Abstract
The expression of human G protein-coupled receptors (GPCRs) in Saccharomyces cerevisiae containing chimeric yeast/mammalian Gα subunits provides a useful tool for the study of GPCR activation. In this study, we used a one-GPCR-one-G protein yeast screening method in combination with molecular modeling and mutagenesis studies to decipher the interaction between GPCRs and the C-terminus of different α-subunits of G proteins. We chose the human adenosine A2B receptor (hA2BR) as a paradigm, a typical class A GPCR that shows promiscuous behavior in G protein coupling in this yeast system. The wild-type hA2BR and five mutant receptors were expressed in 8 yeast strains with different humanized G proteins, covering the four major classes: Gαi, Gαs, Gαq, and Gα12. Our experiments showed that a tyrosine residue (Y) at the C-terminus of the Gα subunit plays an important role in controlling the activation of GPCRs. Receptor residues R1033.50 and I1073.54 are vital too in G protein-coupling and the activation of the hA2BR, whereas L213IL3 is more important in G protein inactivation. Substitution of S2356.36 to alanine provided the most divergent G protein-coupling profile. Finally, L2366.37 substitution decreased receptor activation in all G protein pathways, although to a different extent. In conclusion, our findings shed light on the selectivity of receptor/G protein coupling, which may help in further understanding GPCR signaling.
Keywords: G protein-coupled receptor, G protein coupling, Yeast screening, Adenosine A2B receptor, DRY motif
Introduction
G protein-coupled receptors (GPCRs), also known as seven-transmembrane receptors (7TMRs), are a major class of targets for many of today’s medicines, to combat ailments such as inflammation, cardiac malfunction, asthma, and cancer. Ligands interact with these transmembrane proteins in many different ways, intervening with or mimicking their activation process, which is mediated mostly by a heterotrimeric G protein, composed of α, β, and γ subunits [1, 2]. However, the exact mechanism of GPCR activation at the molecular level is still largely unknown. Here, we used the hA2BR, a typical class A GPCR, as a paradigm to decipher the interaction between receptors and their G proteins.
The adenosine receptors include four subtypes: A1R, A2AR, A2BR, and A3R, which have attracted much attention as therapeutic targets in recent years. All the adenosine receptors are ubiquitously expressed in the human body [3] and can target different intracellular signaling pathways by responding to the same endogenous ligand adenosine. The A2BR has the lowest affinity for adenosine [4] and has been less investigated than other adenosine receptors. Several studies have shown that blocking A2BR signaling reduces experimental autoimmune encephalomyelitis [5] and inhibits growth of prostate cancer cells [6], breast tumors [7], and bladder tumors [8, 9]. On the other hand, stimulation of A2BR protects against trauma-hemorrhagic shock-induced lung injury [10], CHX-induced apoptosis [6] and also vascular injury [11].
Yeast can provide a powerful platform for studying GPCRs and their G protein coupling and selectivity. Saccharomyces cerevisiae (S. cerevisiae) strains, each expressing a specific human Gα/yeast Gpa1 protein chimera, have been used to express heterologous GPCRs, for instance in high-throughput screening assays for drug discovery [12], to perform random mutagenesis screening [13, 14], and to assess the preference of Gα pathways [15] and functional selectivity of agonists and antagonists [16–18]. Several lines of evidence indicate that the C-terminal five amino acid residues of a G protein are sufficient for coupling with many human receptors, including the hA2BR [19]. One of the advantages of the yeast system used is that while these five amino acids of many human G protein α-subunits were “transplanted” to replace these residues on the yeast’s endogenous G protein, Gpa1p (see also Table 1), all other aspects of the system remain the same and intact [20, 21].
Table 1.
| Strain | Genotype | The five C-terminal residues of Gα |
|---|---|---|
| MMY11 | MATahis3 ade2leu2 trp1 ura3can1fus1::FUS1-HIS3FUS1-lacZ::LEU2 far∆::ura3∆gpa1∆ | |
| ::ADE2∆sst2∆::ura3∆ste2∆::G418 R | ||
| MMY12 (GαWT) | MMY11TRP1::GPA1 | KIGIICOOH |
| MMY23 (Gαi1) | MMY11TRP1::Gpa1/G αi1(5) | DCGLFCOOH |
| MMY24 (Gαi3) | MMY11TRP1::Gpa1/G αi3(5) | ECGLYCOOH |
| MMY28 (Gαs) | MMY11TRP1::Gpa1/G αs(5) | QYELLCOOH |
| MMY14 (Gαq) | MMY11TRP1::Gpa1/G αq(5) | EYNLVCOOH |
| MMY21 (Gα14) | MMY11TRP1::Gpa1/G α14(5) | EFNLVCOOH |
| MMY19 (Gα12) | MMY11TRP1::Gpa1/G α12(5) | DIMLQCOOH |
| MMY20 (Gα13) | MMY11TRP1::Gpa1/G α13(5) | QLMLQCOOH |
In the present study, we used this one-GPCR-one-G protein yeast screening method in combination with molecular modeling and mutagenesis studies. Our goal was to provide more information about the mechanism of activation of the hA2BR and the role the binding pocket for the Gα protein’s C-terminus plays in that process. Our findings provide further evidence for the A2B receptor’s G protein preferences, which in itself may be useful for designing and screening selective agonists and antagonists for this receptor. At the same time, our findings have a broader relevance as they reflect on the GPCR-G protein interface.
Materials and methods
hA2B Receptor/G protein homology modeling
A homology model was created using Molsoft’s ICM Homology tool (Version 3.7-2) [22]. The β2 adrenergic receptor (β2AR) in complex with the Gs protein [23] (PDB: 3SN6) was chosen as template since it was the closest (and currently only) homolog of the hA2BR containing the Gαs-protein, with 30 % sequence identity and 48 % sequence similarity using a pair-wise sequence alignment method (EMBOSS_matcher; http://www.ebi.ac.uk/Tools/psa/emboss_matcher/). The hA2BR sequence (uniprot: P29275) was modeled onto 3SN6 and residues were selected to be individually mutated to alanine based on the following two criteria: (1) within 5 Å distance from the last five amino acids of the Gαs protein (QYELL) and (2) oriented towards the Gαs-protein. Final visualizations were created using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC) [24].
Generation of point mutations
The S. cerevisiae expression vectors, the pDT-PGK and pDT-PGK_A2B receptor plasmids, were kindly provided by Dr. Simon Dowell from GSK (Stevenage, UK). The DNA primers of the mutants of the A2B receptor were designed by the QuikChange® Primer Design Program on the website of Agilent Technologies, and contained a single substitution resulting in a codon change for the desired amino acid substitution. The reverse primer sequence of each mutant was the reverse complement of the forward primer. These primers and their complements were synthesized (Eurogentec, the Netherlands) and then used to generate mutant plasmids according to the QuikChange method from Stratagene. The mutagenic reaction contained 40 ng of the pDT-PGK_A2B construct plasmid as dsDNA template, 10 μM of each primer, 1 μl of dNTP mix, 2.5 μl of 10× reaction buffer and 2.5 U PfuUltra HF DNA polymerase. The following thermal cycling parameters were used in the PCR apparatus (T100™ Thermal Cycler, BIO-RAD): 95 °C for 30 s, 55 °C for 1 min, and 68 °C for 10 min. The number of mutagenic PCR cycles was set to 20. Methylated or hemimethylated nonmutated plasmid DNA was removed by adding 5 U Dpn I restriction enzyme for 2 h at 37 °C. The mutated DNA products were transformed into XL-1 Blue supercompetent cells and other details were according to the manual of the QuikChange® II Site-Directed Mutagenesis Kit. Mutant plasmids were isolated from a single clone using a QIAprep midi plasmid purification kit (QIAGEN, the Netherlands). The mutants were confirmed by DNA sequencing (LGTC, Leiden, the Netherlands).
Transformation in S. cerevisiae strains
Each mutant plasmid was transformed according to the Lithium-Acetate procedure [25] into a panel of engineered S. cerevisiae yeast strains expressing different Gpa1p/Gα chimeras. The yeast strains were derived from the MMY11 strain and further adapted to communicate with mammalian GPCRs. The difference between these integrated Gpa1p/Gα chimeras is that the last five amino acids of the endogenous Gpa1p C-terminus have been replaced with the same sequence as that from mammalian Gα proteins [20, 21] (Table 1). To measure the signaling of GPCRs, the pheromone signaling pathway of these strains was coupled via the FUS1 promoter to HIS3 (imidazoleglycerol-phosphate dehydratase), an enzyme catalyzing the sixth step in histidine biosynthesis to produce histidine. 3-AT (3-amino-[1,2,4]-triazole), a competitive inhibitor of imidazoleglycerol-phosphate dehydratase, was added to the assay to reduce background activity caused by endogenous histidine [20]. The degree of receptor activation induced by an agonist of the GPCR was measured by the growth rate of the yeast on histidine-deficient medium.
Liquid growth assay
To measure the efficiency of GPCR-G protein coupling, concentration-growth curves were generated in a liquid growth assay [26]. This assay was carried out in 96-well plates and the growth was determined by measuring the absorption at a wavelength of approx. 600 nm (OD600). To set up an assay, cells were grown to saturation selecting for the transformed plasmid, then seeded at low cell density (2 × 104 cells/ml) into assay medium (YNB + adenine + tryptophan + 10 mM 3-AT) lacking histidine and dispensed into assay plates containing the adenosine receptor agonist NECA (10−9–10−4 M). The 96-well plate was then incubated for 35 h at 30 °C in a Genios plate reader (Tecan, Durham, NC), keeping the cells in suspension by shaking every 10 min at 300 rpm for 1 min. Results were obtained from two independent experiments, performed in duplicate. EC50 values and Emax values of the liquid assay were analyzed using the nonlinear regression package available in Prism 5.0 software (GraphPad Software Inc., San Diego, CA, USA).
Whole cell radioligand-binding experiments
Yeast cells from an overnight culture expressing the wild-type or mutated hA2BR were harvested from rich YAPD medium by centrifuging at 2,000 g for 5 min. The pellet of cells was washed once with 0.9 % NaCl. The cells were centrifuged again using the same speed and diluted in the assay buffer (50 mM Tris–HCl pH7.4 + 1 mM EDTA) to OD600 = 40 (OD600 = 1 ≈ 2.5 × 107 cells/ml). Binding experiments were performed with 1.5 nM of the A2B receptor selective antagonist [3H]PSB-603, and a final cell concentration of 25 × 107 cells/ml in a total volume of 100 μl. Nonspecific binding (NSB) was determined by additionally adding NECA at a final concentration of 1 mM. Samples were incubated for 1 h at 25 ºC keeping the cells in suspension by shaking vigorously. One milliliter of ice-cold assay buffer was added to samples to terminate incubation and the samples were harvested on a Millipore manifold with GF/B filters pre-incubated in 0.1 % polyethylenimine (PEI) at a pressure of 200 mbar, to separate free from receptor-bound radioligand by washing twice with 2 ml buffer (50 mM Tris–HCl pH7.4 + 1 mM EDTA + 0.1%BSA). The filters were transferred into mini-vials and 3.5 ml of PerkinElmer Emulsifier Safe was added, and subsequently incubated for at least 2 h. Filter-bound radioactivity was determined as counts per minute by scintillation spectrometry (Tri-Carb 2900TR; PerkinElmer Life and Analytical Sciences). Results were obtained from at least three independent experiments, performed in duplicate.
Preparation of yeast protein extractions and immunoblotting
Protein extractions were performed with trichloroacetic acid (TCA) according to the Clontech Yeast Protocols Handbook 2001. Yeast transformants were grown in 2 ml YAPD medium and were harvested in mid-exponential phase (1.2 × 108 cells). The yeast cells were collected and washed with cold water. Subsequently, the yeast cells were broken by vigorous vortexing with 20 % TCA and glass beads. The broken yeast cells and the glass beads were washed twice with 200 μl 5 % TCA and centrifuged at 3,000 rpm for 2 min. The supernatant was collected and centrifuged again at 6,000 rpm for 2 min. The pellets were resuspended with cold SDS/PAGE loading buffer (100 mM EDTA, 1 M Tris, 10 % SDS, 0.5 % Bromophenol blue) and 1 M Tris was added to neutralize all remaining TCA. The samples were incubated for 30 min at 37 °C and centrifuged again at 2,000 rpm for 10 min.
Each sample of 4 μl containing 24 μg protein was loaded on 12.5 % SDS/PAGE gel and then blotted on Hybond-ECL membranes (GE Healthcare, the Netherlands) using a semi-automated electrophoresis technique (PhastSystem™, Amersham Pharmacia Biotech). The Hybond-ECL membranes were blocked with TBS containing 5 % milk powder for 1 h and washed three times with TBST (0.05 % Tween-20, TBS pH 7.6). The membranes were incubated with 1:1,250 diluted rabbit anti-human A2B receptor antibody for 1 h. This antibody was directed against the C-terminal region of the A2BR and was kindly provided by Dr. I. Feoktistov (Vanderbilt University, Nashville) [27]. After thorough removal of unbound antibody from the membranes by washing three times with TBST, the membranes were incubated with 1:2,500 diluted HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories) for 1 h. The membranes were washed twice with TBST and once with TBS. The specific signal of the A2B receptor was probed according to the ECL Western blotting analysis system (GE Heathcare, the Netherlands) using enhanced chemiluminescence (ChemiDox XRS, BIO-RAD). The nonspecific band at approximately 45 kDa was used as loading control and the specific hA2BR protein bands were at 29 and 48 kDa. The ratio was determined between the density of the specific bands and density of the nonspecific band that was always present on the blots. MMY12 carrying wild-type or mutant receptor was set as 100 % and MMY12 carrying the empty vector pDT-PGK without receptor was set as 0 %.
Results
G protein selectivity of the wild-type hA2B receptor
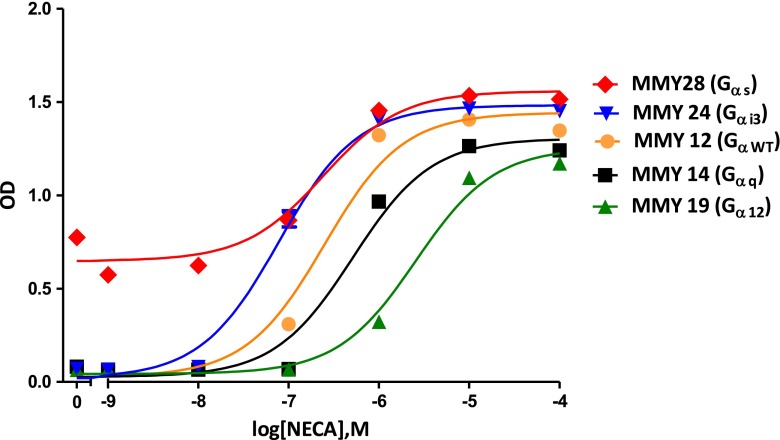
To investigate the activation mechanism of the hA2B receptor at the interface of the C-terminus of the G protein Gα subunit, we expressed the yeast plasmid pDT-PGK_hA2BR in a panel of eight yeast S. cerevisiae strains with humanized G proteins. Corresponding to the replaced last five C-terminal residues of the mammalian Gα subunit, they were classified into five families: GαWT (MMY12), Gαs (MMY28), Gαi (MMY23 and MMY24), Gα12 (MMY19 and MMY20), and Gαq (MMY14 and MMY21) (Table 1) [19]. When the expressed hA2B receptor is activated by an agonist, the yeast pheromone signaling pathway is activated through a chimeric yeast/mammalian G protein leading to subsequent transcription of the HIS3 reporter gene and consequently histidine production [28]. Subsequent growth of the yeast cells on histidine-deficient medium was determined by measuring the absorption at a wavelength of 595 nm, which reflects the activation of the expressed receptor by the adenosine receptor agonist NECA. Concentration-response curves of NECA on the wild-type receptor in MMY28(Gαs), MMY24(Gαi3), MMY12(GαWT), MMY19(Gα12), and MMY14(Gαq) are shown in Fig. 1 and the values of EC50, and Emax of all strains are shown in Table 2.
Fig. 1.
Concentration-effect curves from liquid assay experiments are shown of the A2B wild-type receptor in MMY28 (Gαs) (red filled diamond), MMY24 (Gαi3) (blue filled inverse triangle), MMY12 (GαWT) (filled orange circle), MMY14 (Gαq) (filled black square) and MMY19 (Gα12) (filled green upright triangle) responding to the adenosine receptor agonist NECA. The assay was performed in YNB-ULH medium
Table 2.
EC50, fold of EC50, and E max values of wild-type and mutant A2B receptors in all examined MMY strains. The fold shift of EC50 was calculated by dividing the EC50 of the mutant receptor by the EC50 of the wild-type receptor of the same strain. %E max represents the intrinsic activity of the receptor, where the mean maximal growth level of MMY12 carrying wild-type receptor in response to the agonist NECA (10−4 M) was set as 100 %. Results were mostly obtained from two independent experiments, performed in duplicate (individual values in parentheses)
| EC50 | Fold EC50 | % E max | ||
|---|---|---|---|---|
| MMY 12 (GαWT) | Wild type | 393 ± 92 | 1 | 100 ± 3 |
| R1033.50A | – | – | 13 (17, 9) | |
| I1073.54A | – | – | 0 (0, 1) | |
| L213IL3A | 236 (152, 319) | 0.6 | 100 (104, 96) | |
| S2356.36A | 384 (309, 459) | 1 | 85 (83, 87) | |
| L2366.37A | 3,099 (2,503, 3,694) | 7.9 | 102 (109, 96) | |
| MMY 14 (Gαq) | Wild type | 1,641 ± 552 | 1 | 99 ± 3 |
| R1033.50A | – | – | 1 (1, 1) | |
| I1073.54A | – | – | (0, 0) | |
| L213IL3A | 483 (461, 504) | 0.3 | 76 (75, 77) | |
| S2356.36A | 3,109 (2,102, 4,115) | 1.9 | 50 (48, 51) | |
| L2366.37A | 7,069 (9,827, 4,310) | 4.3 | 83 (83, 82) | |
| MMY 19 (Gα12) | Wild type | 2,843 ± 1,141 | 1 | 91 ± 8 |
| R1033.50A | – | – | 2 (2, 1) | |
| I1073.54A | – | – | 1 (0, 1) | |
| L213IL3A | 272 (199, 345) | 0.1 | 100 (100, 99) | |
| S2356.36A | – | – | 19 (8, 31) | |
| L2366.37A | – | – | 50 (60, 40) | |
| MMY 20 (Gα13) | Wild type | 384 ± 158 | 1 | 99 ± 7 |
| R1033.50A | – | – | 6 (10, 2) | |
| I1073.54A | – | – | 3 (3, 3) | |
| L213IL3A | 90 (42, 137) | 0.2 | 102 (107, 97) | |
| S2356.36A | 437 (501, 372) | 1.1 | 77 (75, 80) | |
| L2366.37A | 5,375 (4,700, 6,050) | 14 | 85 (87, 84) | |
| MMY 21 (Gα14) | Wild type | 212 ± 91 | 1 | 108 ± 7 |
| R1033.50A | – | – | 4 (5, 2) | |
| I1073.54A | – | – | 1 (0, 1) | |
| L213IL3A | 34 (17, 51) | 0.2 | 107 (108, 106) | |
| S2356.36A | 664 (552, 776) | 3.1 | 80 (78, 82) | |
| L2366.37A | 1,335 (783, 1,887) | 6.3 | 99 (104, 94) | |
| MMY 23 (Gαi1) | Wild type | 305 ± 137 | 1 | 91 ± 8 |
| R1033.50A | – | – | 9 (15, 3) | |
| I1073.54A | 7,629 (9,162, 6,095) | 25 | 37 (38, 37) | |
| L213IL3A | 108 (72, 143) | 0.4 | 95 (96, 94) | |
| S2356.36A | 84 (103, 65) | 0.3 | 82 (76, 88) | |
| L2366.37A | 411 (398, 424) | 1.3 | 98 (102, 95) | |
| MMY 24 (Gαi3) | Wild type | 59 ± 10 | 1 | 113 ± 5 |
| R1033.50A | – | – | 9 (7, 10) | |
| I1073.54A | – | – | 2 (2, 1) | |
| L213IL3A | 49 (25, 72) | 0.8 | 104 (113, 96) | |
| S2356.36A | 175 (174, 176) | 3 | 105 (109, 102) | |
| L2366.37A | 680 (504, 855) | 11.6 | 112 (121, 103) | |
| MMY 28 (Gαs) | Wild type | 25 ± 6 | 1 | 112 ± 4 |
| R1033.50A | 57 (53, 61) | 2.2 | 95 (98, 92) | |
| I1073.54A | 47 (25, 69) | 1.9 | 103 (100, 106) | |
| L213IL3A | 23 (31, 15) | 0.9 | 105 (104, 106) | |
| S2356.36A | 33 (42, 24) | 1.3 | 105 (110, 100) | |
| L2366.37A | 538 (755, 321) | 21 | 96 (95, 98) |
We found the five humanized different G protein pathways MMY28(Gαs), MMY24(Gαi3), MMY21(Gα14), MMY23(Gαi1), and MMY20(Gα13) to show varying degrees of enhancement compared to the coupling efficiency of the wild-type hA2BR in the wild-type yeast Gα strain MMY12(GαWT). The most efficient yeast strain MMY28(Gαs) showed a significant 15-fold improvement in coupling efficiency and also MMY24(Gαi3) showed a significant 6.5-fold enhancement in coupling efficiency. Two strains, MMY14(Gαq) and MMY19(Gα12), responded less to NECA, with significantly higher EC50 values for this agonist. In terms of intrinsic activity (Emax values) there was little difference between the strains, however. The one exception was in MMY28(Gαs) with a large degree of constitutive activity (approx. 30 % of the maximal response of MMY12 in response to the agonist NECA (10−4 M)). This may be a feature of the MMY28 strain itself since expression of plasmid pDT-PGK (without any receptor) yielded a similar degree of constitutive activity (data not shown).
Bioinformatics and molecular modeling
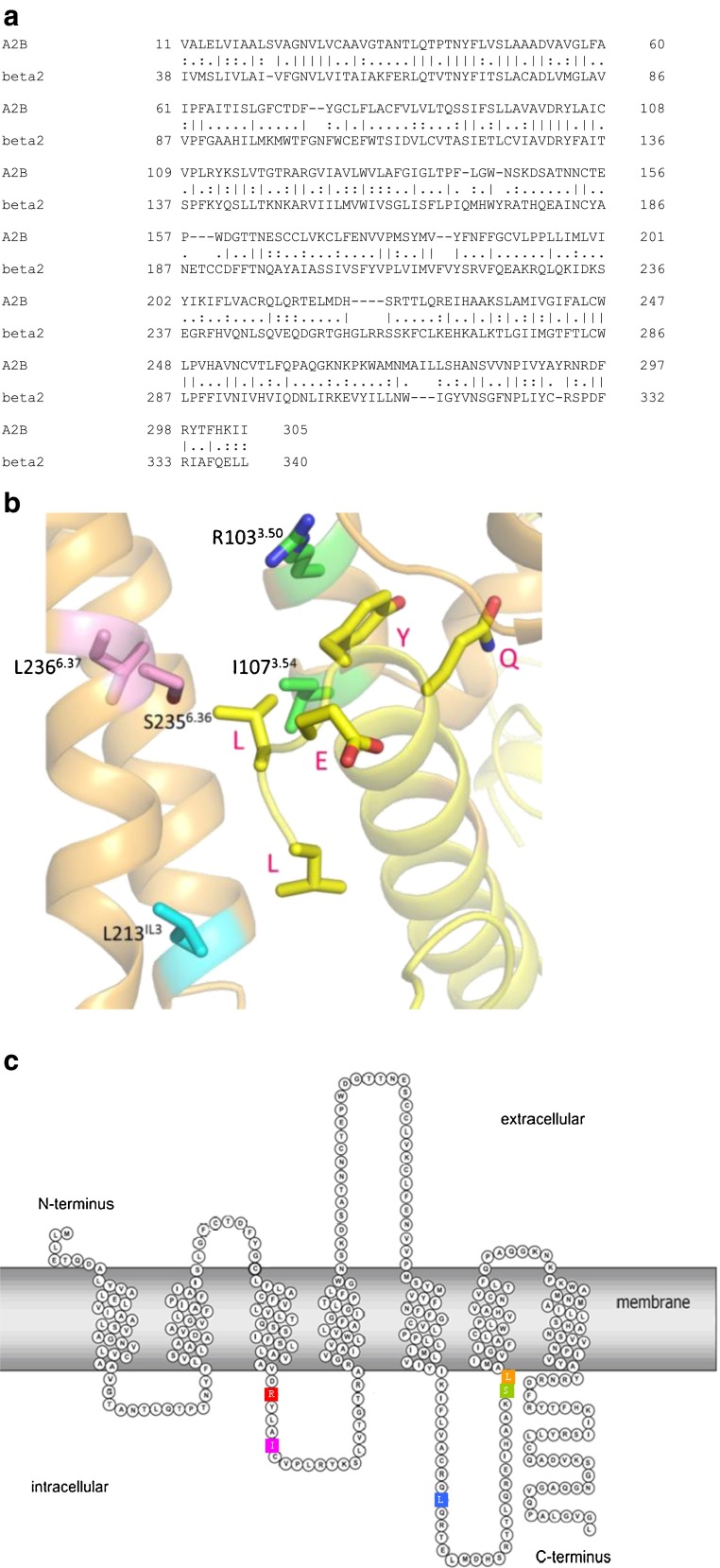
To further decipher the interaction between the hA2BR and G proteins, we predicted a number of amino acid residues as important for activation of G proteins from hA2BR homology modeling. There are more than 75 crystal structures of 18 different class A GPCRs now [29], however, only two receptors, β2AR and opsin (Ops), have been cocrystallised with the Gαs protein [23] and an 11 amino acid synthetic peptide of Gαt [30], respectively. The β2AR-Gs crystal structure (PDB: 3SN6) was chosen as template for mapping amino acid residues of hA2BR that are involved in G protein activation. The hβ2AR shares 30 % sequence identity with the hA2BR (Fig. 2a), compared to 23 % homology with bovine opsin. In many mammalian cells, the hA2BR prefers to couple to Gs next to Gq proteins [31], another indication of the validity of this particular homology modeling approach. The model predicted 16 amino acids to interact with Gαs (i.e., with QYELL, the last five amino acid residues of it): D1023.49, R1033.50, A1063.53, I1073.54, Y1133.60, I2055.61, A2095.65, Q212 IL3, L213IL3, R215 IL3, H2316.32, A2326.33, S2356.36, L2366.37, R2938.46, N2948.47.
Fig. 2.
a Sequence alignment (most similar regions only) between the hA2BR (A2B; uniprot: P29275) and the hβ2AR (beta2; uniprot:P07550). Conserved residues are shown as | between the two sequences. b A hA2B Receptor/Gs protein homology model was generated from the crystal structure of the β2AR in contact with the Gαs protein [23] (PDB: 3SN6) to predict amino acids interacting with Gαs. The last five C-terminal residues of mammalian Gαs subunit are QYELLCOOH, shown in red. c Snake plot of the hA2BR. Five residues (R1033.50, I1073.54, L213 IL3, S2356.36, and L2366.37) were selected to be individually mutated to alanine based on the homology model in b
Arginine (R) 1033.50, isoleucine (I) 1073.54, leucine (L) 213 IL3, serine (S) 2356.36, and leucine (L) 2366.37 were selected for mutation into alanine as they are both closest to the Gαs-protein and with their side chains oriented towards the Gαs-protein as well (Fig. 2b). These selected five residues are also shown in the snake plot of the hA2BR (Fig. 2c). R1033.50 and I1073.54 are located on the intracellular side of TM3 and are included in the consensus sequence (I/L)XXDR3.50YXX(I/V)3.54 [32]. R3.50 is a part of the most conserved motif in the class A GPCRs: Asp-Arg-Tyr (DRY). The residues on positions 3.50, 3.54, and 6.36 according to Ballesteros-Weinstein numbering [33] are also conserved as part of the G protein-binding region in the bovine opsin-Gt peptide crystal structure [23, 29, 30].
G protein coupling profiles of five mutant A2B receptors
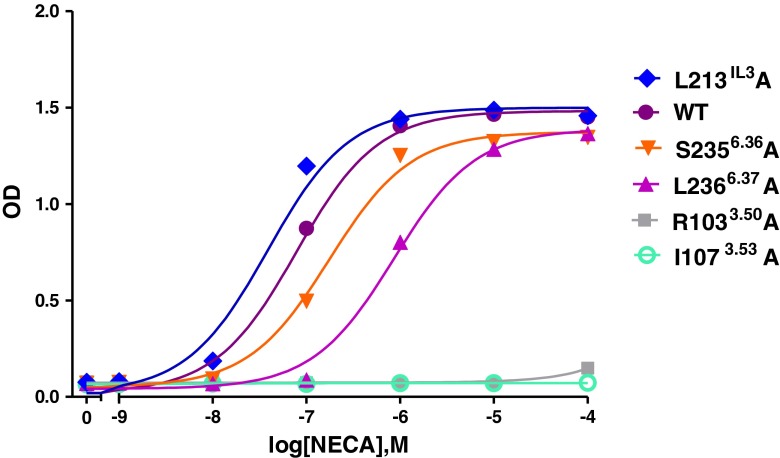
To assess the function of these five residues in G protein activation, we performed a functional yeast liquid assay to determine whether the G protein activation profiles of these mutants had changed or not. NECA concentration-response curves revealed that all five single amino acid changes in the receptor resulted in substantially different humanized G protein coupling profiles of the hA2BR (Table 2; Fig. 3).
Fig. 3.
Concentration-effect curves from liquid assay experiments. Curves are shown of the wild-type (filled purple circle), and five mutant receptors L213 IL3A (filled blue diamond), S2356.36A (filled red inverse triangle), L2366.37A (filled purple upright triangle), R1033.50A (filled gray square), and I1073.53A (open green circle) in the MMY24(Gαi3) strain responding to the adenosine receptor agonist NECA. The assay was performed in YNB-ULH medium
The R1033.50A receptor failed to couple to all humanized G protein pathways except for MMY28(Gαs) with a comparable EC50 value, only twofold higher than wild-type receptor (Table 2). Mutant receptor I1073.54A only responded with higher than wild-type EC50 values for NECA of 7629 nM to MMY23(Gαi1) with a reduced Emax value of 37 %, and 47 nM to MMY28(Gαs) with a maximal activation level. Other humanized G protein pathways did not respond to NECA anymore (Table 2).
The L213 IL3A mutant improved coupling efficiency in all yeast strains compared to the wild-type receptor, but to a different extent. The highest coupling efficiency was found in MMY28(Gαs) and MMY19(Gα12) with a tenfold decrease of NECA’s EC50 values compared to the wild-type receptor in the same strain. This mutant receptor was also able to reach maximal activation levels in each yeast strain upon activation by NECA except in MMY14(Gαq) with a somewhat reduced Emax value of 76 %.
The S2356.36A mutant receptor showed the most divergent humanized G protein coupling profile. In MMY12 (GαWT) and MMY20(Gα13), this mutant did not alter activation compared to the wild-type receptor in the same strain. However, it showed the largest increase of activation in MMY28(Gαs) with a one log-unit shift for the full agonist NECA’s EC50, and a 0.3-fold shift in MMY 23(Gαi1) (Table 2). In contrast, this mutant showed a decrease of activation in MMY24(Gαi3), MMY14(Gαq), and MMY21(Gα14) (up to 3.1-fold in MMY21, Table 2) and a complete loss of activation was observed in MMY19(Gα12). S2356.36A was only able to reach near-maximal activation levels in MMY24(Gαi3) and MMY28(Gαs) strains. In other strains only a partial NECA response of approximately 80 % was observed, except for MMY14(Gαq) (50 %) and MMY19(Gα12) (no activation) (Table 2).
L2366.37A showed a decreased response to NECA in the magnitude of 1.3- to 21-fold in all yeast strains compared to the wild-type receptor in the same strain (Table 2). L2366.37A reached near-maximal activation levels in most strains except for MMY19 (50 %), MMY14 (83 %), and MMY20 (85 %). Interestingly, L2366.37A also induced less constitutive activity in MMY28 than other receptors (wild-type and mutated) in this strain (data not shown).
Determination of expression level of the hA2BR in different yeast strains
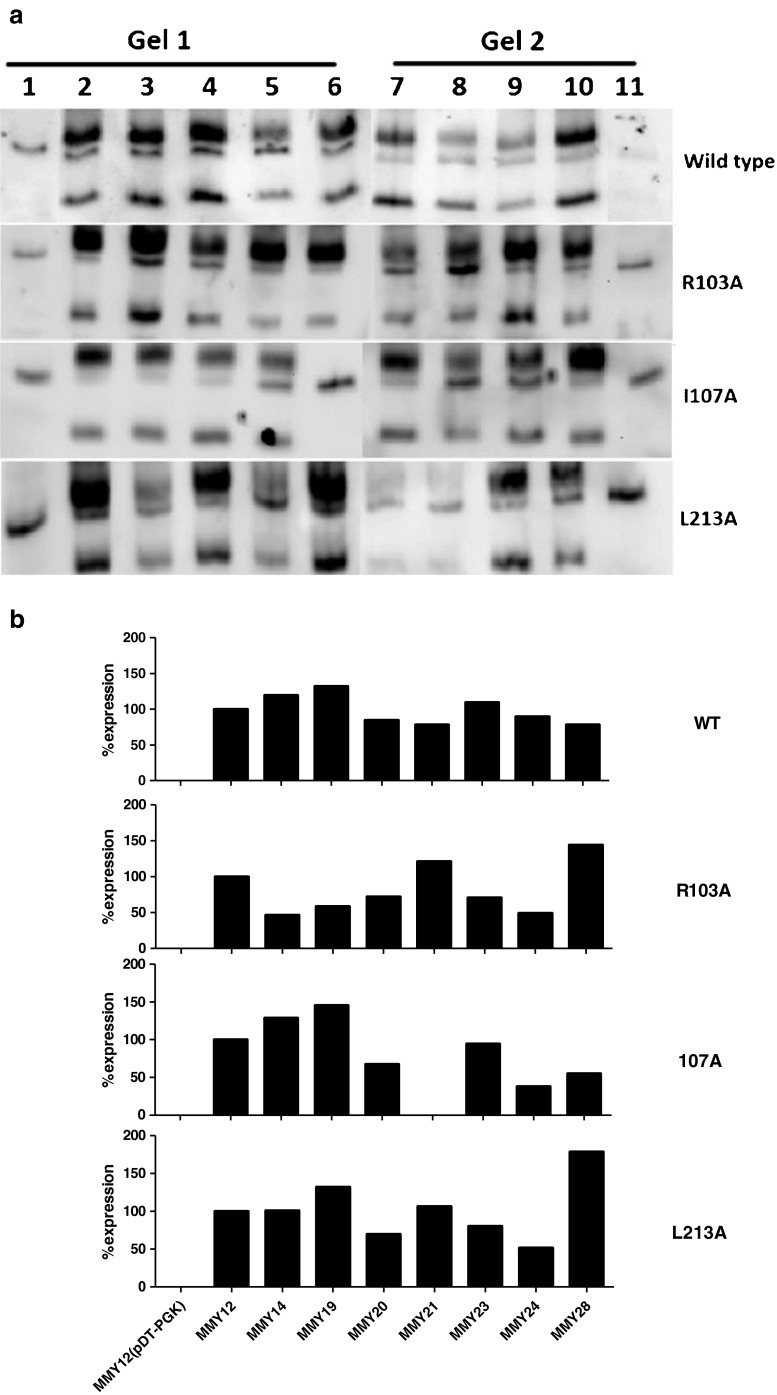
In Fig. 4a, a Western blot analysis of the expression levels of the WT hA2BR and three mutants is shown. In each gel, the antibody used recognized the top and bottom bands as specific bands of the hA2BR while the middle one was used as a reference band, since it is not specific for the receptor as evidenced by the MMY12 strain carrying the plasmid pDT-PGK without receptor. Quantitative bar graphs derived from the data in Fig. 4a are shown in Fig. 4b. Expression levels of the wild-type receptor in most strains were quite comparable (Fig. 4b). Even though mutants R103A and I107 did not respond to NECA in the liquid assay experiments, they did express in all yeast strains, except for MMY21_I107A (Fig. 4b). These data confirm that the transformation protocols used led to robust receptor expression in (almost) all cases and, hence, provide further proof of the validity of the activation profiles established. Moreover, it seems a certain degree of receptor expression is sufficient for receptor activation. As an example, the density of the L213 IL3A mutant was lowest in MMY24 and highest in MMY28. Nevertheless, EC50 and Emax values for NECA were virtually the same in both strains. To save costly (and commercially unavailable) antibody, we did not screen S235A and L236A mutant receptors, since their transferred strains always showed NECA concentration-response curves with high Emax values.
Fig. 4.
Western blot analysis of the wild-type hA2BR and R103A, I107A, and L213A mutations from top to bottom panel as expressed in different MMY strains. a Gel 1: Lane 1 MMY12 carrying pDT-PGK without receptor; Lanes 2–6 MMY12, MMY14, MMY19, MMY20, and MMY21 carrying pDT-PGK_A2B wild-type or mutant receptor. Gel 2: Lane7–10 MMY23, MMY24, MMY28, and MMY12 carrying pDT-PGK_A2B mutant receptor; Lane 11 MMY12 carrying pDT-PGK without receptor. The A2B receptor specific bands are 29 and 50 kDa, which are absent in MMY12 carrying pDT-PGK without receptor; nonspecific band at approximately 45 kDa was used as loading control, which also appeared in MMY12 carrying pDT-PGK without receptor. b Bar graphs were calculated from a densitometric analysis of the blots. The ratio was determined between the density of the specific bands and that of the nonspecific band that is always present on the blots. MMY12 carrying wild-type or mutant receptor was set as 100 % and MMY12 carrying the empty vector pDT-PGK without receptor was set as 0 %
Ligand binding assay of wild-type and mutant hA2BR expressed in MMY24(Gαi3)
To investigate whether the binding affinity of NECA was changed in the mutated receptors, we performed a number of radioligand-binding experiments. Traditionally, this assay is cumbersome in yeast cells, but we managed to obtain sufficient levels of specific [3H]PSB-603 binding to do displacement assays on wild-type, L213A, and L236A receptors. The two mutant receptors had a similar IC50 value for NECA displacing [3H]PSB-603 binding compared to the wild-type receptor (Table 3). However, the binding affinity of the radiolabeled antagonist for the other mutants, R103A, I107A, and S235A, appeared to be decreased. As a consequence, we did not obtain a high-enough window of specific [3H]PSB-603 binding to perform a radioligand displacement assay.
Table 3.
Radioligand-binding experiments of wild-type and mutant hA2B receptors expressed in MMY24(Gαi3) using 1.5 nM [3H]PSB-603. Specific binding of wild-type receptor was set at 100 %. IC50 values were obtained from competition binding curves of five independent experiments, performed in duplicate
| Mutant | % specific binding | IC50 (μM) |
|---|---|---|
| Wild type | 100 | 1.85 ± 0.87 |
| R103A | 0 | nd |
| I107A | 12 | nd |
| L213A | 94 | 1.90 ± 1.64 |
| S235A | 20 | nd |
| L236A | 158 | 1.88 ± 0.6 |
nd not determined
Discussion
Even though the mechanisms of interaction between GPCRs and their G proteins are largely unknown, the α4-helix and α4-β6 loop [34, 35], the N-terminus [36] and C-terminus of Gα subunits [37] have been described to be important for GPCR-Gα protein binding and selectivity. Of those, the C-terminus of the Gα subunit appears most intimately involved in binding the receptor. This was already evident from available crystal structures [23, 30] and further confirmed in molecular dynamics calculations by Kling et al., to show that three residues at the C-terminus of Gαs are in close contact with at least five amino acids of the β2AR [38]. In the present study, we examined the mechanism of interaction between the A2B receptor and the last five amino acid residues in the C-terminus of Gα subunits using a functional yeast system combined with homology modeling and mutagenesis experiments.
G protein-coupling profiles of the wild-type A2B receptor
We found that the wild-type hA2BR can activate many humanized G protein pathways, which is consistent with earlier findings by Brown et al. that the hA2BR is quite promiscuous as it recognizes most strains with a similar rank order of activation [19]. These strains possess different humanized G proteins, in which the last five amino acid residues of the endogenous Gpa1p C-terminal have been replaced with the same sequence from mammalian Gα proteins, covering the four major classes: Gαi, Gαs, Gαq, and Gα12. It is worth noting that the differences in activation profiles found in the present study can only be due to the variation in the five terminal amino acids of the humanized C-terminus of the Gα subunit. This has the advantage of providing a detailed snapshot of G protein activation without confounding factors such as further differences in the composition of the various Gα subunits.
The A2B receptor is preferentially coupled to the Gs pathway and to a lesser degree to the Gq pathway in many cells [39], and these two G proteins also couple well in our yeast system (S. cerevisiae strains MMY28(Gαs), MMY14(Gαq), and MMY21(Gα14/q), respectively). Interestingly, the receptor appears to couple well to two strains with a Gαi protein, MMY24(Gαi3) and MMY23(Gαi1), too, providing proof of the promiscuity mentioned above. However, this is not a “general” GPCR feature. For instance, Stewart et al. used the same yeast system to study functional selectivity of agonists and antagonists of the adenosine A1 receptor and learned that only the Gi/o pathway was addressed [18]. Likewise, the hydroxy-carboxylic acid receptor HCA3 can only activate the Gi pathway through its agonist acifran (preliminary data not shown).
G protein-coupling profiles of mutants of the hA2BR
In the present study, we sought to identify amino acid residues on the hA2BR that are vital in G protein coupling. The β2AR-Gs crystal structure (PDB: 3SN6) was chosen as the template to predict such amino acids in the hA2BR. R1033.50, I1073.54, L213IL3, S2356.36, and L2366.37 were selected to be mutated into alanine because they are interacting with the last five amino acids of the Gαs protein in the β2AR-Gs crystal structure. We will discuss these five amino acids in the light of our findings.
Residues R1033.50 and I1073.54
These two residues (R1033.50 and I1073.54) are located on the intracellular side of TM3 and are included in the consensus sequence (I/L)XXDR3.50YXX(I/V)3.54 [32]. R3.50 is a part of the most conserved motif in the class A GPCRs: Asp-Arg-Tyr (DRY). This arginine is 100 % conserved within the subfamily of adenosine receptors and nucleotide-like receptors, and 97 % of all Class A rhodopsin-like receptors. This is less so for I3.54 with 54 % overall conservation (Table 4).
Table 4.
Sequence conservation of the four helical amino acids involved in G protein interaction, among adenosine receptors, nucleotide-like receptors, and class A rhodopsin-like receptors as found on GMOS (GPCRs Motif Searcher, http://lmc.uab.cat/gmos/)
| Amino acid in A2B | Conservation in adenosine receptors | Most occurring | Conservation in nucleotide-like receptors | Most occurring | Conservation in class A rhodopsin-like receptors | Most occurring |
|---|---|---|---|---|---|---|
| R1033.50 | 100.0 % | R—100 % | 100.0 % | R—100 % | 96.98 % | R—96.98 % |
| I1073.54 | 35.29 % | V—64.70 % | 41.74 % | V—46.60 % | 53.68 % | I—53.68 % |
| S2356.36 | 85.29 % | S—85.29 % | 33.00 % | S—33.00 % | 2.81 % | T—32.25 % |
| L2366.37 | 85.29 % | L—85.29 % | 36.89 % | L—36.89 % | 38.59 % | L—38.59 % |
Nucleotide-like receptors: adenosine A1, A2A, A2B, A3 receptors; P2RY1, 4, 5, 6, 8, 9, 10, 11; GPR23, 35, 91, 92, 174
Not surprisingly, R3.50 has been the subject of many mutation studies, exemplified by their high occurrence in the GPCRDB mutation database [40]. The DRY motif is part of a so-called “ionic lock” [41, 42], consisting of a number of interactions between the DRY motif and amino acids in TM6; the interaction that is most prominent is the interaction between R3.50 and a negatively charged residue in TM6, Asp (D) or Glu (E). These interactions are thought to stabilize the receptor in an inactive conformation and thereby decrease its basal activity [43]. When the receptor is activated, the ionic lock is broken and TM6 is moving outward. Breaking the ionic lock through mutation might thus lead to constitutive activity, which was shown to be the case on the adenosine A3 receptor [44]. While this mechanism seems to hold true for some receptors it does not hold for every GPCR, as on the histamine H4 receptor [45]. R3.50 in this case turned out to be very important for G protein coupling. Our own results comply with data found for the α1b-adrenergic receptor where mutations of R3.50 resulted in a complete loss of receptor-mediated response in the majority of mutant receptors [46].
It has been proposed that there are many other conserved residues that help R3.50 switch the receptor on or off, such as D3.49 and I/V3.54 [32]. The latter position (3.54) is always conserved with a bulky β-branched, hydrophobic residue (Val or Ile). A mutagenesis study in the gonadotropin-releasing hormone (GnRH) receptor offered a hypothesis as to why there is a lack of receptor signaling in a receptor with a mutated I3.54. According to Ballesteros et al., I3.46, A3.49, and I3.54, all highly conserved amino acids, form a layer around R3.50, the so-called arginine-cage motif. I3.54A mutations caused significant reductions in receptor signaling efficacy and reduced the affinity for GnRH as well. In the WT receptor, the bulky side chain of I3.54 cannot move much as it readily clashes with the side chain of R3.50. This does not occur in the I3.54A mutant, which allows R3.50 to take an unfavorable conformation with an orientation to the aqueous cytoplasm. This might prevent R3.50 from taking part in receptor activation. Thus, the purpose of I3.54 appears to be a defined and strictly controlled positioning of R3.50 [32].
The results from our yeast screening assay add a layer of detail to these general findings, in that some G proteins seem more affected than others. We found that mutation to alanine of R1033.50and I1073.54 in the hA2BR abolished many humanized G protein pathways (Table 2) and, hence, we concluded they are very important for interaction with humanized G proteins. R1033.50 is crucial for each pathway, except for Gαs, whereas I1073.54 is vital for most G protein pathways as well, although after mutation some interaction remained with both Gαi1 and Gαs.
Other three residues L213 IL3, S2356.36, and L2366.37
Even though the investigated mutants only have one residue altered at the time, they show substantial differences in ligand activation of the receptor. A prominent enhancement was seen with the L213IL3A mutant; in all cases/strains, NECA was more active than on the wild-type receptor, particularly in MMY19(Gα12), with a tenfold increase in potency. This result shows once more the dependency of agonist potency on amino acids other than those in the ligand-binding site, and may even shed light on pharmacological principles such as receptor reserve. Apparently, the leucine on position 213 in the wild-type receptor acts as a deactivating switch.
The role of S3.36 seems to be somewhat more ambiguous. This residue when mutated to alanine caused mostly a decrease of receptor signaling. S6.36 is much conserved (Table 4) within the adenosine receptor subfamily (85 %), but less so in the nucleotide-like receptors (33 %) and hardly in all class A rhodopsin-like receptors (2.8 %). Several mutations have been made in different receptors on the 6.36 position but none of the original amino acids was a serine. The closest mutation is a T6.36A in the human muscarinic acetylcholine M1 receptor where the mutant did not significantly differ from wild type in PI turnover [47]. In the present study, the S26.36A mutation showed a most divergent G protein profile: improved activation efficiency in MMY28(Gαs) and MMY23(Gαi1); no change in MMY20(Gα13) similar to MMY12(GαWT); decreased activation in MMY14(Gαq), MMY24(Gαi3), MMY21(Gα14), and a complete loss of activation in MMY19(Gα12) (Table 2). Apparently, the change from a hydrophilic (serine) to a hydrophobic (alanine) amino acid is dealt with differently by the G proteins studied.
L6.37 is quite conserved: 85 % in adenosine receptors, 37 % in nucleotide-like receptors, and 39 % in all class A rhodopsin-like receptors. In the A2A receptor, the L6.37A mutation along with several others was used to provide receptor thermostabilization for crystallographic purposes [48]. The mutant caused no effect on ligand pharmacology. In our hands, the L2366.37A mutation decreased activation in all humanized G protein pathways, most outspoken for MMY28. Apparently, the leucine residue is vital for G protein interaction and activation.
Function of hydroxyl-group at C-terminus of Gα subunits for wild-type hA2BR
The slight differences in amino acid composition in some of the Gα subunits allow an almost atomic dissection of the observed effects. There is an eightfold potency difference of NECA in the two Gq pathway strains MMY21(Gα14) with an EC50 value of 212 nM and MMY14(Gαq) with an EC50 value of 1,641 nM. Both have the same amino acid residues at the C-terminus of the Gα protein except for the fourth residue position counting from the end of the C-terminus with a tyrosine in MMY14 and a phenylalanine in MMY21 (Table 1). The only difference between tyrosine and phenylalanine is a hydroxyl-group, which leads to the large decrease in potency for NECA. However, there is an opposite phenomenon at the last amino acid of the C-terminus between two Gαi pathway strains, MMY24(Gαi3) and MMY23(Gαi1). A tyrosine as the last amino acid of the end of the C-terminus position of MMY24 yielded an EC50 value of 59 nM for NECA, whereas the phenylalanine on the same position in MMY23 gave an EC50 value of 305 nM (Tables 1 and 2). This particular tyrosine hydroxyl-group is apparently enough to decrease the EC50 value by sixfold, which is equivalent to an increase of activation and NECA potency. Taken together, subtle changes such as the presence or absence of a hydroxyl-group located in the C-terminus of the Gα protein control activation of GPCRs.
Concluding remarks
We reported on a yeast system that is very well suited for the study of a G protein-coupled receptor (the hA2BR in this case), its activation and its G protein preference. This highly efficient and inexpensive screening system was used to map residues at the cytoplasmic side of the receptor and in the C-terminus of different Gα subunits important for receptor activation. The results provided detailed information about receptor/G protein binding and G protein selectivity.
Acknowledgement
Rongfang Liu thanks the China Scholarship Council (CSC) for her PhD scholarship. NWO provided a TOP grant to A.P. IJ. (714.011.001). The authors are grateful to Prof C.E. Müeller of Bonn University for the generous gift of [3H]PSB-603 and to Dr S. Dowell from GSK for providing the yeast strains and experimental protocols.
References
- 1.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nature structural & molecular biology. 2006;13(9):772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 2.Baltoumas FA, Theodoropoulou MC, Hamodrakas SJ. Interactions of the alpha-subunits of heterotrimeric G-proteins with GPCRs, effectors and RGS proteins: a critical review and analysis of interacting surfaces, conformational shifts, structural diversity and electrostatic potentials. Journal of structural biology. 2013;182(3):209–218. doi: 10.1016/j.jsb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Fredholm BB, Arslan G, Halldner L, Kull B, Schulte G, Wasserman W. Structure and function of adenosine receptors and their genes. Naunyn-Schmiedeberg’s archives of pharmacology. 2000;362(4–5):364–374. doi: 10.1007/s002100000313. [DOI] [PubMed] [Google Scholar]
- 4.Fredholm BB, Irenius E, Kull B, Schulte G. Comparison of the potency of adenosine as an agonist at human adenosine receptors expressed in Chinese hamster ovary cells. Biochem Pharmacol. 2001;61(4):443–448. doi: 10.1016/S0006-2952(00)00570-0. [DOI] [PubMed] [Google Scholar]
- 5.Wei W, Du C, Lv J, Zhao G, Li Z, Wu Z, Haskó G, Xie X. Blocking A2B adenosine receptor alleviates pathogenesis of experimental autoimmune encephalomyelitis via inhibition of IL-6 production and Th17 differentiation. The Journal of Immunology. 2013;190(1):138–146. doi: 10.4049/jimmunol.1103721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei Q, Costanzi S, Balasubramanian R, Gao Z-G, Jacobson KA (2013) A2B adenosine receptor blockade inhibits growth of prostate cancer cells. Purinergic signalling: 1–10 [DOI] [PMC free article] [PubMed]
- 7.Stagg J, Divisekera U, McLaughlin N, Sharkey J, Pommey S, Denoyer D, Dwyer KM, Smyth MJ. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci. 2010;107(4):1547–1552. doi: 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen SJ, Massa HH, Rose’Meyer RB. Loss of adenosine A2B receptor mediated relaxant responses in the aged female rat bladder; effects of dietary phytoestrogens. Naunyn-Schmiedeberg’s archives of pharmacology. 2012;385(5):539–549. doi: 10.1007/s00210-011-0722-y. [DOI] [PubMed] [Google Scholar]
- 9.Cekic C, Sag D, Li Y, Theodorescu D, Strieter RM, Linden J. Adenosine A2B receptor blockade slows growth of bladder and breast tumors. The Journal of Immunology. 2012;188(1):198–205. doi: 10.4049/jimmunol.1101845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koscsó B, Trepakov A, Csóka B, Németh ZH, Pacher P, Eltzschig HK, Haskó G (2013) Stimulation of A2B adenosine receptors protects against trauma–hemorrhagic shock-induced lung injury. Purinergic signalling: 1–6 [DOI] [PMC free article] [PubMed]
- 11.Bot I, De Vries H, Korporaal SJ, Foks AC, Bot M, Van Veldhoven J, Ter Borg MN, Van Santbrink PJ, Van Berkel TJ, Kuiper J, IJzerman AP. Adenosine A2B receptor agonism inhibits neointimal lesion development after arterial injury in apolipoprotein E-deficient mice. Arteriosclerosis, thrombosis, and vascular biology. 2012;32(9):2197–2205. doi: 10.1161/ATVBAHA.112.252924. [DOI] [PubMed] [Google Scholar]
- 12.Pausch MH. G-protein-coupled receptors in Saccharomyces cerevisiae: high-throughput screening assays for drug discovery. Trends in biotechnology. 1997;15(12):487–494. doi: 10.1016/S0167-7799(97)01119-0. [DOI] [PubMed] [Google Scholar]
- 13.Peeters MC, Li Q, van Westen GJP, IJzerman AP (2011) Three “hotspots” important for adenosine A 2B receptor activation: a mutational analysis of transmembrane domains 4 and 5 and the second extracellular loop. Purinergic Signalling:1–16 [DOI] [PMC free article] [PubMed]
- 14.Mathew E, Ding F-X, Naider F, Dumont ME. Functional fusions of T4 lysozyme in the third intracellular loop of a G protein-coupled receptor identified by a random screening approach in yeast. Protein Engineering Design and Selection. 2013;26(1):59–71. doi: 10.1093/protein/gzs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaeschke H, Kleinau G, Sontheimer J, Mueller S, Krause G, Paschke R. Preferences of transmembrane helices for cooperative amplification of G α s and G α q signaling of the thyrotropin receptor. Cellular and molecular life sciences. 2008;65(24):4028–4038. doi: 10.1007/s00018-008-8530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans BJ, Wang Z, Broach JR, Oishi S, Fujii N, Peiper SC. Expression of CXCR4, a G-protein-coupled receptor for CXCL12 in yeast: identification of new-generation inverse agonists. Methods in enzymology. 2009;460:399–412. doi: 10.1016/S0076-6879(09)05220-3. [DOI] [PubMed] [Google Scholar]
- 17.Minic J, Sautel M, Salesse R, Pajot-Augy E. Yeast system as a screening tool for pharmacological assessment of G protein coupled receptors. Current medicinal chemistry. 2005;12(8):961–969. doi: 10.2174/0929867053507261. [DOI] [PubMed] [Google Scholar]
- 18.Stewart GD, Valant C, Dowell SJ, Mijaljica D, Devenish RJ, Scammells PJ, Sexton PM, Christopoulos A. Determination of adenosine A1 receptor agonist and antagonist pharmacology using Saccharomyces cerevisiae: implications for ligand screening and functional selectivity. Journal of Pharmacology and Experimental Therapeutics. 2009;331(1):277–286. doi: 10.1124/jpet.109.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown AJ, Dyos SL, Whiteway MS, White JH, Watson MAE, Marzioch M, Clare JJ, Cousens DJ, Paddon C, Plumpton C. Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein α subunit chimeras. Yeast. 2000;16(1):11–22. doi: 10.1002/(SICI)1097-0061(20000115)16:1<11::AID-YEA502>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 20.Dowell SJ, Brown AJ. Yeast assays for G protein-coupled receptors. G Protein-Coupled Receptors in Drug Discovery. 2009;552:213–229. doi: 10.1007/978-1-60327-317-6_15. [DOI] [PubMed] [Google Scholar]
- 21.Dowell SJ (2002) Yeast assays for G-protein-coupled receptors. receptors and channels 8(343–352) [PubMed]
- 22.Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins: Structure, Function, and Bioinformatics. 2004;23(3):403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477(7366):549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLano DW (The PyMOL Molecular Graphics System, Version 1.5. 0.4 Schrödinger, LLC.)
- 25.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20(6):1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Ye K, Blad CC, den Dulk H, Brouwer J, IJzerman AP, Beukers MW. ZM241385, DPCPX, MRS1706 are inverse agonists with different relative intrinsic efficacies on constitutively active mutants of the human adenosine A2B receptor. Journal of Pharmacology and Experimental Therapeutics. 2007;320(2):637–645. doi: 10.1124/jpet.106.111203. [DOI] [PubMed] [Google Scholar]
- 27.Feoktistov I, Goldstein A, Sheller JR, Schwartz LB, Biaggioni I. Immunological characterization of A2B adenosine receptors in human mast cells. Drug development research. 2003;58(4):461–471. doi: 10.1002/ddr.10180. [DOI] [Google Scholar]
- 28.Peeters M, Wisse L, Dinaj A, Vroling B, Vriend G, IJzerman A. The role of the second and third extracellular loops of the adenosine A1 receptor in activation and allosteric modulation. Biochem Pharmacol. 2012;84:76–87. doi: 10.1016/j.bcp.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Venkatakrishnan A, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494(7436):185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 30.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauß N, Choe H-W, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 31.Fredholm BB, IJzerman AP, Jacobson KA, Linden J, Müller CE (2011) International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors—an update. Pharmacol Rev 63(1):1–34 [DOI] [PMC free article] [PubMed]
- 32.Ballesteros J, Kitanovic S, Guarnieri F, Davies P, Fromme BJ, Konvicka K, Chi L, Millar RP, Davidson JS, Weinstein H. Functional microdomains in G-protein-coupled receptors the conserved arginine-cage motif in the gonadotropin-releasing hormone receptor. Journal of Biological Chemistry. 1998;273(17):10445–10453. doi: 10.1074/jbc.273.17.10445. [DOI] [PubMed] [Google Scholar]
- 33.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in neurosciences. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 34.Bae H, Cabrera-Vera TM, Depree KM, Graber SG, Hamm HE. Two amino acids within the α4 helix of Gαi1 mediate coupling with 5-hydroxytryptamine1B receptors. Journal of Biological Chemistry. 1999;274(21):14963–14971. doi: 10.1074/jbc.274.21.14963. [DOI] [PubMed] [Google Scholar]
- 35.Bae H, Anderson K, Flood LA, Skiba NP, Hamm HE, Graber SG. Molecular determinants of selectivity in 5-hydroxytryptamine1B receptor-G protein interactions. Journal of Biological Chemistry. 1997;272(51):32071–32077. doi: 10.1074/jbc.272.51.32071. [DOI] [PubMed] [Google Scholar]
- 36.Taylor JM, Jacob-Mosier GG, Lawton RG, Remmers AE, Neubig RR. Binding of an alpha 2 adrenergic receptor third intracellular loop peptide to G beta and the amino terminus of G alpha. Journal of Biological Chemistry. 1994;269(44):27618–27624. [PubMed] [Google Scholar]
- 37.Blahos J, Mary S, Perroy J, de Colle C, Brabet I, Bockaert J, Pin J-P. Extreme C terminus of G protein α-subunits contains a site that discriminates between Gi-coupled metabotropic glutamate receptors. Journal of Biological Chemistry. 1998;273(40):25765–25769. doi: 10.1074/jbc.273.40.25765. [DOI] [PubMed] [Google Scholar]
- 38.Kling RC, Lanig H, Clark T, Gmeiner P. Active-state models of ternary GPCR complexes: determinants of selective receptor-G-protein coupling. PLOS ONE. 2013;8(6):e67244. doi: 10.1371/journal.pone.0067244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thimm D, Schiedel AC, Sherbiny FF, Hinz S, Hochheiser K, Bertarelli DC, Maaß A, Müller CE. Ligand-specific binding and activation of the human adenosine A2B receptor. Biochemistry. 2013;52(4):726–740. doi: 10.1021/bi3012065. [DOI] [PubMed] [Google Scholar]
- 40.Vroling B, Sanders M, Baakman C, Borrmann A, Verhoeven S, Klomp J, Oliveira L, de Vlieg J, Vriend G. GPCRDB: information system for G protein-coupled receptors. Nucleic acids research. 2011;39(suppl 1):D309–D319. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel R, Mahalingam M, Lüdeke S, Huber T, Siebert F, Sakmar TP. Functional role of the “ionic lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. Journal of molecular biology. 2008;380(4):648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Scheer A, Fanelli F, Costa T, De Benedetti P, Cotecchia S. Constitutively active mutants of the alpha 1B-adrenergic receptor: role of highly conserved polar amino acids in receptor activation. EMBO J. 1996;15(14):3566. [PMC free article] [PubMed] [Google Scholar]
- 43.Flanagan CA. A GPCR that is not “DRY”. Molecular pharmacology. 2005;68(1):1–3. doi: 10.1124/mol.105.014183. [DOI] [PubMed] [Google Scholar]
- 44.Chen A, Gao Z-G, Barak D, Liang BT, Jacobson KA. Constitutive activation of A3 adenosine receptors by site-directed mutagenesis. Biochemical and biophysical research communications. 2001;284(3):596–601. doi: 10.1006/bbrc.2001.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider EH, Schnell D, Strasser A, Dove S, Seifert R. Impact of the DRY motif and the missing “ionic lock” on constitutive activity and G-protein coupling of the human histamine H4 receptor. Journal of Pharmacology and Experimental Therapeutics. 2010;333(2):382–392. doi: 10.1124/jpet.109.163220. [DOI] [PubMed] [Google Scholar]
- 46.Scheer A, Costa T, Fanelli F, De Benedetti PG, Mhaouty-Kodja S, Abuin L, Nenniger-Tosato M, Cotecchia S. Mutational analysis of the highly conserved arginine within the Glu/Asp-Arg-Tyr motif of the α1b-adrenergic receptor: effects on receptor isomerization and activation. Molecular pharmacology. 2000;57(2):219–231. [PubMed] [Google Scholar]
- 47.Högger P, Shockley MS, Lameh J, Sadée W. Activating and inactivating mutations in N- and C-terminal i3 loop junctions of muscarinic acetylcholine Hm1 receptors. Journal of Biological Chemistry. 1995;270(13):7405–7410. doi: 10.1074/jbc.270.13.7405. [DOI] [PubMed] [Google Scholar]
- 48.Doré AS, Robertson N, Errey JC, Ng I, Hollenstein K, Tehan B, Hurrell E, Bennett K, Congreve M, Magnani F. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19(9):1283–1293. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simon MI, Strathmann MP, Gautam N. Diversity of G proteins in signal transduction. Science. 1991;252(5007):802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]