Historical Perspective
Transcriptional coactivators are defined, broadly, as the family of coregulator molecules which interact with nuclear receptors and other transcription factors to enhance the rate of gene transcription. The existence of coactivator-like proteins was predicted in early 1970’s, as some nuclear, nonhistone receptor-associated proteins were found to bind nuclear receptors and increase their interaction with DNA to enhance their transcription potential (Spelsberg, et al. 1971). This crude fraction was later shown to contain many diverse coactivators; the large number of such proteins was unpredicted at the time and prevented purification. Although it was clear that steroid hormones such as estrogen can rapidly induce the new synthesis of specific mRNA and proteins (Means, et al. 1972), the importance of these nuclear-acceptor molecules in ligand-dependent functions was postulated to enhance NR transcription but the concept was not proven (Yamamoto & Alberts. 1975). In the interim, a series of sophisticated molecular studies unfolded that indicated that ligand binding activates conformational changes in the steroid receptor to promote DNA-binding and transcriptional activity; anti-hormones were shown to effectively oppose such structural alterations (Allan, et al. 1992). In addition to ligand-dependent functions, the steroid receptors were also found to be activated in a ligand-independent manner (Denner, et al. 1990, Power, et al. 1991).
In the 1990’s, studies designed to elucidate the functional roles of the corepressors and coactivators were commenced again, initially in yeast (Baniahmad, et al. 1993, McDonnell, et al. 1991a, McDonnell, et al. 1991b). An inherent negative regulatory function for the steroid receptors was identified in steroid receptors, and analyzed first in yeasts by demonstrating binding of steroid receptors to repressors such as SSN6, which when mutated allowed receptor activation of gene expression (McDonnell, et al. 1992, Vegeto, et al. 1992). Similar yeast studies were carried out to demonstrate ligand-mediated coactivation. These proof-of-principle yeast studies led to the definition of two classes of coregulators: coactivators and corepressors- and were followed by the biochemical discovery of a corepressor activity for TR in mammalian cells and the publications of other receptor-associated proteins in mammals (Baniahmad, et al. 1995, Baniahmad, et al. 1995, Cavailles, et al. 1994, Halachmi, et al. 1994). In aggregate, these studies set the stage for the first cloning of a cDNA encoding a mammalian nuclear receptor interacting coactivator protein. This first authentic NR coactivator, termed Steroid Receptor Coactivator-1 (SRC-1), was identified using a yeast two hybrid genetic screen employing the ligand-binding domain (LBD) of the progesterone receptor (PR) (Onate, et al. 1995, Xu, et al. 1998). SRC-1 was the first member of the p160 family of coactivators cloned, following which two additional family members SRC-2 (NCOA2/GRIP1/TIF2) (Voegel, et al. 1996) and SRC-3 (NCOA3/ACTR/pCIP) (Chen, et al. 1997, Torchia, et al. 1997) were identified. The p160 family members are closely related molecules with ~60% homology, but are functionally distinct. In addition to the full length SRCs, some shorter forms of SRCs were identified as well. SRC-3Δ4 is a splice isoform of SRC-3 with a deletion of exon 4 (SRC-3Δ4) and the protein lacks the N-terminal bHLH (helix-loopl-helix) domain that contains a nuclear localization signal (NLS) (Long, et al. 2010, Reiter, et al. 2001). More recently, a shorter 70kD isoform of SRC-1 was identified and found to be highly elevated in human and mouse endometriotic tissues (Han, et al. 2012). This 70kD isoform of SRC-1 is the C-terminal fragment of the full-length SRC-1which is proteolytically cleaved by MMP-9. Over the last two decades, we gained considerable knowledge about the coactivators and their impact on human health and physiology. These findings together classified a novel family of nuclear receptor coactivators which become known as the master regulators of gene regulation.
Coactivator complexome
After the discovery of the first authentic coactivator SRC-1, it was predicted that cells may have around five to ten coactivators and few corepressors to regulate the gene transcription. Surprisingly, more than 400 coregulators have been reported so far, substantiating their prevalent and critical role in transcriptional regulation (Lonard & O'malley. 2007). Molecular analyses by mass spectrometry identified that SRCs work in tandem with other coregulators in a close association by forming large multi-subunit stable complexes. This proteomics information concerning a coactivator-protein-complex also known as ‘complexome’ - identified that the complexes are in a dynamic rearrangement in an ordered manner to facilitate various reactions and sub-reactions in transcription. These reactions include phosphorylation, ubiquitination, methylation and acetylation of the associated molecules in the coactivator complex, which further defines the specific affinity of the coactivators for NR, transcription factors and other associated molecules (Han, et al. 2009). This multifunctional component of the coactivator-complexome allows them to integrate different upstream environmental stimuli and to transmit to a variety of enzymatic activities at the promoter for regulating transcription.
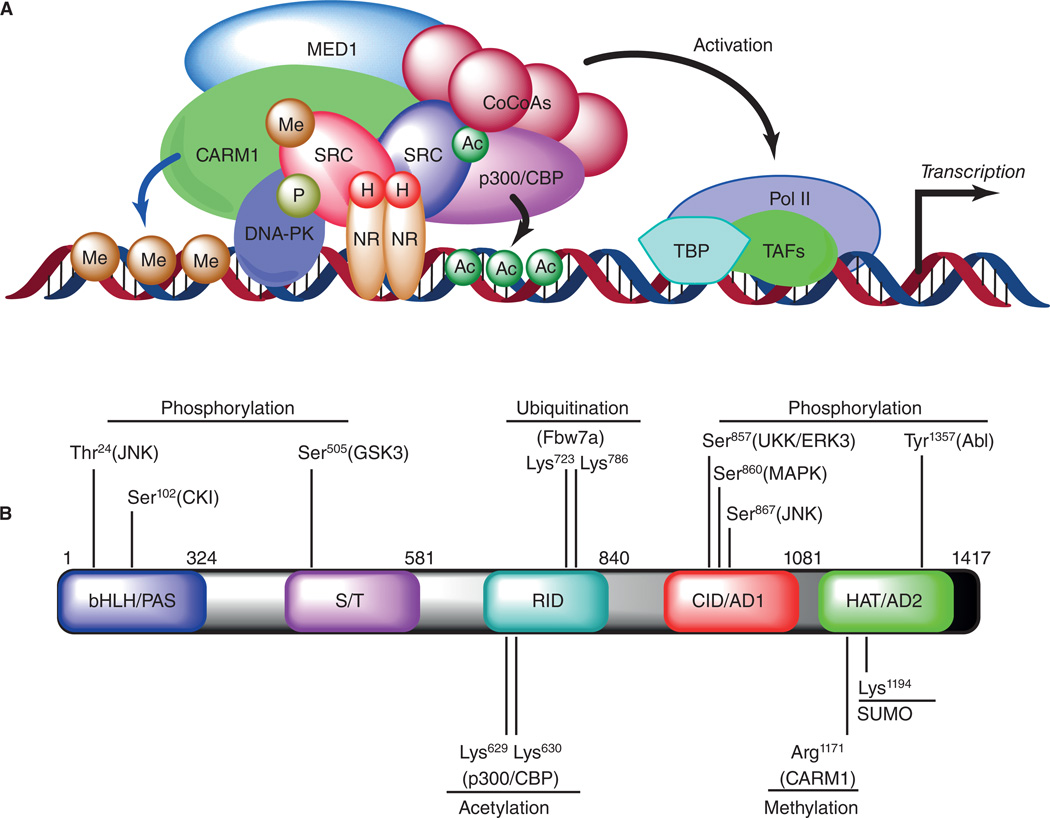
Proteomic investigations identified the dynamic nature of a SRC-3 complex assembled on estrogen response element (ERE) in a ligand dependent manner (Fig. 1A). The SRC-3 complex consists of several interacting partners with enzymatic activities which include kinases, ATPases, acetyl-transferases, methyl-transferases as well as ubiquitin-ligases, all of which contribute to the dynamic functions of the coactivators (Malovannaya, et al. 2010). Recent studies on coregulator dynamics identified some novel mechanisms for ER-regulated gene transcription, and the findings postulated a ‘three-states model’ of coactivator-dependent complex formation (Foulds, et al. 2013). In the first step, ligand-bound ER on canonical EREs forms a biochemically stable ‘poised’ complex by attracting a set of coactivators and certain corepressors. Addition of ATP rapidly converts these complexes into an ‘activated’ state by the kinetic activity of DNA-dependent protein kinase (DNA-PK) which mediates phosphorylation events on coactivators and ER. Finally, DNA-PK promotes ERα-mediated transcription by phosphorylating coactivators SRC-3 and MED1 as well as dismissing corepressors RIP140 from the complex (Foulds, et al. 2013). These studies unravel the dynamic events mediated by kinases on a coactivator complexome to fine-tune transcription.
Figure 1.
(A) Coactivator dependent complex assembly and regulation of gene transcription. Upon hormone (H) binding, the nuclear receptors (NR) interact with steroid receptor coactivators (SRC) and recruit them to the enhancer region of target genes. SRC coactivators then interact with co-activator-associated arginine methyl transferase 1 (CARM1), cyclic AMP response element-binding protein (CBP), p300 (a 300 kDa protein homologous to CBP; also known as EP300), mediator complex (MED1) and recruit other common co-coactivators (CoCoAs) to remodel the chromatin and build up the activated transcription complex. Post-translational modifications (PTM) on SRCs such as phosphorylation (P), acetylation (Ac), and methylation (Me) also regulate the coactivator complex association, and modulate the assembly of general transcription factors such as TBP (TATA-binding protein) and TAF (TBP-associated general transcription factors) along with RNA polymerase II (Pol II). (b) Schematic representation of the molecular structural domains and a comprehensive map of known PTM codes on SRC-3 along with the type of modifications, residues modified, and enzymes imparting the code.
Integrative mass spectrometric-based analysis of affinity purified endogenous coregulator complexes identified a hierarchical organization of protein complexes that exists as three discrete layers in an intrinsically tiered organization of the complexome (Malovannaya, et al. 2011). These include relatively stable minimal endogenous core modules; these combine to form the variable core complex-isoforms; and finally, coregulator complex-complex interactions form networks. Based on the type of protein complexes formed, the coregulators can be broadly classified into two major types: type I classifies relatively stable multi-subunit complexes consisting of conserved coactivator molecules, whereas type II represents context dependent-associated coactivators that are recruited in response to various extra-cellular stimuli (Malovannaya, et al. 2011). Type I coregulators include mediators, CoREST (corepressor-repressor element-1 silencing transcription factor) complex, NCOR (nuclear receptor corepressors), nucleosome remodeling and deacetylase (NURD) complexes and the SWI/SNF (BAF/P-BAF), whereas SRCs are prime-examples of type II complexes. This dynamic regulation of coactivator complex assembly by the SRCs is in-turn regulated by various upstream signaling events that impart post-translational modifications (PTM) onto the coactivators (Dasgupta, et al. 2014).
Signal specific PTM-codes on SRCs
The molecular recognition of the activity of steroid receptor coactivators depends upon the PTM codes on them. Phosphorylation, acetylation, sumoylation, ubiquitination, and methylation of the SRCs (Fig. 1B) intricately coordinate and fine-tune their activity, localization, protein stability and dictate the interacting partner molecules used to build up the complexome.
Phosphorylation
In response to multiple upstream signaling events like growth factors, cytokines, hormones and nutrient signaling, protein kinases phosphorylate SRCs either at a single site or multiple sites. Depending on the pattern of the phoshorylation code(s) on SRCs they attract select binding partners; nuclear receptors or transcription factors along with other coregulator molecules to regulate the gene transcription. In addition to exerting effects on the nuclear genome by binding directly to the NRs, steroid hormones also activate several kinases such as MAPK, JNK, AKT and ERK1/2 which then phosphorylate NRs and coactivators to stimulate gene transcription by non-genomic signaling (Lonard & O'Malley. 2007). Steroid hormone signaling phosphorylates SRC-3 at multiple residues including N-terminal Thr24, several sites in a Serine/Threonine-rich region, and Ser857, Ser860 and Ser867 in the receptor-interacting domain (RID) (Long, et al. 2012, Wu, et al. 2004, Yi, et al. 2005, Yi, et al. 2008). Similarly, SRC-1 is phosphorylated on Thr1179 and Ser1185, and SRC-2 on Ser736 by MAPK thereby increasing coactivator-affinity to NRs (Gregory, et al. 2004, Rowan, et al. 2000). SRC-2 has emerged as a major coactivator for glucocorticoid receptor (GR) and certain phosphorylation events on SRC-2 by casein kinase (CK) and cyclin-dependent kinase 9 (CDK9) dictate GR actions (Dobrovolna, et al. 2012). Four major phosphorylation sites Ser469, Ser487, Ser493 and Ser499 in the N-terminal domain of SRC-2 protein promote GR-dependent transcription by facilitating recruitment of coactivator-complex to native GR targets (Dobrovolna, et al. 2012). SRC-3Δ4, the splicing variant of SRC-3 also is regulated by phosphorylation. But instead of a direct role in nuclear-transcription, the SRC-3Δ4 is localized in the cytosol, and is phosphorylated by PAK kinase, whereupon it then binds to epidermal growth factor receptor (EGFR) and transduces activity to focal adhesion kinase (FAK). Thus, phosphorylated SRC-3Δ4 acts as a critical signaling molecule to regulate the migratory potential of tumor cells by bridging the gap between EGFR and FAK (Long, et al. 2010). In summary, coactivators are molecular integrators of upstream signaling events, and phospho-coded SRCs direct assembly of specific interacting partners for gene transcription.
Acetylation and Methylation
Histone acetylases and deacetylases, along with methylases and demethylases are essential components of coactivator complexes responsible for modifying chromatin. Based on their function of adding or removing histone marks, they are classified as epigenetic ‘writers’ or ‘erasers’. A number of co-coactivators including p300/CBP, GCN5, and PCAF possess intrinsic histone acetyl transferase (HAT) activity (Couture & Trievel. 2006). SRCs recruit the HATs and methyl transferases such as peptidylarginine methyltransferases (PRMTs) to remodel chromatin and regulate gene transcription. Additionally, a coactivator such as SRC-3 is in turn acetylated by p300/CBP and methylated by coactivator-associated arginine methyltransferase 1 (CARM1) at Arg1171 (Feng, et al. 2006). Acetylation of SRC-3 by CBP coincides with the attenuation of hormone induced gene transcription by enforcing the complex disassembly (Chen, et al. 1997, Chen, et al. 1999). Mechanistically, acetylation neutralizes the positive charges of two lysine residues adjacent to the ‘LXLLL’ motif of SRC-3 thereby disrupting the association of HAT complexes with the NR coactivator complex and terminating the gene transcription (Chen, et al. 1999). CARM1, which activates transcription by modifying core histone tails, also promotes dissociation of coactivator complex and terminates hormone-induced transcription by methylating SRC-3 (Feng, et al. 2006). In addition to the acetylases, the family of lysine-deacetylases, histone deacetylases (HDACs) and sirtuin proteins also regulate gene transcription as coregulators (Lahue & Frizzell. 2012). HDACs are recruited to the coregulator complex to repress gene transcription, in particular by corepressors such as NCoR. There are two classes of HDACs, class I and class IIa, the latter being relatively weak in enzymatic activity. Additionally, sirtuins, the NAD-dependent deacetylases, also are recruited to the coregulator-complex and are known to modulate gene transcription.
Ubiquitination and Sumoylation
Activity and stability of coactivators are regulated by ubiquitination, an enzymatic process in which 8.5 kDa small molecules named ubiquitin are systematically added by E3 ubiquitin ligase. Ubiquitination is a highly regulated process, and phosphorylation on coactivators acts as a priming event for this modification by increasing their affinity towards ubiquitin E3 ubiquitin ligase. Phosphorylation by GSK3β on SRC-3-Ser505 increases the coactivator affinity towards Fbw7α, a component of E3-ligase complex which then ubiquinates SRC-3 on Lys723 and Lys786 (Lonard & O'Malley. 2007, Wu, et al. 2007). Mono-ubiquitinated SRC-3 has higher affinity for ERα and stimulates ERα-dependent gene transcription, whereas poly-ubiquitinated SRC-3 is rapidly degraded, thereby decreasing SRC-3 protein stability. SRC-3 protein stability and activity also are regulated by specific phosphorylation-codes that induce degradation of the protein known as “phospho-degron” in the N-terminal domain of the protein; phosphorylation of Ser102 in the degron by CKI (casein kinase I) increases coactivator affinity for speckle-type POZ protein (SPOP)-E3 ligase (Li, et al. 2008). On the contrary, certain mutations in the SPOP protein alter the affinity of SPOP for SRC-3 imposing a SPOP-dependent regulation of SRC-3 activity and gene transcription (Geng, et al. 2013). Similarly, CUL-3, a member of the family of E3-ligase scaffolding proteins also modulates SRC-3 activity by binding to the Ser860-phosphorylated SRC-3 in response to retinoic acid induction (Ferry, et al. 2011). Thus post-translational modifications on SRC-3 by phosphorylation-coupled-ubiquitination modulate the activity and stability of the coactivator to control the dynamics of transcription.
In addition to ubiquitination, covalent modifications by addition of small ubiquitin-like modifier (SUMO) to the lysine-residues of the coactivators have been identified. SRCs are subjected to sumoylation at two conserved lysine residues in the RID motif, which functionally enhance their interaction and affinity for NRs (Wu, et al. 2006). However, sumoylations of SRC-3 on Lys723 and Lys786 were found to have a negative impact on its activity, most likely due to the competitive inhibition of ubiquitinating in these sites. Nevertheless, sumoylation of coactivators provides another degree of dynamic regulation to monitor and manipulate gene transcription.
Coactivators in disease pathophysiology
Coactivators have emerged as cellular integrators of various upstream signaling pathways that transduce these signals into transcriptional outputs to regulate expression of myriad gene targets (Fig. 2). Hence, dysfunctions in coregulators are principal drivers of numerous pathologies (Lonard & O'Malley. 2012). Here we will highlight selected examples of the clinicopathological conditions affected by the transcriptional coactivators.
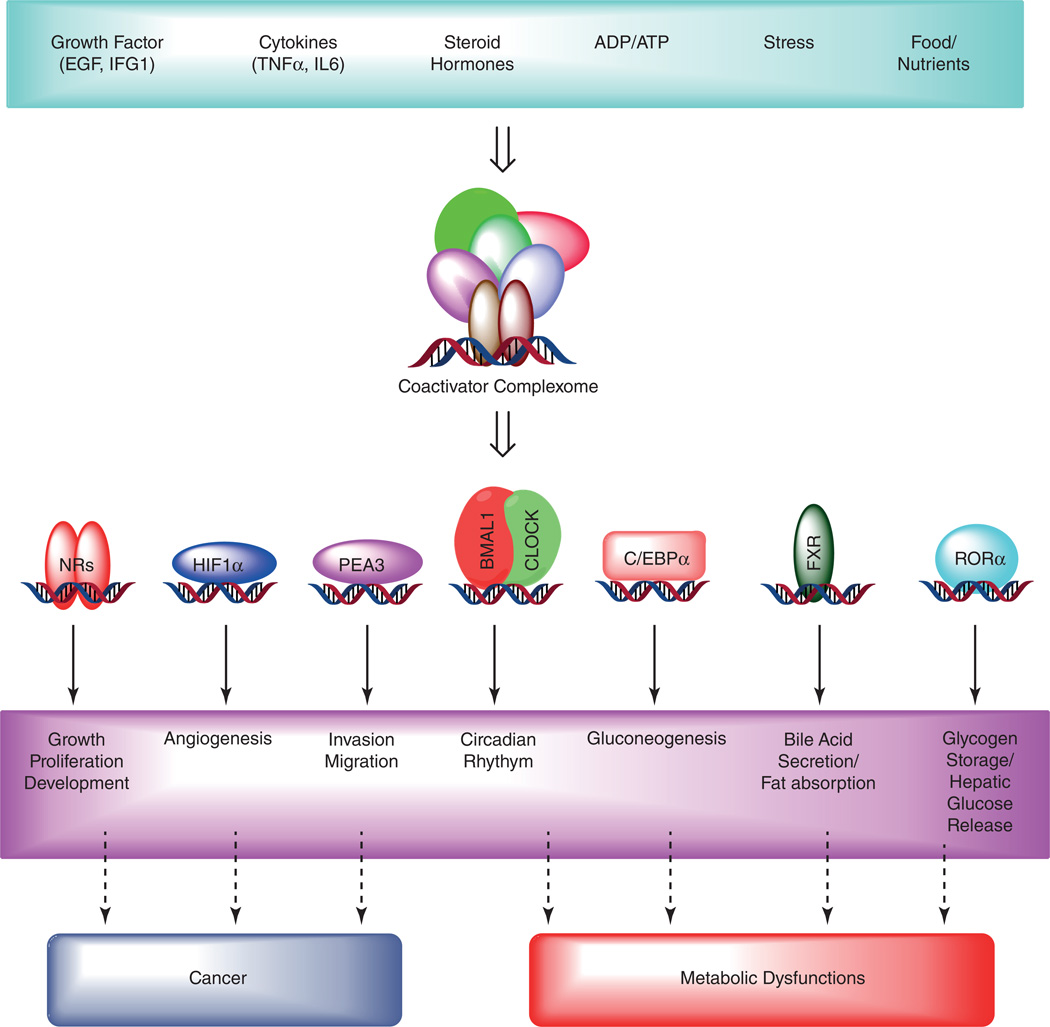
Figure 2.
Coactivator dependent signaling regulates various biological functions, and deregulation causes diseases. Several extracellular stimuli such as growth factors- EGF (epidermal growth factor) and IGF (insulin-like growth factors); cytokines- IL-6 (interlukein-6) and TNFα (tumor necrosis factor-α) and steroid hormones trigger downstream signaling pathway activating coactivator-dependent complex assembly. In addition, alterations in the energy status (ATP/ADP ratio), nutrient signaling, and cellular stress can also promote coactivator recruitment on target gene promoters. Coactivators such as steroid receptor activators (SRCs) then bind to nuclear receptors (NRs) or several other transcription factors to stimulate gene transcription. This coactivator dependent gene activation is highly selective, and intricately regulated by several mechanisms (described in the text) stimulating specific cellular functions. In contrast, deregulated expression and activation of coactivators lead to perturbed signaling pathway resulting in disease pathology.
Neurological disorders
Mutations in certain coregulator genes alter the epigenetic marks on chromosomes, affecting brain development and promoting onset of certain neurodevelopmental disorders (Urdinguio, et al. 2009). These epigenetic dysfunctions cause moderate to severe perturbations in the transcriptomics, disrupting the neuronal growth and differentiation. Mutations in the chromatin remodeling protein ATRX (ATP-dependent helicase ATRX, X-linked helicase II) confer aberrant DNA methylating patterns in the chromatin leading to a neurodegenerative disorder named ATRX syndrome (Gibbons, et al. 2008). This syndrome is an X-linked disorder confined only to the males while the female-carriers manifest limited symptoms. Symptoms include mental retardation often accompanied with alpha-thalassemia, unusual facial appearance and urogenital defects (Gibbons, et al. 1995). ATRX is a member of the Snf2 family of enzymes that maintains nucleosome stability and regulates gene transcription by modulating the functions of chromatin remodeling transcriptional regulators, such as the polycomb group protein EZH2 (Eisen, et al. 1995). Patients with ATRX syndrome have severely comprised genetic defects due to mutated ATRX gene.
Rubinstein-Taybi syndrome (RTS) is another example of a neurological disorder associated with the dysfunction of a histone acetyltransferase (HAT). The majority of the Rubinstein-Taybi cases are associated with mutations in the CBP gene located at chromosome 16p13.3 and some in EP300 (E1A binding protein p300) gene at chromosome 22q13.2 (Lonard & O'Malley. 2012). In 1963, Jack Herbert Rubinstein and Hooshang Taybi described a series of cases with this syndrome demonstrating some typical features which include mental disability; distinctive facial features; broad thumbs and toes; and often associated with cryptorchidism in males. This disease is rare and approximately 1 out of 100,000 to 125,000 children are born with this disorder. CBP is a transcriptional coactivator which has intrinsic HAT-activity, and binds to the transcription factor CREB (cAMP response element-binding protein) to regulate gene transcription (Park, et al. 2014). Mutation or deletion in the CBP gene severely affects HAT activity of CBP and the ability of CBP to transactivate CREB, indicating that loss of the HAT activity of CBP may cause RTS.
In Huntington's disease transcriptional coactivator PGC-1α expression is severely impaired, and mouse genetic studies revealed that loss of PGC-1α severely impairs metabolism and accentuates neurodegeneration. Huntington's disease is an autosomal-dominant disorder characterized by impaired muscle coordination that leads to cognitive malfunctioning and psychiatric problems. PGC-1α is a potent suppressor of reactive oxygen species (ROS) by activating the transcription of ROS defense enzymes superoxide dismutase (SOD1), manganese SOD (SOD2), catalase, and glutathione peroxidase (Chaturvedi, et al. 2009). In absence of PGC-1α coactivator, the neuronal cells are extremely sensitive and vulnerable to neurotoxins leading to apoptotic death of neuronal cells and oxidative damage in the brain.
Studies using SRC knockout animals identified important roles for nuclear receptor coactivators in the coordination of neurobehavioral functions and brain development. SRC-1 is ubiquitously expressed in the human brain with more prominent presence in hippocampus, olfactory bulbs and cortex (Meijer, et al. 2000). SRC-1 is a crucial regulator of sexually dimorphic regions in the brain and coactivates GR functions to coordinate the hypothalamic–pituitary–adrenal (HPA) axis of the brain. Neurobehavioral tests on SRC-1−/− animals compared to wildtype littermates discovered some novel roles of SRC-1 in anxiety response (Stashi, et al. 2013). In comparison, SRC-2−/− females displayed decreased anxiety responses under certain environmental stimuli, whereas males were found to have deficits in sensorimotor gating, a neurological process which is important to understand the functional significance of attentional abnormalities. In contrast, SRC-3−/− males were devoid of any noticeable neurological abnormalities, however the females exhibit reduced exploratory activities and increased anxiety behavior (Stashi, et al. 2013). Collectively, these findings establish the role of SRCs in the regulation of the central nervous system (CNS) and coordination of neurobehavioral phenotypes in a gender-specific manner.
Cardiac development and disease
Transcriptional coactivators can play an essential role in cardiac development by regulating the mitochondrial response of the heart by broadly regulating gene expression from both nuclear and mitochondrial genomes. PGC-1 has been extensively studied with respect to cardiac development and bioenergetics of the heart, and its expression was found to be repressed in numerous models of heart failure with a maladaptive energetic profile (Rowe, et al. 2010). PGC1-α induces expression of numerous genes in cardiac cells regulating major metabolic pathways to maintain a steady supply of ATP production. Genes induced by PGC-1α include the majority of mitochondrial respiratory subunits, ATPase complexes, enzymes of fatty acid biosynthesis and transport, key enzymes of the glycolytic and tricarboxylic acid cycle (TCA) (Banke, et al. 2010). In addition to metabolic pathways, PGC1-α induces angiogenesis in myocytes by directly activating a broad range of angiogenic factors including vascular endothelial growth factor (VEGF) independent of the hypoxia-inducible factor (HIF) pathway (Arany, et al. 2008). Overexpressing PGC-1α in the heart identified univocal roles of the coactivator in mitochondrial biogenesis (Lehman, et al. 2000). PGC-1α activates both mitochondrial as well as nuclear genes by directly transactivating transcription factors nuclear respiratory factor (NRF) and estrogen-related receptor (ERR) (Hock & Kralli. 2009). These findings have clearly placed PGC-1 as a prime regulator of metabolism in heart, both in cardiomyocytes as well as cardiac cells.
In addition to PGC-1, expression of coactivator SRC-2 is found to be repressed in failing hearts. Genetic ablation of SRC-2 identified an activation of a ‘fetal gene program’ in adult mice by altering the expression of metabolic and sarcomeric genes (Reineke, et al. 2012). Mechanistically, SRC-2 depletion reduces the expression of several transcription factors such as GATA as well as coactivators like PGC-1α indicating that SRC-2 is a prime regulator of the steady-state adult cardiac transcriptomic profile (Reineke, et al. 2012). These studies have deciphered the importance of coactivators in cardiac functioning and how subtle changes in their expression can lead to catastrophic medical conditions.
Inflammatory diseases
The most common lung diseases including asthma, chronic obstructive pulmonary disease (COPD), cystic fibrosis and acute respiratory distress involve inflammatory responses coordinated by expression of multiple proinflammatory genes. Several transcriptional coactivators have been linked as the molecular regulators of inflammatory responses, of which HDACs deserves special mention (Barnes, et al. 2005). Patients with asthma exhibit increased expression of HAT with simultaneous reduction in HDAC1 in the brochial and alveolar macrophages compared to normal airways (Cosio, et al. 2004). In patients with COPD, there is a significant decrease in HDAC2 expression with a concomitant increase in HAT activity facilitating activation of NF-κB and transcription of proinflammatory cytokines (Qu, et al. 2013). The alveolar macrophages in COPD patients display increased release of TNF-α and IL-8 in response to stimuli thus contributing to the adversity of the pathology. Traditional therapy includes corticosteroids which effectively suppresses the transcription of proinflammatory genes by inhibiting NF-κB and AP1 transcription factors (Barnes. 2013).
The transcriptional coactivator SRC-3 acts as a protective factor against acute inflammatory response by repressing translation of inflammatory cytokines. SRC-3−/− animals are more susceptible to endotoxic shock compared to their wildtype littermates with enhanced levels of proinflammatory cytokines including TNFα, IL-6 and IL-1β (Yu, et al. 2007). Thus, it is sufficient to conclude that expression of coactivators delicately balances inflammatory responses by modulating expression of interleukins and cytokines.
Metabolic disorders and Circadian biology
Coactivators are essential coordinators of whole body energy homeostasis by modulating the expression of multiple metabolic enzymes. SRC-family coactivators are prime regulators of metabolic pathways in different tissues, and genetic deletion of their expression corresponds to various physiological abnormalities and metabolic disorders (Dasgupta, et al. 2014). SRC-1−/− animals display reduced energy expenditure with an increased risk of developing obesity as well as a defective gluconeogenic program (Louet, et al. 2010, Picard, et al. 2002). Molecularly, SRC-1 coactivates C/EBPα (CCAAT-enhancer-binding proteins) to promote transcription of regulatory enzymes in the gluconeogenic pathways such as pyruvate carboxylase, phosphoenolpyruvate carboxykinase (PEPCK), and fructose-1, 6-bisphosphatase (FBP1) (Picard, et al. 2002). In contrast, SRC-2−/− animals are protected from high-fat-induced obesity and exhibit increased insulin sensitivity, higher lipolysis, and reduced fat uptake (Picard, et al. 2002). Loss of SRC-2−/− also affects the hepatic glucose release due to decreased expression of glucose-6-phosphatase (G6Pase) simulating the phenotypes observed in genetic disorder Von Gierke's disease (Chopra, et al. 2008). SRC-2 also stimulates absorption of fatty acids from the gut by activating the expression of bile salt export pump (BSEP) by coactivating FXR (farnesoid X receptor) under conditions of reduced energy status, thereby coordinating whole-body energy homeostasis (Chopra, et al. 2011). Even in tumor cells, SRC-2 was found to modulate fatty acid biosynthesis by distinct reprogramming of metabolic functions (Dasgupta, et al. 2012a). In contrast, SRC-3 participates in white adipocyte development and supports fatty acid metabolism in skeletal muscle by regulating the expression of the long-chain fatty acid transporter carnitine/acyl-carnitine translocase (CACT) (York, et al. 2012). Thus, alterations in the expression of SRCs promote global changes in numerous metabolic pathways in different tissues (York, et al. 2013) to maintain the energy demands of our body, and genetic loss of their expression can lead to severe metabolic disorders (York & O'Malley. 2010).
In light of this knowledge, recent studies indicated the importance of transcriptional coactivators in circadian biology. Our recent findings indicate that SRC-2 is prime coordinator of circadian activities by regulating the expression of genes that regulate hepatic metabolism and diurnal rhythmicity (Stashi, et al. 2014). Molecularly, SRC-2 coactivates transcription factors Brain and Muscle ARNT-Like 1 (BMAL1/ARNTL) and Circadian Locomotor Output Cycles Kaput (CLOCK), the two core components of the clock machinery (Asher & Schibler. 2011). Cistromic analyses revealed that recruitment of SRC-2 to the genome overlaps with BMAL1 during the light phase targeting expression of core metabolic genes and circadian regulators. In addition, metabolomic profiling of liver metabolites from SRC-2−/− and wildtype littermates identified severe alterations in core metabolic pathways including glycolysis, TCA, and fatty acid biosynthesis (Stashi, et al. 2014). Collectively, these findings uncovered the key role of transcriptional coactivator SRC-2 in circadian biology, and its impact on various metabolic processes.
Coactivators as targets for cancer therapy
Several coactivators including PGC-1, SRC-family members, p300/CBP have been found to be either amplified or overexpressed in different types of cancer (Xu, et al. 2009). SRCs play important roles in endocrine-related cancers such as breast, prostate, ovarian and endometrial cancer (Lonard & O'Malley. 2012) and their functions in other types of cancer are rapidly being decoded (Fig. 3). SRC-1 and SRC-3 promote ER-dependent breast cancer proliferation, as well as facilitate cancer metastasis by upregulating transcription of invasive gene signature coactivating polyoma enhancer activator 3 (PEA3) (Qin, et al. 2009, Qin, et al. 2011). SRC-1 and SRC-3 are overexpressed in endocrine-resistant tumors such as aromatase inhibitor resistant and tamoxifen resistant (McBryan, et al. 2012). In prostate cancer, deep sequencing studies revealed SRC-2 amplification in 8% of primary tumors and 37% metastatic tumors (Taylor, et al. 2010). In addition, SRC-2 expression correlates positively with poor survival of prostate cancer patients (Agoulnik, et al. 2006, Agoulnik & Weigel. 2008), and its expression is an important predictor of time-to-disease relapse (Dasgupta, et al. 2012b). Recent studies have identified coactivators such as SRC-1, SRC-3 and PGC-1α as regulators of bioenergetic pathways in cancer cells (Motamed, et al. 2014, Vazquez, et al. 2013, Zhao, et al. 2014). PGC-1α promotes mitochondrial oxidative phosphorylation to generate sufficient energy supporting the anabolic needs of tumor cells. In addition, recent findings have indicated that coactivators such as p300/CBP along with SRC-3 play critical roles to maintain pluripotency and an embryo stem cell state (Chitilian, et al. 2014, Percharde, et al. 2012, Wu, et al. 2012). SRC-3 coactivates Estrogen-related receptor beta (ESRRB) to enhance the expression of Oct4, Sox2, and the Nanog the master drivers of stem-cellness. Thus it will be important to understand the role of these coactivators in ‘cancer stem cells’.
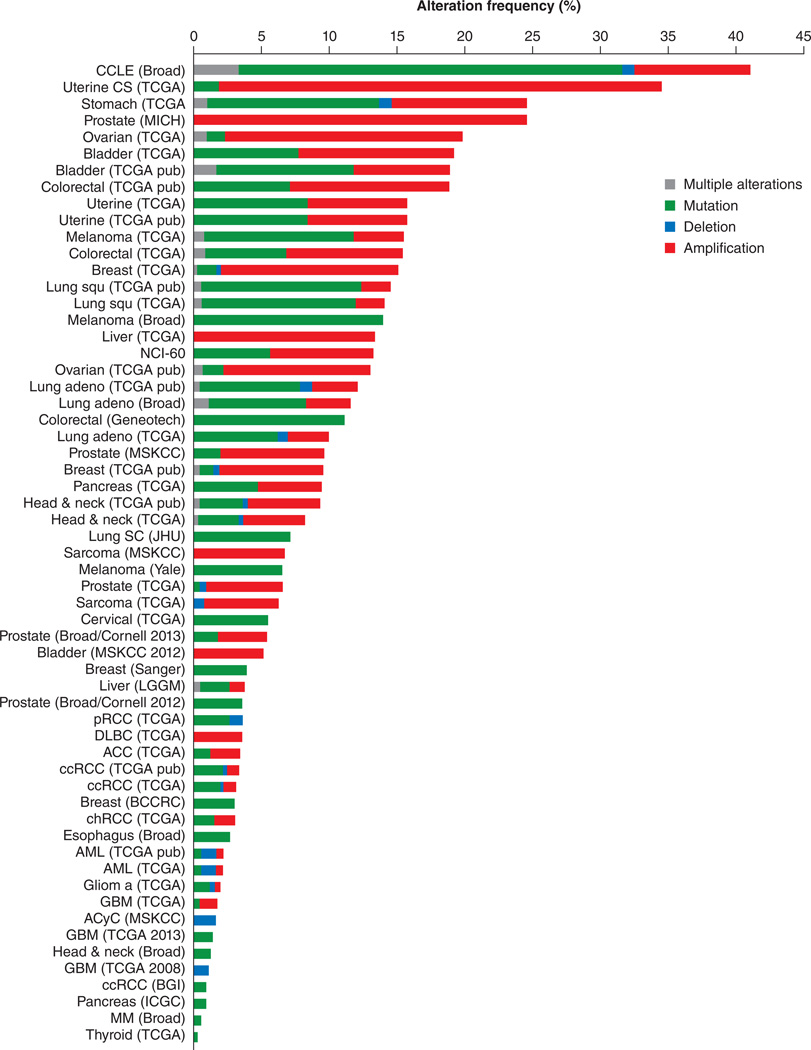
Figure 3.
Graphical representation of percentage of copy number alteration (CNA) frequency of Steroid receptor coactivators (SRC-1, SRC-2 and SRC-3) across different types of cancer. Data represents various types of alterations including gene amplification, mutation, and deletion. Data generated using TCGA datasets from cBIOPortal [Cerami et al. Cancer Discov. 2012 & Gao et al. Sci. Signal. 2013.]
Since SRCs have emerged as ‘master regulators’ of cancer progression and metastasis by integrating various upstream signaling pathways, therapeutic targeting of these molecules may be beneficial for treatment of cancers. High throughput screen (HTS) of a chemical library containing compounds from the NIH-Molecular Libraries Probe Production Centers Network (MLPCN) was used to identify inhibitors blocking the intrinsic transcriptional activity of SRCs (Wang, et al. 2014). The study identified a cardiac glycoside bufalin as a potent small-molecule inhibitor for SRC-3 and SRC-1. Molecularly bufalin and digoxin (a cardiac glycoside) blocked SRC-3 expression by directly binding to it and promoting its rapid degradation in a proteasome-dependent fashion. Bufalin was extremely potent in nanomolar scale to block the growth and proliferation of breast and lung cancer cells (Wang, et al. 2014). In addition, Verrucarin A was also identified as a small molecule inhibitor (SMI) that can selectively promote the degradation of SRC-3 protein, while affecting SRC-1 and SRC-2 to a lesser extent but having no impact on CARM-1 and p300 protein levels. Verrucarin A belongs to a group of sesquiterpene found in toxins of pathogenic fungus, has potent anticancer effects by blocking tumor cell growth, proliferation and migration-invasion (Yan, et al. 2014). Thus, targeting coactivators represents a novel way to block tumor cell growth, and future studies should identify effective small molecule inhibitors to circumvent other pathologies as well.
Conclusion
Transcriptional coactivators have emerged as an important new class of functional proteins that participate with virtually all transcription factors and NRs to intricately regulate gene expression in response to a wide variety of environmental cues. Recent findings have highlighted that coactivators are important for almost all biological functions. Coactivators work in tandem with specific interacting partners to precisely regulate activation of genes, and loss or genetic defects lead to severe pathologies. Future studies will further broaden our understanding about these fascinating molecules in their various biological functions, and drug discovery efforts targeting coactivators may prove valuable for treatment of a variety of diseases.
Acknowledgments
Funding
This work was supported by the National Institutes of Health, USA (R01 HD007857/ NICHD-NIH and 5P01DK059820 from NIDDK-NIH).
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the review reported.
Author contributions
Both authors contributed equally to all aspects of the article.
References
- Agoulnik IU, Weigel NL. Androgen receptor coactivators and prostate cancer. Advances in Experimental Medicine and Biology. 2008;617:245–255. doi: 10.1007/978-0-387-69080-3_23. [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, 3rd, Erdem H, Frolov A, Smith CL, Ayala GE, Ittmann MM, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer research. 2006;66:10594–10602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- Allan GF, Leng X, Tsai SY, Weigel NL, Edwards DP, Tsai MJ, O'Malley BW. Hormone and antihormone induce distinct conformational changes which are central to steroid receptor activation. The Journal of biological chemistry. 1992;267:9513–19520. [PubMed] [Google Scholar]
- Arany Z, Foo SY, Ma Y, Ruas JL, Bommi-Reddy A, Girnun G, Cooper M, Laznik D, Chinsomboon J, Rangwala SM, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell metabolism. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Baniahmad A, Leng X, Burris TP, Tsai SY, Tsai MJ, O'Malley BW. The tau 4 activation domain of the thyroid hormone receptor is required for release of a putative corepressoRs) necessary for transcriptional silencing. Molecular and cellular biology. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai MJ, O'Malley BW. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniahmad C, Nawaz Z, Baniahmad A, Gleeson MA, Tsai MJ, O'Malley BW. Enhancement of human estrogen receptor activity by SPT6: A potential coactivator. Molecular endocrinology (Baltimore, Md.) 1995;9:34–43. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- Banke NH, Wende AR, Leone TC, O'Donnell JM, Abel ED, Kelly DP, Lewandowski ED. Preferential oxidation of triacylglyceride-derived fatty acids in heart is augmented by the nuclear receptor PPARalpha. Circulation research. 2010;107:233–241. doi: 10.1161/CIRCRESAHA.110.221713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. The Journal of allergy and clinical immunology. 2013;131:636–645. doi: 10.1016/j.jaci.2012.12.1564. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Adcock IM, Ito K. Histone acetylation and deacetylation: Importance in inflammatory lung diseases. The European respiratory journal. 2005;25:552–563. doi: 10.1183/09031936.05.00117504. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, Danielian PS, Parker MG. Interaction of proteins with transcriptionally active estrogen receptors. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, et al. Impaired PGC-1alpha function in muscle in huntington's disease. Human molecular genetics. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chitilian JM, Thillainadesan G, Manias JL, Chang WY, Walker E, Isovic M, Stanford WL, Torchia J. Critical components of the pluripotency network are targets for the p300/CBP interacting protein (p/CIP) in embryonic stem cells. Stem cells (Dayton, Ohio) 2014;32:204–215. doi: 10.1002/stem.1564. [DOI] [PubMed] [Google Scholar]
- Chopra AR, Kommagani R, Saha P, Louet JF, Salazar C, Song J, Jeong J, Finegold M, Viollet B, DeMayo F, et al. Cellular energy depletion resets whole-body energy by promoting coactivator-mediated dietary fuel absorption. Cell metabolism. 2011;13:35–43. doi: 10.1016/j.cmet.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra AR, Louet JF, Saha P, An J, Demayo F, Xu J, York B, Karpen S, Finegold M, Moore D, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling von gierke's disease. Science (New York, N.Y.) 2008;322:1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosio BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, Adcock IM. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. American journal of respiratory and critical care medicine. 2004;170:141–147. doi: 10.1164/rccm.200305-659OC. [DOI] [PubMed] [Google Scholar]
- Couture JF, Trievel RC. Histone-modifying enzymes: Encrypting an enigmatic epigenetic code. Current opinion in structural biology. 2006;16:753–760. doi: 10.1016/j.sbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Zhang B, Louet JF, O'Malley BW. Steroid receptor coactivator-2 mediates oncogenic reprogramming of cancer cell metabolism. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research Chicago, IL. Cancer Research. 2012a;72(8 Suppl) 2012; Abstract nr 5153. [Google Scholar]
- Dasgupta S, Srinidhi S, Vishwanatha JK. Oncogenic activation in prostate cancer progression and metastasis: Molecular insights and future challenges. J. Carcinog. 2012b;11:4. doi: 10.4103/1477-3163.93001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O'Malley BW. Nuclear receptor coactivators: Master regulators of human health and disease. Annual Review of Medicine. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner LA, Weigel NL, Maxwell BL, Schrader WT, O'Malley BW. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science (New York, N.Y.) 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- Dobrovolna J, Chinenov Y, Kennedy MA, Liu B, Rogatsky I. Glucocorticoid-dependent phosphorylation of the transcriptional coregulator GRIP1. Molecular and cellular biology. 2012;32:730–739. doi: 10.1128/MCB.06473-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic acids research. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yi P, Wong J, O'Malley BW. Signaling within a coactivator complex: Methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Molecular and cellular biology. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry C, Gaouar S, Fischer B, Boeglin M, Paul N, Samarut E, Piskunov A, Pankotai-Bodo G, Brino L, Rochette-Egly C. Cullin 3 mediates SRC-3 ubiquitination and degradation to control the retinoic acid response. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20603–20608. doi: 10.1073/pnas.1102572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Feng Q, Ding C, Bailey S, Hunsaker TL, Malovannaya A, Hamilton RA, Gates LA, Zhang Z, Li C, et al. Proteomic analysis of coregulators bound to ERalpha on DNA and nucleosomes reveals coregulator dynamics. Molecular cell. 2013;51:185–199. doi: 10.1016/j.molcel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons RJ, Wada T, Fisher CA, Malik N, Mitson MJ, Steensma DP, Fryer A, Goudie DR, Krantz ID, Traeger-Synodinos J. Mutations in the chromatin-associated protein ATRX. Human mutation. 2008;29:796–802. doi: 10.1002/humu.20734. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Brueton L, Buckle VJ, Burn J, Clayton-Smith J, Davison BC, Gardner RJ, Homfray T, Kearney L, Kingston HM. Clinical and hematologic aspects of the X-linked alpha-thalassemia/mental retardation syndrome (ATR-X) American Journal of Medical Genetics. 1995;55:288–299. doi: 10.1002/ajmg.1320550309. [DOI] [PubMed] [Google Scholar]
- Gregory CW, Fei X, Ponguta LA, He B, Bill HM, French FS, Wilson EM. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. The Journal of biological chemistry. 2004;279:7119–7130. doi: 10.1074/jbc.M307649200. [DOI] [PubMed] [Google Scholar]
- Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: Possible mediators of hormone-induced transcription. Science (New York, N.Y.) 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Han SJ, Lonard DM, O'Malley BW. Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends in endocrinology and metabolism: TEM. 2009;20:8–15. doi: 10.1016/j.tem.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Hawkins SM, Begum K, Jung SY, Kovanci E, Qin J, Lydon JP, DeMayo FJ, O'Malley BW. A new isoform of steroid receptor coactivator-1 is crucial for pathogenic progression of endometriosis. Nature medicine. 2012;18:1102–1111. doi: 10.1038/nm.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annual Review of Physiology. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Frizzell A. Histone deacetylase complexes as caretakers of genome stability. Epigenetics : official journal of the DNA Methylation Society. 2012;7:806–810. doi: 10.4161/epi.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. The Journal of clinical investigation. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liang YY, Feng XH, Tsai SY, Tsai MJ, O'Malley BW. Essential phosphatases and a phospho-degron are critical for regulation of SRC-3/AIB1 coactivator function and turnover. Molecular cell. 2008;31:835–849. doi: 10.1016/j.molcel.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. Nuclear receptor coregulators: Modulators of pathology and therapeutic targets. Nature reviews.Endocrinology. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'malley BW. Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Molecular cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, Mancini MA, Tsai SY, Tsai MJ, O'Malley BW. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Molecular cell. 2010;37:321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba II, Qin J, Tsai SY, et al. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. The Journal of clinical investigation. 2012;122:1869–1880. doi: 10.1172/JCI61492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, Chopra AR, Sagen JV, An J, York B, Tannour-Louet M, Saha PK, Stevens RD, Wenner BR, Ilkayeva OR, et al. The coactivator SRC-1 is an essential coordinator of hepatic glucose production. Cell metabolism. 2010;12:606–618. doi: 10.1016/j.cmet.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Li Y, Bulynko Y, Jung SY, Wang Y, Lanz RB, O'Malley BW, Qin J. Streamlined analysis schema for high-throughput identification of endogenous protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2431–2436. doi: 10.1073/pnas.0912599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryan J, Theissen SM, Byrne C, Hughes E, Cocchiglia S, Sande S, O'Hara J, Tibbitts P, Hill AD, Young LS. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC-1. Cancer research. 2012;72:548–559. doi: 10.1158/0008-5472.CAN-11-2073. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Vegeto E, O'Malley BW. Identification of a negative regulatory function for steroid receptors. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Nawaz Z, O'Malley BW. In situ distinction between steroid receptor binding and transactivation at a target gene. Molecular and cellular biology. 1991a;11:4350–4355. doi: 10.1128/mcb.11.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Nawaz Z, Densmore C, Weigel NL, Pham TA, Clark JH, O'Malley BW. High level expression of biologically active estrogen receptor in saccharomyces cerevisiae. The Journal of steroid biochemistry and molecular biology. 1991b;39:291–297. doi: 10.1016/0960-0760(91)90038-7. [DOI] [PubMed] [Google Scholar]
- Means AR, Comstock JP, Rosenfeld GC, O'Malley BW. Ovalbumin messenger RNA of chick oviduct: Partial characterization, estrogen dependence, and translation in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1972;69:1146–1150. doi: 10.1073/pnas.69.5.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer OC, Steenbergen PJ, De Kloet ER. Differential expression and regional distribution of steroid receptor coactivators SRC-1 and SRC-2 in brain and pituitary. Endocrinology. 2000;141:2192–2199. doi: 10.1210/endo.141.6.7489. [DOI] [PubMed] [Google Scholar]
- Motamed M, Rajapakshe KI, Hartig SM, Coarfa C, Moses RE, Lonard DM, O'Malley BW. Steroid receptor coactivator 1 is an integrator of glucose and NAD(+)/NADH homeostasis. Molecular endocrinology (Baltimore, Md.) 2014;28:395–405. doi: 10.1210/me.2013-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science (New York, N.Y.) 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Park E, Kim Y, Ryu H, Kowall NW, Lee J, Ryu H. Epigenetic mechanisms of rubinstein-taybi syndrome. Neuromolecular medicine. 2014;16:16–24. doi: 10.1007/s12017-013-8285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, Martin N, Yeo JC, Gil J, Prabhakar S, Ng HH, et al. Ncoa3 functions as an essential esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes & development. 2012;26:2286–2298. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Power RF, Mani SK, Codina J, Conneely OM, O'Malley BW. Dopaminergic and ligand-independent activation of steroid hormone receptors. Science (New York, N.Y.) 1991;254:1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer research. 2009;69:3819–3827. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Chen X, Wu Y, Feng Z, He T, Wang L, Liao L, Xu J. Steroid receptor coactivator-1 upregulates integrin alpha(5) expression to promote breast cancer cell adhesion and migration. Cancer research. 2011;71:1742–1751. doi: 10.1158/0008-5472.CAN-10-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Yang Y, Ma D, He L, Xiao W. Expression level of histone deacetylase 2 correlates with occurring of chronic obstructive pulmonary diseases. Molecular biology reports. 2013;40:3995–4000. doi: 10.1007/s11033-012-2477-z. [DOI] [PubMed] [Google Scholar]
- Reineke EL, York B, Stashi E, Chen X, Tsimelzon A, Xu J, Newgard CB, Taffet GE, Taegtmeyer H, Entman ML, et al. SRC-2 coactivator deficiency decreases functional reserve in response to pressure overload of mouse heart. PloS one. 2012;7:e53395. doi: 10.1371/journal.pone.0053395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R, Wellstein A, Riegel AT. An isoform of the coactivator AIB1 that increases hormone and growth factor sensitivity is overexpressed in breast cancer. The Journal of biological chemistry. 2001;276:39736–39741. doi: 10.1074/jbc.M104744200. [DOI] [PubMed] [Google Scholar]
- Rowan BG, Garrison N, Weigel NL, O'Malley BW. 8-bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Molecular and cellular biology. 2000;20:8720–8730. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, Jiang A, Arany Z. PGC-1 coactivators in cardiac development and disease. Circulation research. 2010;107:825–838. doi: 10.1161/CIRCRESAHA.110.223818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelsberg TC, Steggles AW, O'Malley BW. Progesterone-binding components of chick oviduct. 3. chromatin acceptor sites. The Journal of biological chemistry. 1971;246:4188–4197. [PubMed] [Google Scholar]
- Stashi E, Wang L, Mani SK, York B, O'Malley BW. Research resource: Loss of the steroid receptor coactivators confers neurobehavioral consequences. Molecular endocrinology (Baltimore, Md.) 2013;27:1776–1787. doi: 10.1210/me.2013-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Reineke LC, Dasgupta S, et al. SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell reports. 2014;6:633–645. doi: 10.1016/j.celrep.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: Genes, syndromes, and therapies. Lancet neurology. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegeto E, Allan GF, Schrader WT, Tsai MJ, McDonnell DP, O'Malley BW. The mechanism of RU486 antagonism is dependent on the conformation of the carboxy-terminal tail of the human progesterone receptor. Cell. 1992;69:703–713. doi: 10.1016/0092-8674(92)90234-4. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. The EMBO journal. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lonard DM, Yu Y, Chow DC, Palzkill TG, Wang J, Qi R, Matzuk AJ, Song X, Madoux F, et al. Bufalin is a potent small-molecule inhibitor of the steroid receptor coactivators SRC-3 and SRC-1. Cancer research. 2014;74:1506–1517. doi: 10.1158/0008-5472.CAN-13-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun L, Zhang Y, Chen Y, Shi B, Li R, Wang Y, Liang J, Fan D, Wu G, et al. Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. The Journal of biological chemistry. 2006;281:21848–21856. doi: 10.1074/jbc.M603772200. [DOI] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Molecular cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yang M, Liu H, Guo H, Wang Y, Cheng H, Chen L. Role of nuclear receptor coactivator 3 (ncoa3) in pluripotency maintenance. The Journal of biological chemistry. 2012;287:38295–38304. doi: 10.1074/jbc.M112.373092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nature reviews.Cancer. 2009;9:615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O'Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science (New York, N.Y.) 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Alberts B. The interaction of estradiol-receptor protein with the genome: An argument for the existence of undetected specific sites. Cell. 1975;4:301–310. doi: 10.1016/0092-8674(75)90150-6. [DOI] [PubMed] [Google Scholar]
- Yan F, Yu Y, Chow DC, Palzkill T, Madoux F, Hodder P, Chase P, Griffin PR, O'Malley BW, Lonard DM. Identification of verrucarin a as a potent and selective steroid receptor coactivator-3 small molecule inhibitor. PLoS One. 2014;9:e95243. doi: 10.1371/journal.pone.0095243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Feng Q, Amazit L, Lonard DM, Tsai SY, Tsai MJ, O'Malley BW. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Molecular cell. 2008;29:465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Wu RC, Sandquist J, Wong J, Tsai SY, Tsai MJ, Means AR, O'Malley BW. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1) Molecular and cellular biology. 2005;25:9687–9699. doi: 10.1128/MCB.25.21.9687-9699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, O'Malley BW. Steroid receptor coactivator (SRC) family: Masters of systems biology. The Journal of biological chemistry. 2010;285:38743–38750. doi: 10.1074/jbc.R110.193367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York B, Reineke EL, Sagen JV, Nikolai BC, Zhou S, Louet JF, Chopra AR, Chen X, Reed G, Noebels J, et al. Ablation of steroid receptor coactivator-3 resembles the human CACT metabolic myopathy. Cell metabolism. 2012;15:752–763. doi: 10.1016/j.cmet.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, York B, Wang S, Feng Q, Xu J, O'Malley BW. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Molecular cell. 2007;25:765–778. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Chang C, Cui Y, Zhao X, Yang J, Shen L, Zhou J, Hou Z, Zhang Z, Ye C, et al. Steroid receptor coactivator-3 regulates glucose metabolism in bladder cancer cells through coactivation of hypoxia inducible-factor 1alpha. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M113.535989. [DOI] [PMC free article] [PubMed] [Google Scholar]