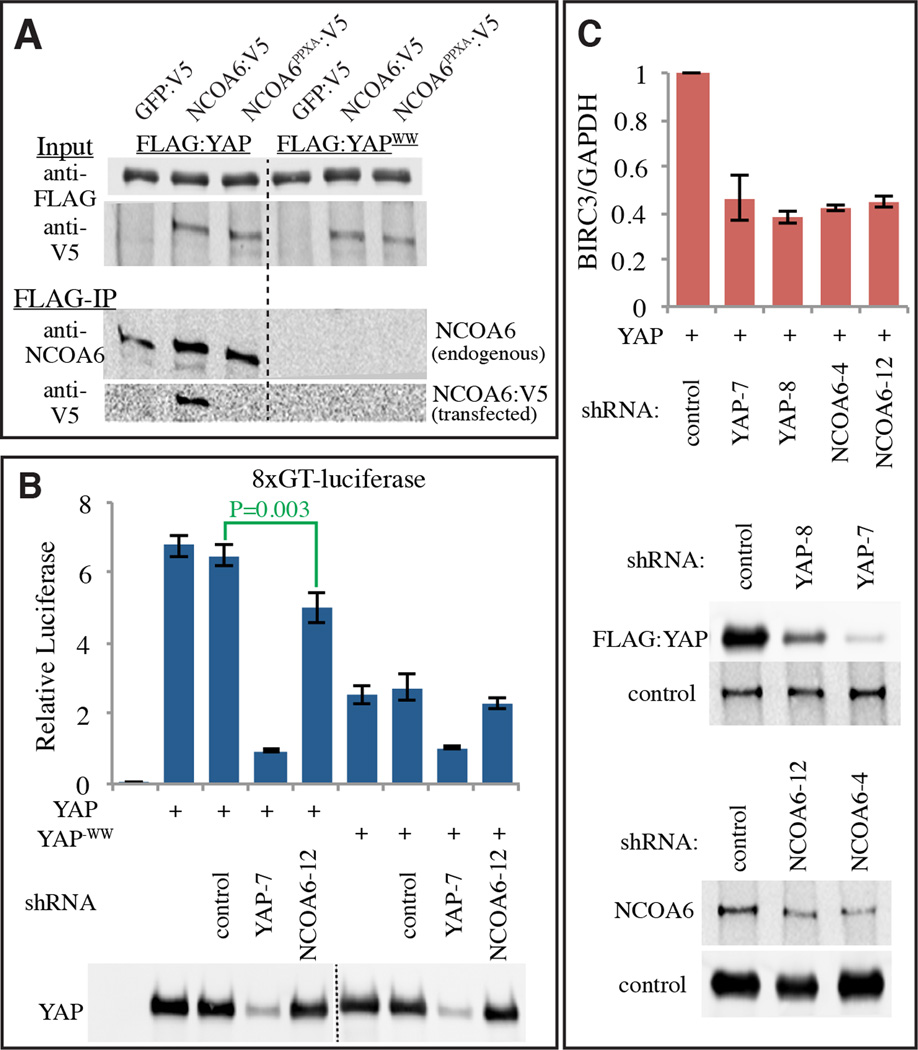
To investigate whether Yki’s mammalian homologue YAP could interact with human NCOA6, we expressed epitope-tagged proteins in HEK-293T cells. Physical association of NCOA6 with YAP was confirmed by their co-precipitation. Moreover, this association depends upon both the WW domains of YAP, and the PPxY motifs of NCOA6 (Fig. 6A). Thus, Yki/YAP transcriptional coactivators have an evolutionarily conserved ability to interact directly with NcoA6, a subunit of a conserved H3K4 HMT complex (Drosophila Trr, mammalian MLL2/3). YAP and TAZ also have WW domains that can contribute to transcriptional activation, although prior studies have yielded mixed results in their assessment of the requirement for these WW domains (Chan et al., 2011; Zhang et al., 2012; Zhang et al., 2009; Zhao et al., 2009). We found that in HEK-293T cells under non-confluent conditions, wild-type YAP stimulates transcription more than a YAP−WW mutant does (Fig 6B). The lack of an absolute requirement for the WW domains in YAP might reflect the presence of an additional activation domain in YAP that is not conserved in Drosophila Yki, and which appears to be partially redundant with the WW domains (Zhang et al., 2012). Nonetheless, reduction of NCOA6 by co-transfection of shRNAs decreased transcriptional activation by YAP, both as assayed using a luciferase reporter construct (Fig. 6B), and by examination of mRNA levels of a known YAP target (Fig. 6C). We also note that a recent ChIP-seq study (Lee et al., 2013) identified binding sites for TEAD4 (a mammalian homologue of Sd and major binding partner for YAP) as enriched at genomic loci bound by the Trr homologue MLL2 (also known as MLL4). Together these observations suggest that direct interaction with MLL HMT complexes contributes to transcriptional activation by YAP in mammals.
Figure 6. Mammalian NcoA6 interacts with YAP.
(A) Western blots showing coimmunoprecipitation of Flag:YAP or Flag:YAP-WW with hNcoA6:V5 or hNcoA6-3PPxA:V5 from HEK-293T total extracts (GFP:V5 is a negative control). Two upper panels (Input) show blots on total lysates (anti-Flag and anti-V5), and two lower panels (V5-IP) show blots (anti-hNcoA6 and anti-V5) on material precipitated by anti-V5 beads. (B) Histogram showing results of luciferase assays (depicted as average firefly/renilla ratio from triplicate experiments, where error bars indicate SD) using TEAD/YAP-luciferase reporter (8XGTIIC-luc) in HEK-293T cells transfected to express Flag:YAP or Flag:YAP-WW, as indicated. shRNA constructs for RNAi against the specific genes were also added as indicated. P values for statistical significance obtained from pairwise t tests for sample comparisons are indicated in green. C) Histogram showing results of quantitative RT-PCR analysis of a YAP target gene, BIRC3, (depicted as average BIRC3/GAPDH ratio from triplicate experiments, normalized to the ratio in control samples, where error bars indicate SD) in HEK-293T cells transfected to express Flag:YAP and shRNA constructs for RNAi against the indicated genes. Western blots in lower panels show effectiveness of shRNAs in reducing protein levels.
DISCUSSION
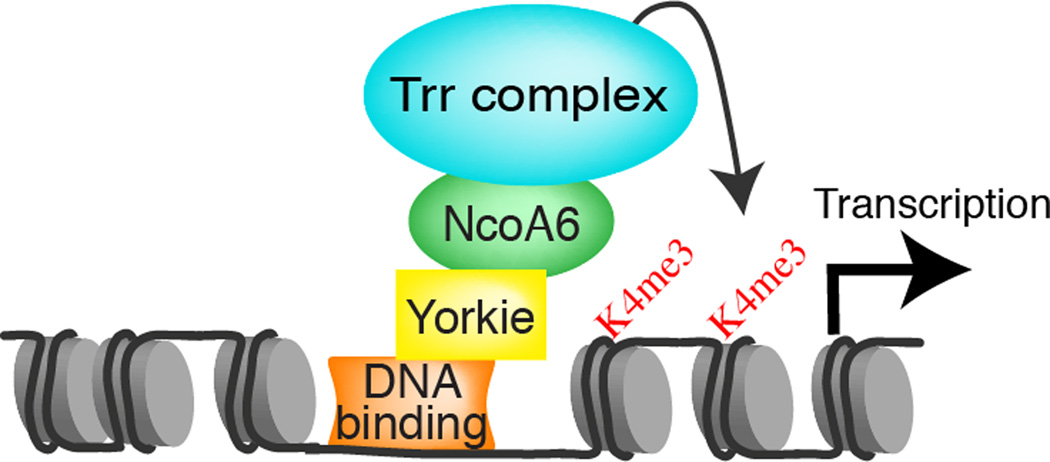
Transcriptional activators increase transcription through recruitment of transcriptional proteins or through chromatin modification. Each of these encompasses a wide range of specific mechanisms, including interaction with core subunits of RNA polymerase, interaction with Mediator proteins, interaction with chromatin remodeling complexes, or interaction with complexes that influence post-translational modifications of histones, such as acetylation or methylation. We and others have previously observed that Yki and YAP could interact with Mediator subunits, ATP-dependent chromatin remodeling complexes, and other transcription factors such as GAGA (Bayarmagnai et al., 2012; Oh et al., 2013; Varelas et al., 2008). Nonetheless, based on the results described here we argue that a key mechanism by which Yki activates transcription is increasing H3K4 methylation through recruitment of the Trr HMT complex (Fig. 7). Most notably, point mutations in Yki that specifically impair its ability to interact with NcoA6 abolish its transcriptional activity, and this transcriptional activity is restored by fusion with NcoA6. Moreover, the essential role of Yki as a transcriptional coactivator for its DNA binding partner Sd can be bypassed by fusing NcoA6 directly with Sd.
Figure 7. Schematic of the Yki transcriptional activation complex.
A diagram depicting recruitment of Trr HMT complexes to the promoter region of Yki target genes, which is mediated by direct physical interaction between Yki and NcoA6. As discussed in the text, this results in increased H3K4 methylation, and consequently increased transcription.
Our observations tie Yki’s transcriptional activity most directly to NcoA6, and the argument that this reflects a necessary and sufficient role for H3K4 methylation in transcriptional activation by Yki rests in part on the identity of NcoA6 as a component of the Trr HMT complex (Mahajan and Samuels, 2008; Shilatifard, 2012). This argument receives further support from several additional observations: the strong, genome wide correlation between Yki’s association with chromatin and H3K4 methylation, the increased H3K4 methylation when Yki competent to interact with NcoA6 is targeted to a novel chromosomal location, the similar decreases in expression of Yki target genes when either NcoA6 or Trr are reduced by RNAi in cultured cells or in vivo, and the recent biochemical demonstration that H3K4 methylation of chromatin by MLL2, a Trr-homologous complex in mammals, could increase transcription in in vitro assays (Jiang et al., 2013).
NcoA6 and Trr have previously been linked to transcriptional activation by nuclear hormone receptors (Mahajan and Samuels, 2008; Sedkov et al., 2003; Shilatifard, 2012). NcoA6 is believed to play an analogous role in transcriptional activation by nuclear receptors, i.e. its direct binding to these transcription factors recruits the Trr HMT complex, or its mammalian homologues. However, a distinct structural motif (LxxLL, Fig. S1) within NcoA6 mediates interactions with nuclear receptors (Mahajan and Samuels, 2008). Thus, NcoA6 appears to act as a multi-functional adapter protein that can link different classes of transcriptional activators to Trr/MLL2/3 HMT complexes, which as we establish here are involved not only in transcriptional activation induced by nuclear receptors, but also by Yki and its mammalian homologues (Fig. S1). Crosstalk between Hippo signaling and other pathways has been observed at the level of transcription factors (Attisano and Wrana, 2013; Irvine, 2012), including physical interactions between Yki, YAP and TAZ, and β-catenin and SMADs, which are transcriptional effectors of Wnt and BMP signaling, respectively. Thus, our observations raise the possibility that Trr-dependent H3K4 methylation could also contribute to transcriptional activation by these pathways.
In humans, NCOA6 has been identified as a gene commonly amplified and overexpressed in breast, colon, and lung cancers (it is also known as Amplified in breast cancer 3)(Guan et al., 1996; Lee et al., 1999; Mahajan and Samuels, 2008). In mice, gene targeted mutations have implicated NcoA6 in promoting growth during development and wound healing (Antonson et al., 2003; Kuang et al., 2002; Mahajan et al., 2004; Zhu et al., 2003). These roles in promoting growth are reminiscent of YAP, which is similarly required for growth during embryonic development and wound repair, and linked to these cancers when amplified or activated (Harvey et al., 2013; Hong and Guan, 2012). Thus, while functional studies linking mammalian NCOA6 to cell survival, growth, and cancer have previously been interpreted as a reflection of its role as a coactivator of transcription mediated by nuclear hormone receptors, our results, together with analysis of MLL2 binding by Chip-Seq (Lee et al., 2013), argue that at least part of its effects reflect its role as a cofactor of YAP.
A notable feature of Hippo signaling is the recurrence of WW domains or PPxY motifs in multiple pathway components (Oh and Irvine, 2010; Salah and Aqeilan, 2011). Within Yki, YAP and TAZ, the WW domains serve a dual role. They facilitate inhibition, as major negative regulators, including Warts/Lats, Expanded (in Drosophila) and Angiomotin (in mammals), utilize PPxY motifs to bind Yki, YAP and TAZ and promote their cytoplasmic localization. Conversely, they also facilitate activation, both through binding to Wbp2 (Chan et al., 2011; Zhang et al., 2011), and as we show here, NcoA6. It seems unlikely to be coincidental that key positive and negative partners of Yki/YAP/TAZ bind the same structural motifs. Rather, this shared recognition of the same motifs may have evolved to ensure tight on/off regulation of Yki/YAP/TAZ-dependent transcription.
EXPERIMENTAL PROCEDURES
Fly Stocks and crosses
Previously described mutations and transgenes used include tub-EGFP:2Xanti-bantam (bs-GFP) (Brennecke et al., 2003), ex-lacZ (Blaumueller and Mlodzik, 2000), UAS-YkiS168A:GFP (Oh and Irvine, 2008), UAS-YkiS250A:V5 (Oh and Irvine, 2009), GMR-Gal4 (Bloomington 1104), en-Gal4 (Bloomington), UAS-Dcr2 (VDRC 60008 and 60009), ET40-QF (Potter et al., 2010), UAS-wts-RNAi (VDRC 9928), UAS-RNAi-NcoA6 (Bloomington 34964 and VDRC 36480), UAS-RNAi-trr (Bloomington 29563 and 36916), UAS-trx-RNAi (Bloomington 33703 and 31092) and UAS-RNAi-Set1 (Bloomington 33704), Df(2L)BSC653 (Bloomington 25743) and CH321-83D07 (Bloomington 38673). QUAST-Gal4DB:Yki:3XFL and QUAST-Gal4DB:Yki-WW:3XFL flies were generated by phiC31-mediated site-specific transformation, using the attpP2 site at 68A (Groth et al., 2004) (Genetic Services, Inc.). 3XHA:gNcoA6 and 3XHA:gNcoA63PPXA flies were created by inserting 3 tandem HA tags and 3PPXA mutations into the CH322-05H20 (BPRC) by recombineering (Venken et al., 2009), and then flies were transformed with these BAC clones at the attpP2 site at 68A (Groth et al., 2004) using phiC31-mediated site-specific transformation (Genetic Services, Inc.). HA antibody staining (Fig. S4) confirmed that both wild-type and PPxA mutant forms of NcoA6 were expressed similarly. Flies were cultured at 25°C, or, when stronger expression of UAS transgenes was desired, at 29°C. Ectopic expression was also induced in eyes using GMR-Gal4 and posterior cells using en-Gal4 UAS-GFP, en-Gal4 UAS-GFP ex-LacZ or en-Gal4 bs-GFP with or without UAS-dcr2. For flip-out ectopic expression clones, UAS-transgenes with y w hs-FLP[122] were crossed to w; Act> y+>Gal4 bs-GFP (AyGal4-bs-GFP) or AyGal4,UAS-GFP.
ChIP-PCR
Late 3rd instar larvae overexpressing Gal4DB:Yki:3XFLAG and Gal4DB:YkiWW:3XFLAG with UAS-GFP under the control of M2ET40-QF (Potter et al., 2010) were dissected and anterior parts were collected in PBS on ice. Chromatin preparation and immunoprecipitation were performed as described in the protocol of EZ ChIP Chromatin Immunoprecipitation Kit (Millipore). About ten larval anterior parts were used for an IP reaction. Mouse anti-Flag M2 (1 µg, Sigma), rabbit anti-H3K4Me3 (1 µg, Abcam), guinea pig anti-NcoA6 (1 µg affinity purified IgG) or bulk guinea pig IgG (1 µg, negative control) were used for immunoprecipitation. No specific signal was detected in the ChIP-PCR negative control. After the immunoprecipitation, the recovered DNA was analyzed by qPCR with QuantiTect SYBR Green PCR Kit (Qiagen) using SmartCyclerII (Cepheid). The following primers were used in qPCRs: pka (5’-agccgcactcgcgcttctac/ 5’-caatcagcagattctccggct), UAS-primer (5’-gcatgcctgcaggtcggag / 5’-cgcttagcgacgtgttcact) and promoter-primer (5’-caagcgcagctgaacaagcta / 5’-gaaaagttcttctcctttactcat) were used.
Histology and imaging
Imaginal discs were fixed and stained as described previously (Cho et al., 2006), using as primary antibodies rabbit anti-Yki (1:400) (Oh and Irvine, 2008), guinea pig anti-NcoA6 (1:1000), rabbit anti-H3K4Me3 (1:10000 abcam), rabbit antiTrr (1: 400, a gift of Ali Shilatifard), rabbit Trx (1:400, a gift of Ali Shilatifard), rabbit anti-Dcr2 (1:1600, abcam), goat anti-β-gal (1:400, Biogenesis), active caspase3 (1: 400, Cell Signaling), and mouse anti-Diap1 (1:400, a gift of Bruce Hay). Fluorescent stains were captured on a Leica TCS SP5 confocal microscope.
Tissue culture assays and Protein interactions
S2 cells were cultured with Schneider’s Drosophila medium (Invitrogen) and 10% FBS (Sigma). RNAi and luciferase reporter assays were performed using the Dual-Glo Luciferase Assay System (Promega) as described previously (Oh and Irvine, 2011) using pAc-hRluc (Potter et al., 2010) or Copia-RLuc (Oh and Irvine, 2009) as a transfection control. The HEK-293T cell line was maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. For RNAi of mammalian genes, shRNA constructs (200 ng) were transiently transfected in the reporter assays along with 8XGTIIC-luc (100 ng), pCMV2-RLuc (0.25 ng) and p2xFlag CMV2-YAP2 (50 ng), CMV2-YAP2-WW (50 ng), and the shRNA constucts used are pGIPZ- RHS4346 (non-silencing control), V3LHS-306099 (shYAP-7), V3LHS_306101 (shYAP-8), V2LHS_248718 (shNCoA6-4) and V3LHS-322282 (shNcoA6-12) (Thermo Scientific). For coimmunoprecipitations, transient transfections were performed with equal amounts of DNA using Cellfectin (Invitrogen) for S2 cells or Lipofectamine 2000 (Invitrogen) for mammalian cells in six-well plates according to the manufacturer’s protocol. Coimmunoprecipitation assays were performed as described previously (Oh and Irvine, 2009). For Western blotting mouse anti-V5 (1:5000, Invitrogen), mouse anti-Flag (1:10000, Sigma), rabbit anti-GST (1:5000, Millipore), rabbit anti-Trr (1: 1000, a gift from Ali Shilatifard), rabbit anti-Trx (1: 1000, a gift from Ali Shilatifard), rabbit anti-dSET1 (1: 1000, a gift from Ali Shilatifard), rabbit anti-hNCOA6 (1:2000, Novus Biologicals), and guinea pig anti-dNcoA6 (1:2000). IR dye 680 or 800 conjugated secondary anti-mouse and anti-rabbit (1:10000, Odyssey) were used. Blots were scanned and analyzed using the Odyssey Infrared Imaging system (LiCor biosciences). For the V5-pull down experiments, GFP:V5, Yki:V5, YkiWW:V5, YkiN:V5, GST-YkiC:V5 and GST were expressed in BL21(DE3)pLysS (Invitrogen), induced with 0.5 mM IPTG and then pull down assays were performed as described previously (Oh et al., 2013).
Supplementary Material
Highlights.
-
-
Yki binding to chromatin promotes H3K4 methylation
-
-
Yki directly associates with NcoA6, a subunit of the Trr methyltransferase complex
-
-
Recruitment of NcoA6 is necessary and sufficient for transcriptional activation
-
-
Yki-NcoA6 interaction is conserved in mammals
ACKNOWLEDGMENTS
We thank Ali Shilatifard, the DSHB and the Bloomington Stock Center for antibodies and Drosophila stocks, Flybase, Dibyendu Kumar, and the Waksman Genomics Core for bioinformatics and sequencing support. This research was supported by HHMI, and NIH grants GM078620 (KDI), 5R01GM054510 (RSM), 5P50GM081892 and 3U01HG004264 (KPW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional details are in the Supplemental Material
AUTHOR CONTRIBUTIONS
HO Designed, performed, and analyzed experiments, and wrote manuscript. MS and LM performed and interpreted bioinformatic analysis. KPW and RSM supervised research and analyzed experiments. KDI supervised research, designed and analyzed experiments, wrote manuscript.
REFERENCES
- Antonson P, Schuster GU, Wang L, Rozell B, Holter E, Flodby P, Treuter E, Holmgren L, Gustafsson JA. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Signal integration in TGF-β, WNT, and Hippo pathways. F1000Prime Reports. 2013;5 doi: 10.12703/P5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayarmagnai B, Nicolay BN, Islam ABMMK, Lopez-Bigas N, Frolov MV. Drosophila GAGA factor is required for full activation of the dE2f1-Yki/Sd transcriptional program. Cell cycle (Georgetown, Tex) 2012;11:4191–4202. doi: 10.4161/cc.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaumueller CM, Mlodzik M. The Drosophila tumor suppressor expanded regulates growth, apoptosis, and patterning during development. Mech Dev. 2000;92:251–262. doi: 10.1016/s0925-4773(00)00246-x. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner D, Stark A, Russell R, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Huang C, Chong YF, Gunaratne HJ, Hogue KA, Blackstock WP, Harvey KF, Hong W. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan XY, Xu J, Anzick SL, Zhang H, Trent JM, Meltzer PS. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res. 1996;58:5009–5013. [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature Reviews Cancer. 2013 doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes and Development. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Guan KL. The YAP and TAZ transcription coactivators: key downstream effectors of the mammalian Hippo pathway. Seminars in Cell & Developmental Biology. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Irvine KD. Integration of intercellular signaling through the Hippo pathway. Seminars in Cell & Developmental Biology. 2012;23:812–817. doi: 10.1016/j.semcdb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Lu X, Shimada M, Dou Y, Tang Z, Roeder RG. Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95. Nature structural & molecular biology. 2013 doi: 10.1038/nsmb.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A. Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell. 2011;44:51–61. doi: 10.1016/j.molcel.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H, Nguyen A, Chen L, Okano H, Hariharan IK. The Drosophila Ortholog of MLL3 and MLL4, trithorax related, Functions as a Negative Regulator of Tissue Growth. Molecular and cellular biology. 2013;33:1702–1710. doi: 10.1128/MCB.01585-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2010;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz LM, Liu-Chittenden Y, Yin F, Zheng Y, Yu J, Huang B, Chen Q, Wu S, D P. The hippo effector yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Developmental Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Pereira FA, Yuan Y, DeMayo FJ, Ko L, Xu J. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem. 2002;277:45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- Lee J-E, Wang C, Xu S, Cho Y-W, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife. 2013;2:e01503. doi: 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. PNAS. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Anzick SL, Choi JE, Bubendorf L, Guan XY, Jung YK, Kallioniemi OP, Kononen J, Trent JM, Azorsa D, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Mahajan MA, Das S, Zhu H, Tomic-Canic M, Samuels HH. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan MA, Samuels HH. Nuclear receptor coactivator/coregulator NCoA6(NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Lucler Receptor Signaling. 2008;6:1–19. doi: 10.1621/nrs.06002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol. 2011:21. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends in Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad thgrough bantam. Developmental Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Slattery M, Ma L, Crofts A, White KP, Mann RS, Irvine KD. Genome-wide Association of Yorkie with Chromatin and Chromatin-Remodeling Complexes. Cell Reports. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell. 2010;141:536–548. doi: 10.1016/j.cell.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell death & disease. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Developmental Dynamics. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballaré C, Beato M. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes and Development. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes and Development. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Grusche FA, Harvey KF. Control of tissue growth and cell transformation by the Salvador/Warts/Hippo pathway. PLoS One. 2012;7:e31994. doi: 10.1371/journal.pone.0031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Poon CL, Hong W, Harvey KF. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]
- Zhu YJ, Crawford SE, Stellmach V, Dwivedi RS, Rao MS, Gonzalez FJ, Qi C, Reddy JK. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.