Abstract
While the adult human heart has very limited regenerative potential, the adult zebrafish heart can fully regenerate after 20% ventricular resection. Although previous reports suggest that developmental signaling pathways such as FGF and PDGF are reused in adult heart regeneration, the underlying intracellular mechanisms remain largely unknown. Here we show that H2O2 acts as a novel epicardial and myocardial signal to prime the heart for regeneration in adult zebrafish. Live imaging of intact hearts revealed highly localized H2O2 (∼30 μM) production in the epicardium and adjacent compact myocardium at the resection site. Decreasing H2O2 formation with the Duox inhibitors diphenyleneiodonium (DPI) or apocynin, or scavenging H2O2 by catalase overexpression markedly impaired cardiac regeneration while exogenous H2O2 rescued the inhibitory effects of DPI on cardiac regeneration, indicating that H2O2 is an essential and sufficient signal in this process. Mechanistically, elevated H2O2 destabilized the redox-sensitive phosphatase Dusp6 and hence increased the phosphorylation of Erk1/2. The Dusp6 inhibitor BCI achieved similar pro-regenerative effects while transgenic overexpression of dusp6 impaired cardiac regeneration. H2O2 plays a dual role in recruiting immune cells and promoting heart regeneration through two relatively independent pathways. We conclude that H2O2 potentially generated from Duox/Nox2 promotes heart regeneration in zebrafish by unleashing MAP kinase signaling through a derepression mechanism involving Dusp6.
Keywords: heart regeneration, zebrafish, Duox, H2O2, Dusp6
Introduction
The ability to reverse heart failure, which accounts for > 10% annual mortality in humans, is the most challenging task in cardiac regenerative medicine. Many efforts are devoted to cardiac stem cell generation and tissue engineering, although making cardiac stem cells or tissue scaffolds functionally integrated into their host cardiac tissues remains difficult in clinical settings1,2. A number of studies have shown that the adult mammalian heart has limited regenerative capacity, although not sufficient to fully restore heart function after substantial myocardial injury2. On the other hand, the adult zebrafish and newt hearts have full capacity of cardiac regeneration after ventricular resection or cryoinjury, making zebrafish and newt as powerful animal models for studying heart regeneration in particular and organ regeneration in general3,4,5,6. The neonatal mouse heart in the first week of their postnatal life can fully regenerate after ventricular resection, suggesting that mammals gradually lose their regenerative potential during postnatal development10. It remains mysterious how a species gains or loses the regenerative potential during evolution.
Zebrafish has become a well-recognized model system for studying organ regeneration due to incredible regenerative potential in all of its adult organs tested and the availability of transgenesis, genetics and genomic resources7. The dedifferentiation model for the cellular sources of cardiac repair/regeneration was supported by the fact that cardiac muscles mainly regenerate from pre-existing cardiomyocytes after injury in zebrafish8,9 and mammals10,11,12. The stem cell/progenitor cell model for cardiac regeneration in mammals was also recognized by the existence of Sca1+ and c-Kit+ cardiac stem cells in mice13,14, while the transdifferentiation or lineage reprogramming model was proposed partly based on the ability of cardiac fibroblast transdifferentiation into myocytes upon induction by reprogramming factors or microRNA in mice15,16,17 and reprogramming of embryonic atrial into ventricular myocytes after injury in zebrafish18. In spite of great efforts, the molecular mechanisms underlying heart regeneration remain incompletely understood. Previous reports support that developmental signaling pathways such as FGF, PDGF, retinoid acid, TGFβ and integrin are essential for adult zebrafish heart regeneration19,20,21,22,23. Ventricular resection induces expression of fgfr2/4 in the epicardium and their ligand fgf17b in the myocardium, and conditional blocking of the FGF signaling by overexpression of dominant-negative FGFR1 substantially increases fibrin/collagen deposits and diminishes myocardial regeneration, supporting a notion that the FGF signaling promotes myocardial regeneration through epicardial-myocardial interaction21. A similar strategy revealed that the PDGF signaling is also required for zebrafish heart regeneration20. The exact molecular mechanisms downstream of the FGF/PDGF signaling during adult heart regeneration are currently unknown.
Duox is a member of NADPH-oxidase (Nox) and related dual oxidase (Duox) family, which has seven members, Nox1, Nox2, Nox3, Nox4, Nox5, Duox1 and Duox2 in humans, but only five (Nox1, Nox2, Nox4, Nox5 and Duox) identified in zebrafish24. Functionally, the Duox or Nox enzyme complex in the plasma membrane catalyzes single-electron transfers from the electron donor NADPH to molecular oxygen, forming superoxide anions, which are quickly converted into hydrogen peroxide (H2O2) by spontaneous dismutation or catalytic reactions via Cu2+Zn2+-superoxide dismutase. Recently, it has been shown that Duox-generated H2O2 in the tail-fin epithelium, which responds within ∼3 min and peaks around 20 min after wounding, is critical for the rapid recruitment of leukocytes to initiate inflammation in zebrafish larvae25,26. Sustained H2O2 production would shift the intracellular redox homeostasis from a more reduced to a more oxidized state, affecting numerous redox-sensitive signaling pathways. Ataxia-telangiectasia mutated (ATM), a PI3K-like serine/threonine protein kinase preferentially activated by DNA double-strand breaks, serves as a sensor of oxidative stress (H2O2). Oxidative stress (H2O2) elicits the ATM-homodimer-Chk2 pathway to induce cell growth arrest or apoptosis27. On the other hand, H2O2 acts through a redox sensor Lyn to mediate leukocyte wound attraction25,26. Here we report that, after ventricular resection, H2O2 levels are locally elevated in the epicardium and adjacent myocardium to promote heart regeneration by a derepression mechanism, whereby H2O2 destabilizes the redox-sensitive phosphatase Dusp6 to enhance growth factor signaling.
Results
Duox and Nox2 are spatio-temporally associated with heart regeneration
In a genome-wide search for genes critical for cardiac regeneration by mRNA deep sequencing (Supplementary information, Figures S1 and S2), we found that the expression levels of a group of genes related to immune system diseases were upregulated, including p22phox, p47phox and p67phox of the Nox2 complex (Supplementary information, Tables S1-S3; GEO repository accession number GSE50203). Combining with other public microarray databases, we identified 37 genes in the reactive oxygen species (ROS) system that were differentially expressed in the injured zebrafish heart at 7 days post amputation (dpa) (Supplementary information, Table S4), when injury-induced inflammation started to fade and active myocardial regeneration began3,28,29. RNA in situ hybridization was then applied at multiple time points from 3 to 30 dpa for validation. We found that duox was particularly interesting because of the spatial and temporal patterns of its expression during regeneration. RNA in situ hybridization with two independent duox probes revealed that duox expression was weak at sham, peaked and was highly localized beneath the surface of the amputation at around 3, 7 and 14 dpa, and returned to the basal level at 30 dpa when regeneration was nearly complete (Supplementary information, Figures S3A-S3E and S5A-S5E). Each of the two sense duox probes had no detectable signals (Supplementary information, Figures S4 and S5F), indicating the specificity of the two duox probes. RNA in situ hybridization on Tg(tcf21:nucEGFP) transgenic hearts revealed that duox expression largely overlapped with the EGFP signal (Figure 1A-1C and Supplementaryinformation, Figure S6), which primarily labels the epicardium30, suggesting that duox was enriched in the epicardium. Thus, both the time course and epicardial localization of duox expression correlated with those of active myocardial regeneration after injury.
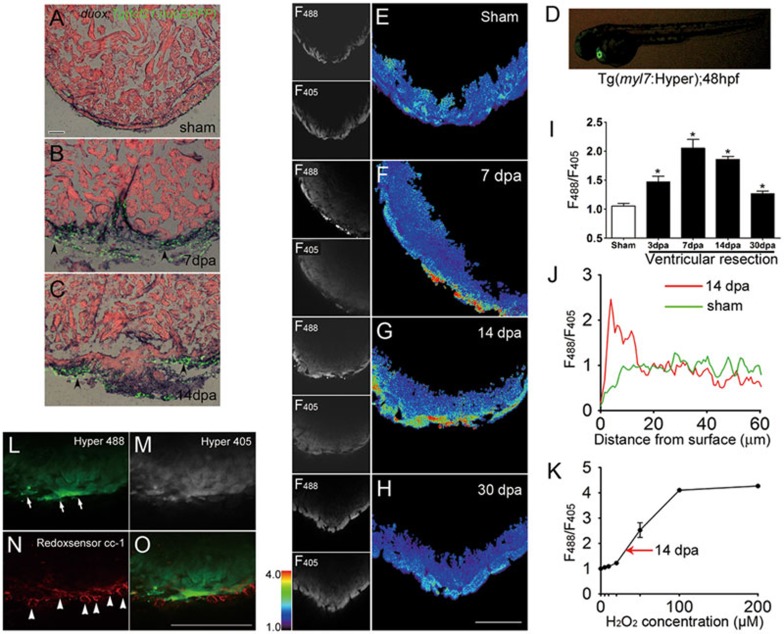
Figure 1.
Epicardial Duox-generated H2O2 associated with myocardial regeneration. (A-C) After imaging of Tg(tcf21:nucEGFP), we performed RNA in situ hybridization with duox probes on 10-μm cryosections of Tg(tcf21:nucEGFP) hearts at sham, 7 and 14 dpa. Note the induced expression of duox and its colocalization with epicardial reporter Tg(tcf21:nucEGFP) in injured areas (arrowheads) at 7 and 14 dpa (B, C) while little staining at sham (A). Scale bar for A-C, 50 μm. (D) Cardiac-specific expression of Hyper in a zebrafish embryo. Hyper was restricted in the Tg(myl7:Hyper) transgenic heart at 48 hpf. (E-H) Ex vivo Hyper heart images from sham control (E), 7 dpa (F), 14 dpa (G) and 30 dpa (H). Spatially resolved H2O2 image, indexed by the ratio between the F488 and F405 images of Hyper (left panels), is presented in pseudocolor. (I) Ratiometric Hyper signals (F488/F405) averaged over the regenerative zone of injured heart during the first month of regeneration after resection. n = 3. (J) Representative transmural spatial profiles of the Hyper signal at 14 dpa (red line) and sham (green line) hearts. (K) Ex vivo calibration of Hyper F488/F405 ratio as a function of ambient H2O2 concentration. n = 3. Arrow denotes the average F488/F405 ratio seen in regenerative zones at 14 dpa. (L-O) Redox signal showing greater epicardial and myocardial oxidization during regeneration. A 14 dpa Tg(myl7:hyper) heart was labeled with Redox sensor cc-1. Images show Hyper signals at 488 (L) and 405 nm (M), and Redox sensor cc-1 signals at 555 nm excitation (N), and the merging of L and N (O). Note that the Redox sensor cc-1 signal (arrowheads), indicative of intracellular oxidization, was most conspicuous in epicardial cells lacking Hyper expression, and also overlapped with the Hyper signal (arrows) in adjacent myocardium. Scale bars, 100 μm.
Systematic examination revealed that the expression of nox2, but none of the other nox genes, was moderately induced at 3, 7 and 14 dpa in a pattern similar to that of duox (Supplementary information, Figure S7). RT-PCR confirmed expression of duox and nox2 in 14 dpa hearts (Supplementary information, Figure S3F). These results indicate that the expression of duox and nox2, with duox being dominant, is spatially and temporally associated with myocardial regeneration after ventricular resection.
H2O2 signal is essential for heart regeneration
To visualize the spatiotemporal dynamics of H2O2 in the intact heart during regeneration, we used Tol2-based transgenesis and zebrafish myosin light chain 7 (myl7) promoter to generate transgenic zebrafish with cardiac-specific expression of Hyper (Figure 1D), a fluorescent protein-based H2O2 sensor25,31. High-resolution confocal imaging was performed in intact adult hearts excised from Tg(myl7:Hyper) transgenic animals, and H2O2 levels were directly measured as the ratio between the fluorescent signals dually excited at 488 and 405 nm (R=F488/F405). By ex vivo titration on the Hyper-expressing hearts freshly excised from uninjured animals, we obtained a calibration curve (Figure 1K and Supplementary information, Figure S9A-S9G) similar to that obtained in vitro31. Compared to sham controls (Figure 1E), local H2O2 concentration was clearly elevated in injured hearts at 3, 7 and 14 dpa (Figure 1F-1G and 1I), and returned to the basal level after nearly full regeneration at 30 dpa (Figure 1H and 1I). In contrast to the rapid (on the scale of minutes) inflammatory H2O2 response in the tail-fin epithelium of zebrafish larvae25,26, there was a slow onset of ventricular resection-induced H2O2 production, as no detectable increase in the Hyper fluorescence ratio occurred within ∼1 h after heart injury (data not shown). As seen on confocal optical sections (Figure 1E-1H) and from line plots of the spatial profiles (Figure 1J), the H2O2 gradients spanned ∼20 μm depth from the epicardium to the myocardium, reaching a peak concentration of ∼30 μM (Figure 1K and Supplementary information, Figure S9A-S9G), which is within the range of concentrations for H2O2 to act as an intracellular messenger32. In an independent measurement, we stained 14 dpa Tg(myl7:Hyper) transgenic hearts with Redox sensor cc-1 and visualized the H2O2 signal in both the epicardium that lacked Hyper expression (Figure 1O) and adjacent compact myocardium (Figure 1L-1O). Therefore, direct visualization of H2O2 with either a genetically encoded probe or a small-molecule indicator revealed that H2O2 was elevated primarily in the epicardium and the adjacent myocardium at the resection site, highly correlated with myocardial regeneration.
To investigate whether there is a causal relationship between elevated Duox/Nox2 and H2O2 production and cardiac regeneration, we applied diphenyleneiodonium (DPI) (10 μM) or apocynin (100 μM), widely used inhibitors of Duox/Nox enzymes33 and examined subsequent changes in H2O2 production and cardiac regeneration. We found that Hyper fluorescent signals were decreased by 50% within ∼ 40 min after soaking 14 dpa Tg(myl7:Hyper) hearts in DPI-containing medium (Figure 2A-2B), while the Hyper signals remained stable in control medium containing DMSO only (Figure 2B and Supplementary information, Figure S8A), suggesting that high concentration of H2O2 likely resulted from elevated duox/nox2 expression but was not produced from mitochondria. Functionality of ex vivo cultured hearts was validated by their rhythmic contractions under 1 Hz electrical field stimulation after 1 h culture (Supplementary information, Figure S8B-S8C). Based on the previous report that myocardial cells enter mitotic phases in injured areas of the heart upon ventricular resection in zebrafish or myocardial infarction in mice3,34, we quantified injury-induced myocyte proliferation by counting BrdU+/Mef2C+ double-positive myocytes during heart regeneration. After ventricular resection, zebrafish were allowed to recover for 7 days and BrdU was then co-injected with either DPI or apocynin into the abdominal cavity once daily until the hearts were collected for examination. Compared with sham-operated controls, we found that BrdU+/Mef2C+ myocyte proliferation was induced in injured hearts at 14 dpa (Figure 2C, 2C' and 2E), but their numbers were significantly reduced by treatment with either DPI (Figure 2D, 2D' and 2E) or apocynin (Figure 2E and data not shown). In addition, extensive cardiac fibrosis (Figure 2G) and reduced Myl7+ myocardial regeneration were evident in injured hearts with DPI (Figure 2I) or apocynin treatment (data not shown), compared with DMSO-treated hearts (Figure 2F and 2H). Although injection of H2O2 did not induce myocyte proliferation in sham-operated animals (Supplementaryinformation, Figure S10), it rescued the inhibitory effects of DPI on the numbers of BrdU+/Mef2C+ myocytes and Gata4+ cells (Supplementary information, Figure S11). Importantly, DPI-treated hearts failed to regenerate even at 60 dpa (Supplementary information, Figure S12), suggesting a permanent arrest rather than a delay of regenerative process. In addition, exogenous H2O2 partially rescued the inhibitory effect of DPI on embryonic heart regeneration (Supplementary information, Figure S13A and S13B) after a large number of cardiomyocytes were damaged by incubating Tg(myl7:CFP-NTR) transgenic embryos with metronidazole (Mtz), as Mtz can be converted into a toxic DNA crosslinking agent by nitroreductase (NTR) as previously described35. We further validated the penetration of H2O2 by imaging the 72 hours post fertilization (hpf) embryonic Tg(myl7:Hyper) hearts and found that Hyper signals were increased upon elevated external H2O2 concentration (Supplementary information, Figure S14). These results strongly support the hypothesis that H2O2 is required and sufficient for cardiac regeneration.
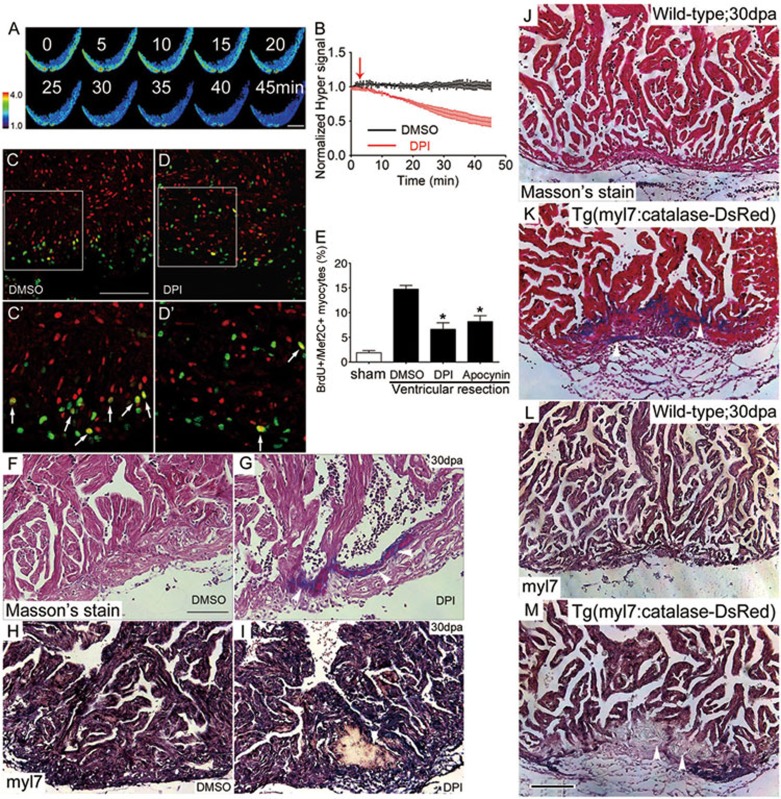
Figure 2.
H2O2 signaling is required for heart regeneration. (A-B) Inhibiting Duox/Nox NADPH oxidases by DPI decreased H2O2 generation. Time-lapse confocal images (A) and statistics (B) of the Hyper ratio in Tg(myl7:Hyper) hearts prior to and after application of DPI (10 μM). DMSO (0.1%) was used as control. n = 3-5. (C-E) Treatment with DPI or apocynin impaired cardiac myocyte regeneration. Proliferating myocytes were identified by double-staining with anti-Mef2C (red) and anti-BrdU (green) (C', D', arrows). Note that there were fewer double-positive cells (yellow) in DPI-treated heart (D, D') than in DMSO control heart (C, C'). Quantitative results with sham, DMSO, DPI or apocynin treatment are shown in E. n = 5 to 7. See Materials and Methods for details of treatment. (F-I) Accumulated fibrin/collagens (white arrowheads in G, Masson's staining) and compromised myocardial regeneration (white arrowhead in I, in situ hybridization with myl7 probes) after DPI treatment, compared with DMSO control (F, H). (J-M) Cardiac-specific overexpression of catalase retarded heart regeneration. Tg(myl7:Catalase-DsRed) heart displayed larger amounts of fibrin/collagens (white arrowheads in K, Masson's staining) and compromised myocardial regeneration (white arrowheads in M, in situ hybridization with myl7 probes), as compared with respective non-transgenic sibling hearts at 30 dpa (J, L). n = 5-8. Scale bars, 100 μm.
If H2O2 is causally linked to cardiac regeneration as suggested by its inhibition upon treatment with DPI or apocynin, limiting H2O2 signaling by augmenting antioxidant capacity of the heart should decrease cardiac regeneration. To directly test this hypothesis, we generated Tg(myl7:catalase-DsRed) transgenic zebrafish with cardiac-specific expression of catalase. The efficacy of catalase-DsRed was validated by its ability to scavenge H2O236 (Supplementary information, Figure S13C-S13E). Similar to the Duox/Nox inhibition, there were fewer Myl7+ regenerated myocytes (Figure 2M), and extensive cardiac fibrosis (Figure 2K) at 30 dpa in catalase-transgenic hearts, compared to non-transgenic sibling hearts (Figure 2L and 2J). However, we were not able to generate ubiquitously heat-inducible Tg(hsp70:catalase) transgenic lines, due to lethality of these transgenic embryos from leaky expression of catalase without heat shock. Moreover, cardiac-specific overexpression of catalase-DsRed suppressed embryonic heart regeneration in Mtz-induced heart injury of Tg(myl7:CFP-NTR) transgenic embryos (Supplementary information, Figure S13F-S13H). Previous studies suggest that myocardial dedifferentiation is a critical step for heart regeneration in zebrafish8,9. Interestingly, inhibiting H2O2 signaling by either DPI treatment or catalase overexpression had no effects on myocardial sarcomere disassembly, an indicator of myocardial dedifferentiation, in injured hearts at 14 dpa (Supplementary information, Figure S15). These results suggest that the high concentration of H2O2, likely caused by elevated duox/nox2 expression, is obligatory for cardiac regeneration after injury without affecting injury-induced myocardial dedifferentiation.
The Duox-H2O2-Dusp6 pathway primes myocardial regeneration by elevating Erk1/2 activity via a derepression mechanism
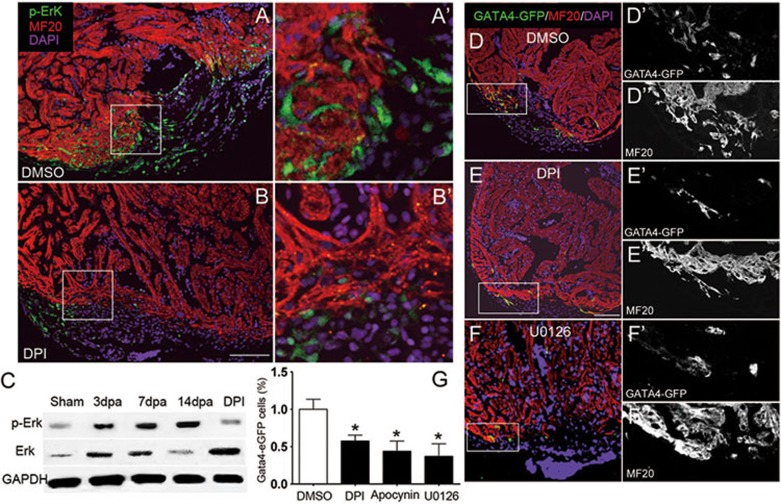
Because developmental signaling pathways of FGF and PDGF are reused during adult zebrafish heart regeneration20,21, we investigated whether H2O2 modulated the common MAP kinase pathway downstream of the growth factors. We found that phosphorylation of Erk1/2 (pErk) was increased in injured hearts at 3, 7 and 14 dpa (Figure 3C and Supplementary information, Figure S16), with the pErk1/2+ cells primarily located in non-myocardium such as the epicardium (Figure 3A and 3A'), evidenced by overlapping with epicardial Raldh2, a molecular marker for both the epicardium and endocardium (Supplementary information, Figure S18A-S18C). The total erk1 and erk2 mRNA, as well as total Erk proteins were slightly increased during heart regeneration (Figure 3C and Supplementary information, Figure S17). Using Tg(gata4:EGFP) transgenic reporter zebrafish to track newly regenerated myocardium in injured hearts8, we found that decreasing pErk1/2 with U0126 (a Mek1/2 inhibitor) interfered with the regeneration of myocardial cells (Figure 3F, 3F' and 3G), as evidenced by reduced EGFP-positive cells compared with DMSO controls (Figure 3D, 3D' and 3G). More importantly, the formation of pErk1/2+ cells (Figure 3B, 3B' and data not shown) and the generation of GATA4-EGFP+ myocardial cells (Figure 3E, 3E', 3G and data not shown) were sensitive to inhibition of Duox/Nox2 by treatment of either DPI or apocynin. Unfortunately, strong autofluorescence in Tg(myl7:catalase-DsRed) prevented us from performing any fluorescence-based immunostaining. These results showed that epicardial Erk1/2- and H2O2-dependent signals are interlinked in promoting regeneration in the adult zebrafish heart.
Figure 3.
H2O2 activates Erk1/2 for myocardial regeneration. (A-B) Phosphorylation of Erk1/2 (pErk1/2) in epicardial cells (A, A') in 14 dpa heart, as detected by anti-pErk1/2 antibody. Decrease of pErk1/2-positive cells by DPI treatment (B, B'). Scale bar for A and B, 100 μm. (C) Western blot showing greater pErk1/2 in injured hearts at 3, 7 and 14 dpa, and its blockade by DPI treatment at 14 dpa. (D-F) Gata4+ myocardial cells in injured areas in control Tg(gata4:EGFP) heart (DMSO treatment, D, D', D'') and their disappearance after inhibition of Duox (DPI treatment, E, E', E”), or Mek1/2 (U0126 treatment, F, F', F”). Cardiomyocytes were co-labeled with anti-EGFP and anti-MF20 antibodies and nuclei were labeled by DAPI. Scale bar for D-F, 100 μm. (G) Statistics of Gata4+ cardiomyocytes at 14 dpa.
Of the potential redox-sensitive targets of H2O2, the dual-specificity MAPK phosphatase 6 (Dusp6) is distinct in that it is stimulated by pErk1/2, but reciprocally dephosphorylates and deactivates pErk1/2, forming a negative feedback that limits Erk1/2 activity. Previous reports have shown that H2O2 directly oxidizes Dusp6 in TNFα-induced cell death and ovarian cancers, and that oxidized Dusp6 is prone to degradation, leading to aberrant Erk1/2 activation37,38. Dusp6-knockout mice show greater proliferation of myocytes39, but the function of Dusp6 in cardiac regeneration is unknown. We thus formulated a hypothesis with two seemingly paradoxical predictions. First, Erk1/2 activation would elevate Dusp6 expression during regeneration, activating a negative feedback (i.e., a repression). However, if H2O2 can oxidize and thereby destabilize Dusp6, ventricular resection-induced H2O2 production would unleash the pro-regenerative pErk1/2 signal (i.e., a derepression).
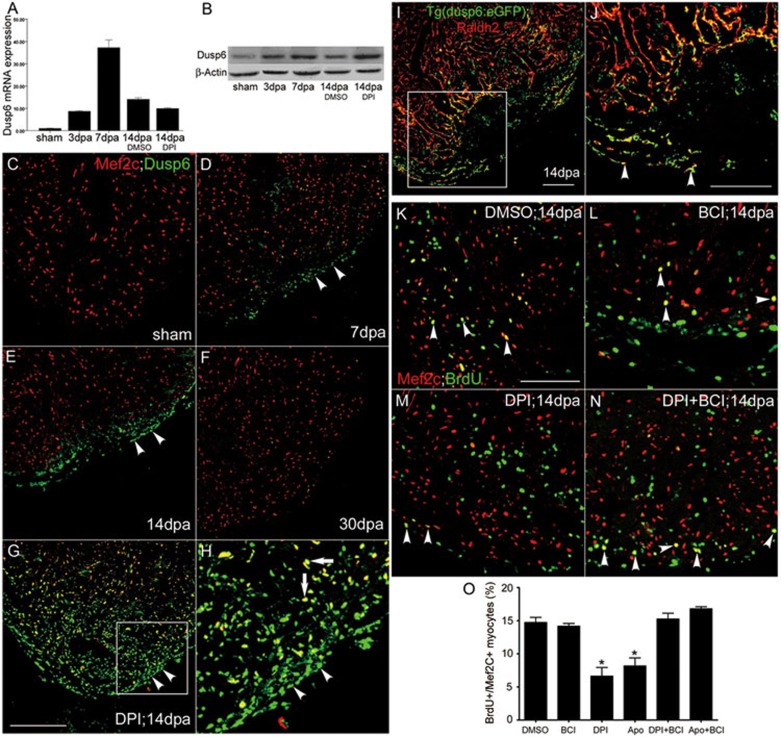
To test the above predictions, we examined Dusp6 expression at both the mRNA and protein levels. Upon injury, dusp6 mRNA level was elevated, peaking at 7 dpa (Figure 4A and Supplementary information, Figure S19), and Dusp6 protein levels were increased (Figure 4B) and Dusp6 was particularly localized in the epicardium at 7 and 14 dpa (Figure 4D-4E and Supplementary information, Figures S18D-S18F and S20B-S20D) and decreased at 30 dpa (Figure 4F and Supplementary information, Figure S20G). The most intriguing finding is that in injured hearts at 14 dpa after DPI or apocynin treatment, Dusp6 protein, but not dusp6 mRNA, was dramatically further increased in both the epicardium and myocardium with clear nuclear enrichment (Figure 4A, 4B, 4G and 4H), unmasking an inhibitory effect of H2O2 on Dusp6 protein stability in the epicardium and myocardium undergoing active regeneration. We also confirmed this finding on Tg(dusp6:EGFP), where EGFP expression was previously shown to recapitulate the endogenous dusp6 mRNA expression pattern during embryogenesis40. Epicardial EGFP expression was evident in injured areas of adult hearts at 14 dpa (Figure 4I-4J). Furthermore, pErk1/2 was largely co-localized with Dusp6 (Supplementary information, Figure S21C) and inhibiting pErk1/2 by U0126 reduced Dusp6 expression (Supplementary information, Figure S21A-S21D), suggesting a close interlink between them. These data substantiate the idea that Dusp6 mRNA and protein are initially induced as a negative feedback for pErk1/2, but subsequently H2O2, likely produced by Duox/Nox2, destabilizes Dusp6 to impart a derepression mechanism by suppressing this negative feedback in the epicardium.
Figure 4.
Suppressing Dusp6 by H2O2 or BCI promotes cardiac regeneration. (A) Quantitative RT-PCR showing induced dusp6 expression during regeneration. Note that dusp6 mRNA expression was unaffected by DPI treatment. n = 5. (B) Western blotting of Dusp6 proteins at 3, 7 and 14 dpa. Note that DPI treatment further augmented Dusp6 abundance at 14 dpa. (C-F) Dusp6 immunostaining in sham control (C) and at 7, 14 and 30 dpa (D-F). (G-H) DPI treatment induced widespread intense Dusp6 protein expression in the epicardium (arrowheads) and myocardium (arrows) at 14 dpa. Similar results were obtained in apocynin-treated hearts (data not shown). (I-J) Immunostaining showing colocalization of reporter EGFP (green) in Tg(dusp6:EGFP) heart and the epicardial and endocardial marker Raldh2 protein (red) in the injured area at 14 dpa. White arrowheads in J: epicardial cells positive for both Raldh2 and EGFP. (K-N) DPI treatment diminished proliferating myocytes identified as Mef2C (red) and BrdU (green) double-positive cells at 14 dpa (M) as compared to DMSO control (K). This inhibitory effect was reversed by BCI treatment (N), while BCI alone exerted no significant effect (L). (O) Statistics of the BrdU+/Mef2C+ proliferating myocytes. Scale bars, 100 μm.
To further evaluate the role of the Duox-H2O2-Dusp6 derepression signaling pathway in heart regeneration, we used the small molecule BCI, recently developed in a transgenic zebrafish chemical screen40, which directly inhibits the phosphatase activity of Dusp6. We found that BCI counteracted the inhibitory effect of DPI or apocynin on the generation of the double-positive BrdU+/Mef2C+ myocytes, but had no effects on DPI- or apocynin-enhanced cardiac fibrosis at 14 dpa (Figure 4K, 4M, 4N, 4O and Supplementary information, Figure S24A-S24C). Moreover, BCI also reversed the effects of DPI or apocynin on MF20 staining (which indicates myocardial regeneration), and cardiac fibrosis at 30 dpa (Supplementary information, Figure S24D-S24I). These results indicate that BCI mimics the endogenous H2O2 signal when the latter is abrogated. However, in hearts untreated with DPI or apocynin, BCI did not alter the generation of BrdU+/Mef2C+ myocytes at 14 dpa (Figure 4K, 4L and 4O), consistent with the idea that BCI is redundant in the presence of an intact endogenous H2O2 signal. Like exogenous H2O2 (Supplementary information, Figure S10), BCI had no effects on myocardial proliferation in sham-operated hearts (Supplementary information, Figure S23), which is consistent with the relatively low levels of Dusp6 proteins in sham hearts (Figure 4C and Supplementary information, Figure S20A). To examine whether H2O2 is involved in coronary vessel regeneration, we used the Tg(flk1:EGFP) zebrafish to visualize coronary vessels in the adult heart. We found that DPI treatment reduced the amount of Flk1+ vessels, which was rescued by co-injection of BCI (Supplementary information, Figure S22), suggesting an additional role of H2O2 in coronary vessel repair upon ventricular resection. Collectively, interventions at the levels of Duox/Nox2, H2O2 and Dusp6 corroborated the hypothesis that the Duox-H2O2-Dusp6 pathway plays an essential role in priming myocardial and blood vessel regeneration, by elevating Erk1/2 activity via a derepression mechanism.
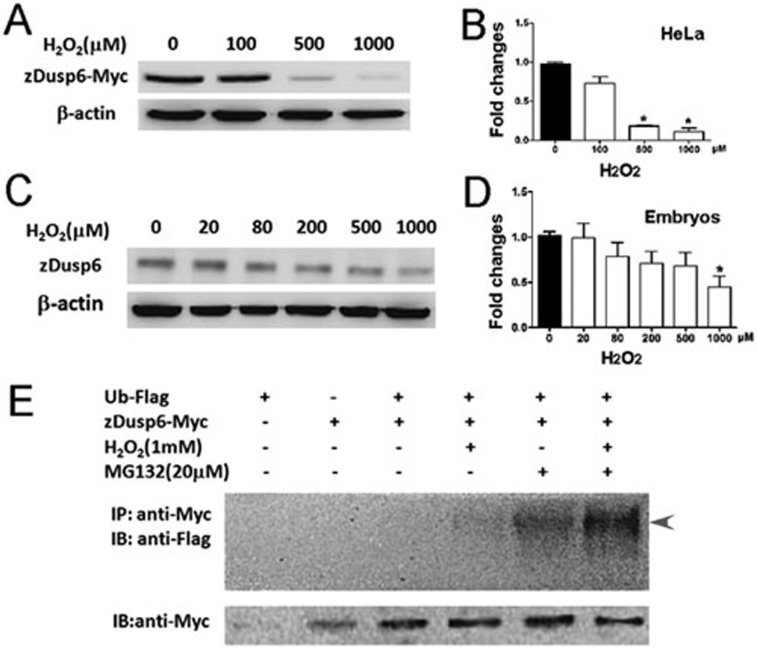
To directly test whether zebrafish Dusp6 (zDusp6) is subjected to H2O2-dependent ubiquitination, we measured the ubiquitinylated zDusp6 (Ub-zDusp6) protein levels with or without H2O2 stimulation in HeLa cells and zebrafish embryos, given that adult zebrafish heart is hardly accessible for these molecular and biochemical methods. We found that exogenous H2O2 treatment decreased, in a dose-dependent manner, the levels of Myc-tagged zDusp6 proteins in HeLa cells overexpressing zDusp6-Myc (Figure 5A and 5B). Similarly, endogenous zDusp6 underwent degradation in zebrafish embryos after H2O2 treatment (Figure 5C and 5D). Furthermore, Ub-zDusp6 was hardly detectable, but was detected after treatment with 1 mM H2O2 in HeLa cells overexpressing zDusp6-Myc and Flag-tagged ubiquitin (Ub-Flag) (Figure 5E), suggesting the existence of unstable Ub-zDusp6 that is dependent on H2O2. Proteasome inhibitor MG132 increased the amount of Ub-zDusp6 in the absence or presence of H2O2, supporting the notion that H2O2 treatment enhances the ubiquitination of zDusp6. On the other hand, we failed to detect Ub-zDusp6 in the presence of H2O2 and MG132 in zebrafish embryos overexpressing zDusp6-Myc and Ub-Flag (data not shown), suggesting that Ub-zDusp6 is highly unstable in zebrafish embryos. Therefore, H2O2 regulates the ubiquitination and proteasome degradation of zebrafish Dusp6.
Figure 5.
H2O2 induces ubiquitination and proteasome degradation of zebrafish Dusp6. (A-B) Exogenous H2O2 decreased the expression level of Myc-tagged zDusp6 in a dose-dependent manner in HeLa cells as evidenced by western blotting (A). The statistics of A is shown in B. β-actin was used for normalizing protein loading. (C-D) Exogenous H2O2 decreased the expression level of endogenous zDusp6 in a dose-dependent manner in zebrafish embryos at 24 hpf (C), and the statistics of C is shown in D. β-actin was used for normalizing protein loading. (E) Ub-zDusp6 (arrowhead) was detected after H2O2 and/or MG132 treatments in HeLa cells overexpressing Myc-tagged zDusp6 and Flag-tagged ubiquitin (Ub-Flag). Myc-tagged Dusp6 proteins were detected for all groups except the group without zDusp6-Myc overexpression.
H2O2 promotes heart regeneration independent of immune cell recruitment
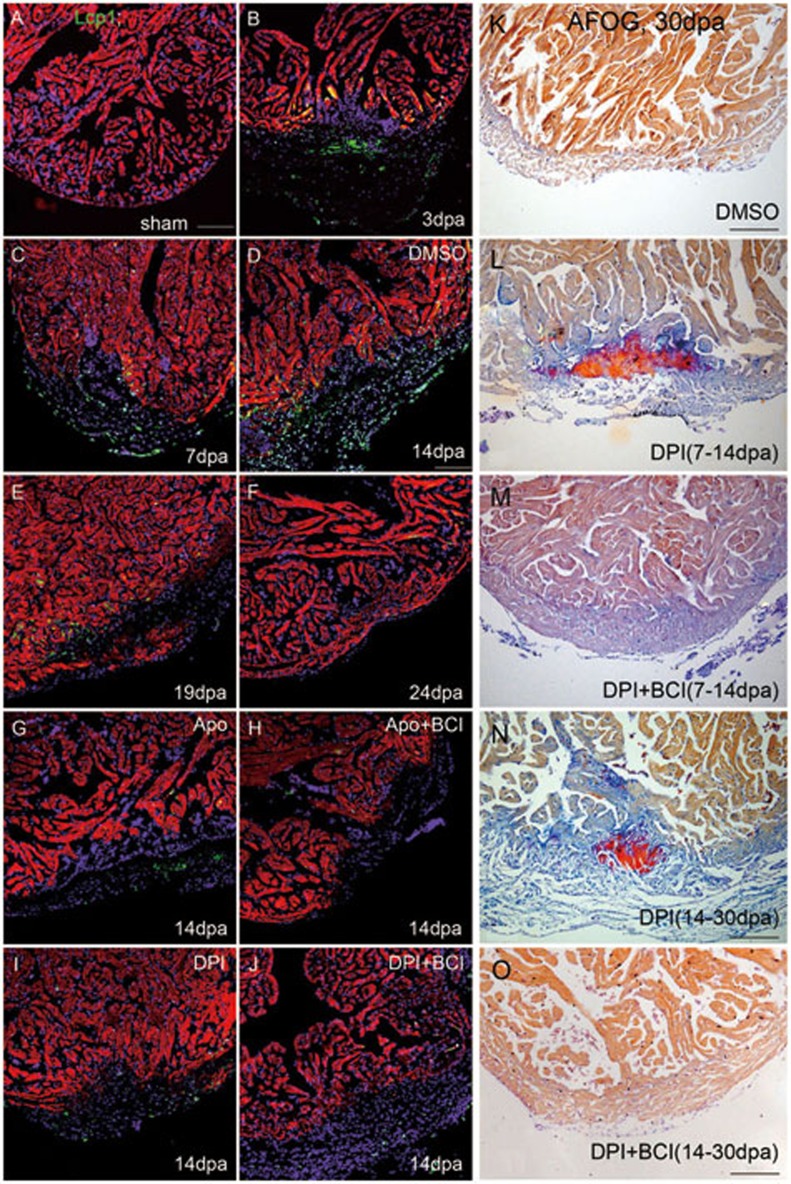
Previous studies have shown that H2O2 signal is required for the rapid inflammatory response in the tail-fin epithelium of zebrafish larvae25,26. To differentiate potentially dual function of H2O2 in recruiting inflammatory cells and priming heart regeneration, we examined temporal and spatial distribution of immune cells during heart regeneration by labeling macrophages and neutrophils with either anti-lymphocyte cytosolic protein 1 (Lcp1) or Tg(coro1a:EGFP)41. We found that Lcp1+ leukocytes appeared around 3 dpa, peaked at 7 and 14 dpa, decreased at 19 dpa and almost disappeared at 24 dpa (Figure 6A-6F), and the recruitment occurred in a H2O2-dependent manner (Figure 6G and 6I). Quantification of Lcp1+ leukocytes during heart regeneration in Figure 6 was included in Supplementary information, Figure S25. Similar dynamics of leukocyte recruitment was obtained by labeling leukocytes with an independent Tg(coro1a:EGFP) transgenic reporter (Supplementary information, Figure S26A-S26F). Furthermore, DPI treatment either from 7 to 14 dpa (when immune cells peaked, DPI timely inhibited leukocyte recruitment) or from 14 to 30 dpa (when immune cells gradually disappeared, DPI should not affect normal leukocyte recruitment) led to increased cardiac fibrosis and compromised myocardial regeneration (Figure 6L and 6N), suggesting that both the early- (7-14 dpa) and late-phase H2O2 production (14-30 dpa) are required for cardiac regeneration. Importantly, co-injection of BCI with DPI or apocynin largely restored myocardial regeneration (Figure 6M and 6O), but failed to rescue immune cell recruitment (Figure 6H, 6J and Supplementary information, Figure S26G-S26H), indicating that Dusp6 inhibition by BCI suffices to initiate regeneration independently of immune cell recruitment. Taken together, these data suggest that while H2O2 plays a dual role in recruiting immune cells and promoting heart regeneration, the two pathways are apparently separable and relatively independent of each other.
Figure 6.
H2O2 has dual function in recruiting Lcp1+ leukocytes and promoting heart regeneration. (A-F) Immunostaining of Lcp1 showing that leukocytes (macrophages and neutrophils) were undetectable at sham (A), appeared at 3 dpa (B), peaked between 7 and 14 dpa (C, D) and gradually disappeared from 19 to 24 dpa (E, F). Either apocynin (Apo) (G) or DPI (I) inhibited leukocyte recruitment, which could not be rescued by BCI (H, J) at 14 dpa. Myocytes were labeled by anti-MF20, and nuclei were labeled by DAPI. (K-M) All AFOG staining analyses were performed at 30 dpa and DPI and/or BCI treatment was applied as indicated. Myosin was visualized as orange, fibrin as red and collagen as bright blue. AFOG staining showed that DPI treatment from 7 to 14 dpa increased cardiac fibrosis (L), which was rescued by BCI (M), compared with DMSO control (K) at 30 dpa. (N-O) AFOG staining showed that DPI treatment from 14 to 30 dpa increased cardiac fibrosis (N), which was rescued by BCI (O). Scale bars, 100 μm.
Dusp6 is a repressor for zebrafish heart regeneration
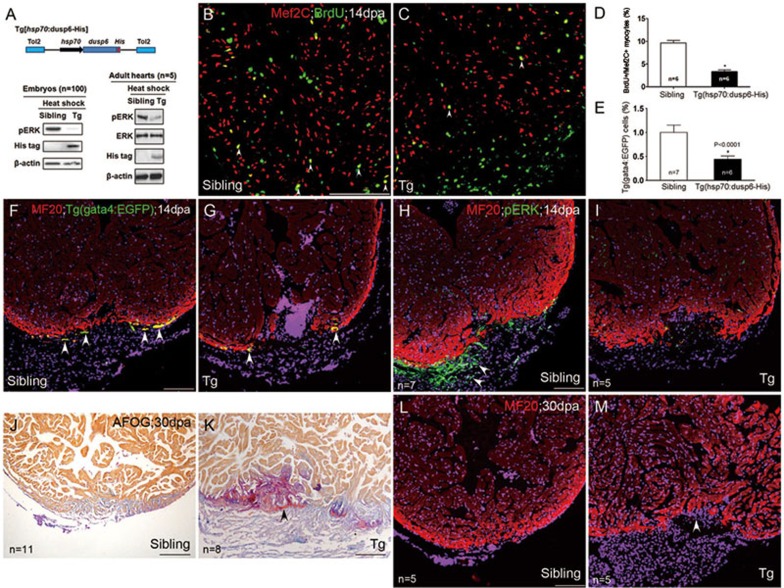
To directly examine the role of dusp6 in heart regeneration, we established Tg(hsp70:dusp6-His) transgenic zebrafish, where overexpression of Dusp6 in both epicardium and myocardium was conditionally driven by heat shock21. Transgenic expression of dusp6 was confirmed to decrease pErk in both embryos and adult hearts (Figure 7A). Compared with non-transgenic siblings (Figure 7B, 7F, 7H, 7J and 7L), injured transgenic hearts displayed fewer Mef2C+/BrdU+ proliferating myocytes (Figure 7C-7D), Gata4+ myocytes (Figure 7E and 7G) as well as pERK+ cells (Figure 7I) at the injury site, leading to increased cardiac fibrosis (Figure 7K) and diminished cardiac regeneration (Figure 7M). Together with the BCI data (Figures 4 and 6), these results establish a repressive role of dusp6 in heart regeneration, acting through dephosphorylation of pErk in the epicardium.
Figure 7.
The repressive effects of Dusp6 overexpression on heart regeneration through dephosphorylation of pERK. (A) Top: a scheme showing that in Tg(hsp70:dusp6-His), His-tagged Dusp6 is driven by the zebrafish hsp70 promoter. Transgenesis was mediated by Tol2 transponase. Bottom: heat shock-induced expression of His-tagged Dusp6 proteins diminished pErk in transgenic embryos (Tg) at 72 hpf or transgenic adult hearts (Tg), compared with their sibling embryos or adult hearts. β-actin was used to normalize protein loading. (B-D) Mef2C+/BrdU+ proliferating myocytes (arrowheads) were decreased in dusp6 transgenic hearts (C) compared with those in non-transgenic siblings (B), with the statistics shown in D. (E-G) Representative images showing that Tg(gata4:EGFP) myocytes (arrowheads), which colocalized with cardiac marker MF20, were diminished in dusp6 transgenic hearts (G) compared with their siblings (F), with the statistics shown in E. (H-I) Reduction of pErk+ cells (arrowheads), not overlapping with cardiac marker MF20, in transgenic hearts (I), compared with non-transgenic siblings (H). (J-M) Transgenic overexpression of Dusp6 led to cardiac fibrosis (assayed by AFOG staining, K) and compromised cardiac regeneration (assayed by MF20 staining, M), compared with their siblings (J, L). Nuclei were co-stained with DAPI. Scale bars, 100 μm.
Discussion
In summary, we have presented three major interrelated findings. First, upon injury, H2O2 is produced, likely by Duox/Nox2, up to 30 μM in the epicardium and adjacent myocardium during the entire course of active myocardial regeneration. Second, H2O2 is required for promoting cardiac myocyte proliferation and suppressing fibrosis. Third, the pro-regeneration effect of H2O2 is mediated through destabilizing the redox-sensitive and ubiquitin-conjugated phosphatase Dusp6, which provides a derepression of the common MAP kinase signaling. Furthermore, we have identified BCI as the first small molecule for promoting cardiac regeneration when H2O2 generation was defective.
In spite of previous findings that H2O2 signal is required for the rapid inflammatory response in the tail-fin epithelium of zebrafish larvae25,26, our data show that the H2O2-guided inflammatory response occurs around 3 dpa in adult zebrafish hearts (Figure 6 and Supplementary information, Figure S26). This time course, when compared with that of Duox/Nox2 induction and H2O2 production, supports the idea that it likely involves de novo synthesis of duox transcripts and possibly proteins for the production of H2O2 and subsequent recruitment of leukocytes in heart regeneration. In contrast, because the injured larvae tail fin is directly exposed to the environment, it needs immediate immune defense after injury and therefore only takes 3 min to recruit neutrophils to the injured site, through activation of Duox without transcription and translation in the tail fin epithelium25,26. In addition to its function in recruiting inflammatory leukocytes, our data suggest that H2O2 is also required for promoting adult heart regeneration, which is consistent with previous reports on H2O2 function in promoting injury-induced peripheral sensory axon regeneration in zebrafish larvae42 and tail regeneration in Xenopus tadpole43. H2O2 acts through a redox sensor Lyn to mediate injury-induced leukocyte wound attraction in zebrafish larvae tail fin25,26, but direct targets of H2O2 in zebrafish sensory neurons or Xenopus tadpole tail epithelium are not known42,43. Our data show that H2O2 not only destabilizes Dusp6 to elevate growth factor signaling in promoting adult heart regeneration, but also acts through an unknown, independent mechanism to recruit inflammatory leukocytes during heart regeneration. Future studies need to examine whether H2O2 acts through Lyn or other redox sensors to induce inflammatory leukocyte recruitment during adult heart regeneration.
Growth factor signaling pathways such as FGF and PDGF pathways are essential for adult heart regeneration20,21. In contrast to previous reports on a negative role of p38 MAPK in mammalian and zebrafish heart regeneration44,45, our data support that H2O2 in the epicardium (Figure 1) is essential for elevating phosphorylation of Erk1/2 MAPK through repression of Dusp6 during zebrafish heart regeneration (Figures 3,4,5 and 7). Our data are aligned well with the previous reports that H2O2 directly oxidizes Dusp6 in TNFα-induced cell death and ovarian cancers, leading to ubiquitination and subsequent proteasome degradation of Dusp6 and thus aberrant Erk1/2 activation37,38 (Figure 5). The elevated pErk1/2 in the epicardium might then activate soluble factors at transcriptional and/or post-transcriptional levels, which in turn bind to their receptors in the myocardium and promote myocardial proliferation and regeneration. However, we failed to detect the changes in Dusp6 and pErk protein levels in the heart by western blotting (data not shown). It is likely that the subtle changes in Dusp6 and pErk protein levels in the epicardium of injured hearts might not be well detected by western blotting. On the other hand, cardiac-specific overexpression of catalase, the H2O2-specific scavenger, also led to regenerative defects in the heart (Figure 2J-2M). New transgenic lines, such as Tg(tcf21:catalase) without autofluorescence or heat-inducible Tg(hsp70:catalase) without leaky transgene expression are needed for determining the levels of Dusp6 and pErk by immunostaining after scavenging H2O2 by catalase in the future.
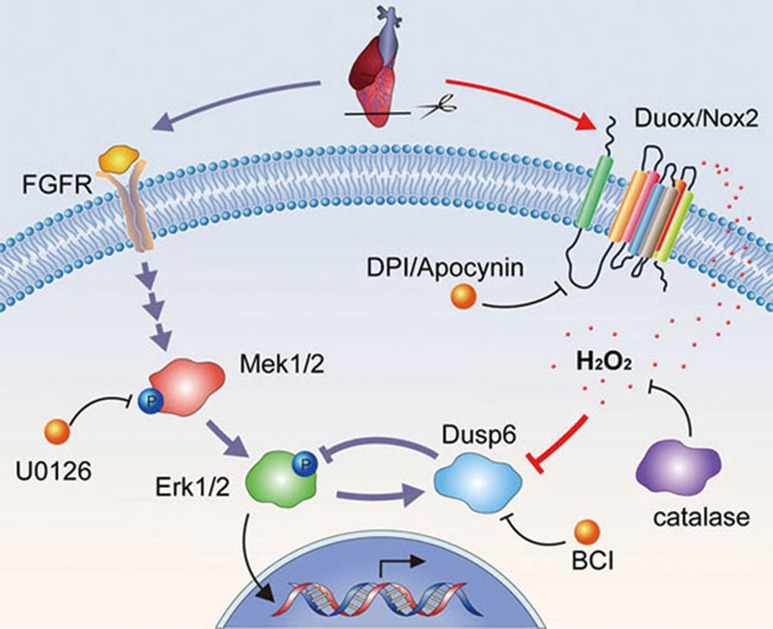
Based on the data from others and ours, we propose that the working model for cardiac regenerative signaling comprises two main branches: the forward pro-regenerative MAPK signaling triggered by FGF and other growth factors, as recently elucidated20,21; and the derepression mechanism through the Duox/Nox2-H2O2-Dusp6 pathway that converges on Erk1/2 MAPK signaling primarily in the epicardium, which then activate putative soluble factors to promote myocardial regeneration (Figure 8). Such epicardial soluble factors might include Wnt1 and IGF2 that were previously shown to play an essential function in the epicardium/cardiac fibroblasts and myocardium46,47,48,49. During heart regeneration, the growth factor-MAPK signaling (pErk) is gradually enhanced in injured hearts from 3 to 14 dpa with the peak around 14 dpa, which induces dusp6 expression (positive induction) that in turn dephosphorylates pErk (negative feedback) (Figures 3 and 4). Simultaneously, H2O2 increases and destabilizes Dusp6 with the peak from 7 to 14 dpa, which results in decreased Dusp6 expresssion and thus increased pErk levels at 14 dpa when myocardial regeneration actively takes place. In addition, we have shown that H2O2/Dusp6 is also involved in coronary vessel regeneration upon ventricular resection. This is consistent with previous studies that FGF and PDGF signaling pathways are essential for coronary vessel formation during heart regeneration20,21. Therefore, this work provides the first intracellular H2O2-Dusp6-pErk mechanisms downstream of growth factor signaling in adult myocardial and coronary vessel regeneration. As there are highly conserved mammalian homologs for all components of the Duox/Nox2-H2O2-Dusp6-Erk1/2 axis, this derepression mechanism for cardiac regeneration, as demonstrated here in zebrafish, may also operate in mammals, and if so, future efforts on activating the mammalian counterpart may offer opportunities for therapeutic myocardial regeneration in heart failure.
Figure 8.
H2O2 promotes heart regeneration through a derepression mechanism in zebrafish. Upon ventricular resection, FGF and other growth factor receptors are activated and resultant phosphorylation of Mek1/2 (pMek1/2) and Erk1/2 (pErk1/2) induces expression of dusp6, which in turn, exerts a negative feedback on pErk1/2 in non-myocardial cells (primarily epicardial cells). Concurrently, injury elicits a time-dependent, enduring localized duox and, to a minor extent, nox2 expression that supports intense H2O2 formation primarily in the epicardium close to the injury site. High-concentration intracellular H2O2 oxidizes and thereby destabilizes the Dusp6 phosphatase, diminishing its inhibition of pErk1/2. Thus, the newly identified Duox/Nox2-H2O2-Dusp6 signaling serves as a derepression mechanism and, when activated, unleashes the pro-regenerative pErk1/2 signaling likely through generation of soluble factors that would eventually act to enhance myocardial regeneration. DPI and apocynin (Apo), inhibitors of Duox/Nox2; catalase, catalytic scavenger of H2O2; BCI, inhibitor of Dusp6; U0126, inhibitor of Mek1/2.
Materials and Methods
Zebrafish lines
Zebrafish were raised and handled in accordance with the Peking University Institutional Animal Care and Use Committee accredited by the AAALAC International. Wild-type TL, Tg(myl7:HyPer)pku327, (myl7:catalase-DsRed), Tg(dusp6:EGFP) and Tg(gata4:EGFP) transgenic zebrafish lines at 6-12 months of age were used for heart surgery. Several Tg(myl7:HyPer)pku327, Tg(myl7:catalase-DsRed)pku343, Tg(myl7:CFP-NTR)pku360 and Tg(hsp70:dusp6-His)pku344 transgenic zebrafish lines were generated in TL background by using Tol2-based transgenesis31,50. Tg(tcf21:nucEGFP) was previously reported30. Tg(coro1a:EGFP) was provided by Dr Zilong Wen41, Tg(dusp6:EGFP) transgenic zebrafish were obtained from Dr Michael Tsang40 and Tg(gata4:EGFP) transgenic zebrafish were kindly provided by Dr Todd Evans51. The zebrafish myl7 promoter was provided by Dr C Geoff Burns52; pHyPer-Cyto cDNA clone was purchased from Evrogen (Cat No. FP941); zebrafish catalase cDNA (CDS) was isolated from 2-day embryos according to catalase sequence (NM_130912); rat Dusp6 cDNA was obtained from Dr Zhi-Xin Wang53; and CFP-NTR was provided by Dr Ling-Fei Luo35. These expression constructs were cloned into the Tol2 backbone50. Each of the transgenic lines made in our laboratory was characterized and/or confirmed by more than two independent founders.
Mtz-induced embryonic heart injury in Tg(myl7:CFP-NTR) embryos
A large number of cardiomyocytes were damaged by incubating Tg(myl7:CFP-NTR) transgenic embryos with Mtz essentially as previously described35. Briefly, transgenic embryos at 70 hpf were incubated with 2 mM Mtz, and Mtz was then washed off and DPI and/or H2O2 were added at 72 hpf. In this condition, about 80% control transgenic hearts restored normal morphology and function at 124 hpf.
Adult zebrafish heart resection
A ventricular resection procedure was carried out essentially as described by Poss and colleagues3. Briefly, adult zebrafish were anesthetized with tricaine and placed with ventral side up into the groove of a sponge. The pericardial sac was exposed by removing surface scales and a small piece of skin with iridectomy scissors. Opening the pericardial sac was aided by tweezers, and the apex of the ventricle was gently pulled up and removed with Vannas scissors. A small piece of wet tissue was placed on the top of the ventricular wound. The heart stopped bleeding very quickly (within 10-15 s) due to the formation of blood clots. The fish was placed back into a water tank, where water was puffed over the gills with a plastic pipette until the fish was breathing and swimming regularly. The surface opening sealed automatically within a few days.
Library preparation for a strand-specific poly(A)-positive RNA-seq
Sham and 7 dpa hearts were freshly excised and rinsed with pre-cooled PBS. The outflow tract and atrium were removed immediately. The total RNA of each group was extracted from about 25 hearts by using QIAGEN RNeasy Microarray Tissue Mini Kit (Cat No. 73304). Poly(A)-positive RNA was purified from 5 μg of total RNA (RNA integrity number ≥ 7.5) with the Dynabeads mRNA purification kit (Invitrogen), following the manufacturer's instructions. A strand-specific RNA-seq library was then prepared according to the deoxy-UTP approach as reported previously54. Amplified materials were loaded onto a flow cell and sequencing was carried out on the Illumina HiSeq2000 platform by running 90 cycles (paired-end design) according to the manufacturer's instructions.
RNA-Seq and microarray data analysis and mining
Paired-end mRNA sequence tags were mapped on the Zebrafish Genome (Zv9) by Tophat (v1.4.1). Multiple alignment reads were discarded. The RPKM scores for each gene were calculated as previously described55. A series of Perl (v5.12.2) and R (v2.13.1) scripts were implemented to evaluate the quality of the RNA-seq experiments. Overall, high-quality RNA-seq data were obtained with high base quality, unbiased read distribution on transcripts, correct strand information, little DNA contamination and low mutation rate across the reads (Supplementary information, Figure S1). Eight hundred and forty-five upregulated genes between sham and 7 dpa hearts were identified using Cuffdiff (1.3.0) (Supplementary information, Figure S2), on the basis of three criteria: (1) P (un-corrected) < 0.05; (2) fold change > 2; and (3) RPKM > 0.2 in at least one sample. Protein sequences of upregulated genes were then subjected to KOBAS (v2.0) to identify enriched biological pathways, with the P-value calculated using a hypergeometric distribution. FDR correction was performed to adjust for multiple testing and pathways with FDR-corrected P-value < 0.05 were considered statistically significantly enriched. The RNA-seq data entitled “Transcriptome analysis of heart regeneration in adult zebrafish” were deposited to the GEO repository with accession number GSE50203 (GSM1215619 for sham heart and GSM1215618 for 7 dpa heart).
A list of previous known ROS genes were manually selected and evaluated for their relative expression levels between sham and 7 dpa hearts, based on a publicly available microarray of zebrafish heart regeneration29 and the above RNA-seq data on zebrafish heart regeneration. We found that 23 of them were changed between sham and 7 dpa hearts in the microarray data that were expressed as log (7dpa_log2FC_Belmonte), and 32 of them were changed between sham and 7 dpa hearts in our RNA-seq data that were expressed as log (7dpa_logFC_RNAseq) in Supplementary information, Table S4. We wrote in-house R scripts to analyze the microarray data, using package Limma from Bioconductor56.
Ex vivo intact heart imaging
Adult Tg(myl7:Hyper) hearts were removed and rinsed in modified Tyrode's solution (in mM: 150 NaCl, 5.4 KCl, 1.5 MgSO4, 0.4 NaH2PO4, 2 CaCl2, 10 glucose, 10 HEPES, pH 7.4). The atrium was removed to diminish spontaneous beatings. Hearts were viable for up to 1 h in such conditions, manifested by responses to 1 Hz electric field stimulation. Viability of the hearts was confirmed by their rhythmic contractions upon stimulation. A high speed camera was used to capture the contractions of the hearts and then kymograph was generated from the movies. Hearts were imaged under the 20× water objective lens of a Zeiss LSM700 upright confocal microscope with 405- and 488-nm lasers for dual excitation of Hyper, and 505-565 nm emission lights were collected with band-pass filter. For calculating Hyper 488/405 ratio, F488 and F405 images were smoothed, background subtracted and divided. For the Hyper ratiometric calibration curve, Hyper transgenic hearts were treated and imaged ex vivo under designated concentrations (5, 10, 20, 50, 100 and 200 μM) of H2O2 for 2 min. For time-lapse recordings with 10 μM DPI or DMSO treatment, Hyper transgenic hearts were imaged at 30-s intervals for 3 min in normal Tyrode's solution before application of DPI or DMSO; the total recording time was 45 min. To detect redox signals with chemical indicators, 30 μM of Redox sensor cc-1 (R-14060; Invitrogen) was injected into the thoracic cavity of 14 dpa Tg(myl7:Hyper) or wild-type zebrafish 30 min before heart isolation. Redox sensor cc-1 was excited at 555 nm. Image processing and data analysis were performed using the IDL program (Research Systems).
BrdU incorporation and small molecule treatment
After ventricular resection, zebrafish were allowed to recover for 7 days and 50 μl of 2.5 mg/ml BrdU (B5002; Sigma) was then co-injected with 10 μM DPI (D2926; Sigma), 100 μM apocynin (W508454; Sigma), 10 μM BCI (B4313; Sigma), or 10 μM U0126 (U120; Sigma) into the thoracic cavity once daily until the hearts were collected. In pilot experiments, BCI was kindly provided by Dr Michael Tsang of the University of Pittsburg40.
Western blotting and immunoprecipitation
Zebrafish tissues were treated in cell lysis buffer (P0013; Beyotime) in the presence of a protease inhibitor cocktail (P8340; Sigma) and PhosSTOP phosphatase inhibitor cocktail (04906845001; Roche). Proteins were quantified with a BCA kit (PA115; Tiangen Biotech), resolved on 12% SDS-PAGE gels, and transferred onto PVDF membranes (162-0177; Bio-Rad). Membranes were incubated with the appropriate primary antibody followed by the incubation with the corresponding HRP-conjugated secondary antibody, and proteins were detected with ECL western blotting substrates (W1001; Promega).
HeLa cells were grown in DMEM high glucose medium (Thermo HyClone, SH30022.01B) containing 10% FBS (Thermo HyClone, SV30087.02). The zDusp6 expression construct pcDNA4-zDusp6-Myc and the Flag-tagged ubiquitin expression construct pcDNA-Ub-Flag57 were cotransfected into HeLa cells using Lipofectamine 2000 (Invitrogen, 11668). Cells were collected and lysed using RIPA buffer (Applygen, C1053) with 0.1 mM PMSF, 5 mM EDTA and 1 mM Complete TM protease inhibitors (Roche, 04693132001) in the presence or absence of 10 mM MG132 (Sigma, M7449). Cell lysates were then incubated with rabbit anti-Myc mAb (Cell Signaling, 2278) at 4 °C overnight. After incubation, 40 μl of Protein A+G beads (Abmart, A10001) was added to each sample and the mixture was rotated at 4 °C for 6 h. After centrifugation, the beads were washed five times with RIPA buffer and then boiled for 10 min with 30 μl of 2× sample buffer. Mouse anti-Flag antibody (Sigma, F1804) and mouse anti-Myc (HRP) mAb (Cowin Bioscience, CW0300) were used for immunoblotting.
In situ hybridization, immunostaining and histological methods
Adult hearts were removed and fixed in 4% paraformaldehyde at room temperature for 2 h, and then went through an ethanol series for dehydration followed by paraffin embedding and sectioning (4 μm). Two duox cDNA clones were isolated from an embryonic zebrafish cDNA library made in Xiong's lab, and the primer sequences are shown as duox 1F and 1R, or duox 2F and 2R in Supplementary information, Table S5. Duox clone 1 and duox clone 2 cDNAs were used to make two independent duox probe 1 (Supplementary information, Figure S3) and duox probe 2 (Supplementary information, Figure S5). Dusp6 cDNA was kindly provided by Dr Michael Tsang40 and tcf21 cDNA by Dr Fabrizio Serluca. In situ hybridization on paraffin sections was performed using digoxygenin-labeled probes. Primary antibodies were anti-BrdU (B8434; Sigma), anti-Mef2c (sc-313; Santa Cruz), anti-tErk (4695; Cell Signaling), anti-pErk (9101; Cell Signaling), anti-tErk (MA5-15134; Pierce), anti-GFP (A-11122; Invitrogen), anti-GAPDH (sc-166545; Santa Cruz), anti-MF20 (Developmental Studies Hybridoma Bank), anti-Dusp6 (WH0001848M1; Sigma), anti-Lcp1 (124420; Genetex), anti-β-actin (4967; Cell Signaling) and anti-Raldh2 (P30011; Abmart). Secondary antibodies used for immunostaining were Alexa Fluor 488 goat anti-mouse IgG (A21121; Invitrogen), Alexa Fluor 488 goat anti-rabbit IgG (A11034; Invitrogen), Alexa Fluor 555 goat anti-mouse IgG (A21424; Invitrogen) and Alexa Fluor 555 goat anti-rabbit IgG (A21428; Invitrogen). Secondary antibodies for western blotting were HRP goat anti-rabbit IgG (sc-2004; Santa Cruz) and HRP goat anti-mouse IgG (sc-2005; Santa Cruz).
Masson's and AFOG staining
For AFOG staining, paraffin sections were incubated in Bouin's solution (HT10132; Sigma) preheated at 56 °C for 2.5 h, incubated at room temperature for 1 h, briefly washed in tap water, incubated in 1% phosphomolybdic acid (P4869; Sigma) for 5 min, rinsed with distilled water and stained with AFOG staining solution (3 g of acid fuchsin (F8129; Sigma), 2 g of orange G (O3756, Sigma) and 1 g of anilin blue (AB0083; BBI) that were dissolved in 200 ml of acidified distilled water, pH 1.1) for 10 min. Sections were rinsed with distilled water, dehydrated with ethanol gradients and mounted. Myosin was visualized as orange, fibrin as red and collagen as bright blue.
Masson's staining was performed on paraffin sections using BASO Masson's Trichrome Stain Kit (BA-4079A; Baso Diagnostics Inc). Myosin was visualized as red and collagen as bright blue.
Transmission electron microscopy
DMSO- and DPI-treated wild-type adult hearts, as well as Tg(myl7:catalase-DsRed) transgenic adult hearts after injury at 14 dpa were fixed with glutaraldehyde at a final concentration of 2.5% (v/v) in 0.1 M sodium cacodylate buffer. Samples were rinsed and post-fixed with 1.0% OsO4. Next, the samples were embedded in the Spur resin and sectioned. The sections were stained with uranyl acetate and lead citrate and observed under a transmission electron microscope JEOL 1010 (JEOL Ltd., Japan).
RT-PCR analysis
Total RNA from sham and injured hearts (15-20 hearts per sample) was extracted and purified using RNeasy@ mini kit (74106; Qiagen), and RT-PCR was performed with OneStep RT-PCR kit (210212; Qiagen) using 40 cycles on 50 ng of total RNA per reaction. Semi-quantitative RT-PCR of ERK1 and ERK2 was performed using 25 PCR cycles. Primer sequences were shown in Supplementary information, Table S5.
Statistical analysis
Statistical analysis was performed on relevant data using Student's two-tailed t-test and the GraphPad Prism software package or SPSS18.0. Data were reported as mean ± SEM. For Supplementary information, Figure S12B and S12F, statistic significance between the two groups was determined with χ2-test. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Drs RP Xiao, Y Rao, JR Peng, C Wei, K Ouyang, YM Wang, ZL Huang and JC Luo for critical comments on the manuscript, Dr IC Bruce for language editing, Dr Yingchun Hu for help on transmission electromicroscopy, and the Pathology Core of Institute of Molecular Medicine for histology. We also thank Drs J Wang and KD Poss for their kind help in performing in situ hybridization with duox probes on Tg(tcf21:nucEGFP) ventricles and Drs M Tsang, ZL Wen, T Evans, ZX Wang, LF Luo, GC Burns, J Chen, CM Cao and F Serluca for generous gifts of protocols, plasmid clones and/or transgenic zebrafish lines. This work was supported by the National Basic Research Program of China (2012CB944501, 2010CB529503 and 2013CB531200) and the National Natural Science Foundation of China (31271549, 31130067, 31000644 and 31221002).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Strand-specific RNA-Seq of sham and 7 dpa adult hearts.
Scatter plot showing expression values of Ensembl genes in sham and 7dpa hearts.
Duox expression during heart regeneration.
The sense duox probe 1 detected no signals during heart regeneration.
Duox expression in the wound area during heart regeneration.
Partial overlapping of duox with epicardial Tg(tcf21:nucEGFP) during heart regeneration.
Nox2, but not nox1, nox4 and nox5, was expressed during heart regeneration.
Ex vivo cultured zebrafish hearts are functionally intact.
(A-G) Ex vivo calibration of Hyper F488/F405 ratio as a function of ambient H2O2 concentration.
Exogenous H2O2 do not stimulate myocyte proliferation in adult hearts.
H2O2 rescues heart regeneration in the presence of DPI.
DPI irreversibly inhibited heart regeneration.
Exogenous H2O2 rescued, while transgenic over-expression of catalase-DsRed suppressed, embryonic heart regeneration.
H2O2 penetrate freely into zebrafish cardiac tissue.
Inhibiting H2O2 signaling has no effect on cardiac sarcomere disassembly during heart regeneration.
pErk are induced in non-myocardium during heart regeneration.
Total Erk mRNA are slightly induced during heart regeneration.
Both pErk and Dusp6 are induced in the epicardium during heart regeneration.
Dusp6 expression after ventricular resection.
Dusp6 are induced primarily in non-myocardium during heart regeneration.
Inhibiting pErk by U0126 decreases Dusp6 expression during heart regeneration.
H2O2 is essential for coronary vessel formation during heart regeneration.
BCI does not stimulate myocyte proliferation in adult hearts.
BCI rescues heart regeneration when H2O2 signal is defective.
Quantification of DPI or Apo effects on Lcp1+ leukocytes during heart regeneration in Figure 6.
H2O2, but not dusp6, is required for Tg(coro1a:EGFP) leukocyte recruitment during heart regeneration.
Up-regulated genes in 7 dpa hearts by RNA-Seq.
The KEGG enrichment pathways in 7 dpa hearts.
The immune system disease genes enriched in 7 dpa hearts.
A number of ROS genes are up- or down-regulated in 7 dpa hearts.
Gene-specific primer sequences
References
- Yi BA, Wernet O, Chien KR. Pregenerative medicine: developmental paradigms in the biology of cardiovascular regeneration. J Clin Invest. 2010;120:20–28. doi: 10.1172/JCI40820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JW, Chang NN. Recent advances in heart regeneration. Birth Defects Res C Embryo Today. 2013;99:160–169. doi: 10.1002/bdrc.21039. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Raya A, Koth CM, Büscher D, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci USA. 2003;100:11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rosa JM, Martin V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development. 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyoctes is induced by tissue regeneration. J Cell Sci. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Annu Rev Genet. 2010;11:710–722. doi: 10.1038/nrg2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Dal Ferro M, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- Ellison GM, Vicinanza C, Smith AJ, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- Uchida S, De Gaspari P, Kostin S, et al. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Reports. 2013;1:397–410. doi: 10.1016/j.stemcr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Han P, Yang H, et al. In vivo cardiac reprogramming contributes to zebrafish heart regeneration. Nature. 2013;498:497–501. doi: 10.1038/nature12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wu Q, Zhang Y, et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci USA. 2010;107:17206–17210. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development. 2012;139:1921–1930. doi: 10.1242/dev.078543. [DOI] [PubMed] [Google Scholar]
- Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC Evol Biol. 2007;7:109. doi: 10.1186/1471-2148-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Starnes TW, Deng Q, Huttenlocher A. Lyn is a redox sensor that mediates leukocyte wound attraction in vivo. Nature. 2011;480:109–112. doi: 10.1038/nature10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal. 2012;24:981–990. doi: 10.1016/j.cellsig.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLos Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleep E, Boué S, Jopling C, et al. Transcriptomics approach to investigate zebrafish heart regeneration. J Cardiovasc Med (Hagerstown) 2010;11:369–380. doi: 10.2459/JCM.0b013e3283375900. [DOI] [PubMed] [Google Scholar]
- Wang J, Panáková D, Kikuchi K, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- Jaquet V, Scapozza L, Clark RA, Krause KH, Lambeth JD. Small-molecule NOX inhibitors: ROS-generating NADPH oxidases as therapeutic targets. Antioxid Redox Signal. 2009;11:2535–2552. doi: 10.1089/ars.2009.2585. [DOI] [PubMed] [Google Scholar]
- Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Kirkman HN, Gaetani GF. Mammalian catalase: a venerable enzyme with new mysteries. Trends Biomed Sci. 2006;32:44050. doi: 10.1016/j.tibs.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Kamata H, Honda S, Maeda S, et al. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Chan DW, Liu VW, Tsao GS, et al. Loss of MKP3 mediated by oxidative stress enhances tumorigenicity and chemoresistance of ovarian cancer cells. Carcinogenesis. 2008;29:1742–1750. doi: 10.1093/carcin/bgn167. [DOI] [PubMed] [Google Scholar]
- Maillet M, Purcell NH, Sargent MA, et al. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J Biol Chem. 2008;283:31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina G, Vogt A, Bakan A, et al. Zebrafish chemical screening reveals an inhibitor of Dusp6 that expands cardiac cell lineages. Nat Chem Biol. 2009;5:680–687. doi: 10.1038/nchembio.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yan B, Shi YQ, Zhang WQ, Wen ZL. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J Biol Chem. 2012;287:25353–25360. doi: 10.1074/jbc.M112.349126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger S, Sagasti A. Hydrogen peroxide promotes injury-induced peripheral sensory axon regeneration in the zebrafish skin. PLoS Biol. 2011;9:e1000621. doi: 10.1371/journal.pbio.1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love NR, Chen Y, Ishibashi S, et al. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel FB, Hsieh PC, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci USA. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sune G, Morera C, Izpisua Belmonte JC. p38alpha MAPK regulates myocardial regeneration in zebrafish. Cell Cycle. 2012;11:1195–201. doi: 10.4161/cc.11.6.19637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Cavallero S, Gu Y, et al. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Gherghe C, Liu D, et al. Wnt1/βcatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31:429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Gemberling M, Wang J, et al. In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development. 2013;140:660–666. doi: 10.1242/dev.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Harrison MR, Osorio A, et al. Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One. 2013;8:e67266. doi: 10.1371/journal.pone.0067266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Heicklen-Klein A, Evans T. T-box binding sites are required for activity of a cardiac GATA-4 enhancer. Dev Biol. 2004;267:490–504. doi: 10.1016/j.ydbio.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Burns CG, Milan DJ, Grande EJ, et al. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Wu JW, Wang ZX. Mitogen-activated protein kinase (MAPK) phosphatase 3-mediated cross-talk between MAPKs ERK2 and p38alpha. J Biol Chem. 2011;286:16150–16162. doi: 10.1074/jbc.M110.203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK.Limma: linear models for microarray dataIn: Gentleman R , Carey V, Dudoit S, Irizarry R, Huber W, eds. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer New York, 2005397–420.
- Song R, Peng W, Zhang Y, et al. Central role of E3 ubiquitin ligase MG53 in insulin resistance and metabolic disorders. Nature. 2013;494:375–379. doi: 10.1038/nature11834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strand-specific RNA-Seq of sham and 7 dpa adult hearts.
Scatter plot showing expression values of Ensembl genes in sham and 7dpa hearts.
Duox expression during heart regeneration.
The sense duox probe 1 detected no signals during heart regeneration.
Duox expression in the wound area during heart regeneration.
Partial overlapping of duox with epicardial Tg(tcf21:nucEGFP) during heart regeneration.
Nox2, but not nox1, nox4 and nox5, was expressed during heart regeneration.
Ex vivo cultured zebrafish hearts are functionally intact.
(A-G) Ex vivo calibration of Hyper F488/F405 ratio as a function of ambient H2O2 concentration.
Exogenous H2O2 do not stimulate myocyte proliferation in adult hearts.
H2O2 rescues heart regeneration in the presence of DPI.
DPI irreversibly inhibited heart regeneration.
Exogenous H2O2 rescued, while transgenic over-expression of catalase-DsRed suppressed, embryonic heart regeneration.
H2O2 penetrate freely into zebrafish cardiac tissue.
Inhibiting H2O2 signaling has no effect on cardiac sarcomere disassembly during heart regeneration.
pErk are induced in non-myocardium during heart regeneration.
Total Erk mRNA are slightly induced during heart regeneration.
Both pErk and Dusp6 are induced in the epicardium during heart regeneration.
Dusp6 expression after ventricular resection.
Dusp6 are induced primarily in non-myocardium during heart regeneration.
Inhibiting pErk by U0126 decreases Dusp6 expression during heart regeneration.
H2O2 is essential for coronary vessel formation during heart regeneration.
BCI does not stimulate myocyte proliferation in adult hearts.
BCI rescues heart regeneration when H2O2 signal is defective.
Quantification of DPI or Apo effects on Lcp1+ leukocytes during heart regeneration in Figure 6.
H2O2, but not dusp6, is required for Tg(coro1a:EGFP) leukocyte recruitment during heart regeneration.
Up-regulated genes in 7 dpa hearts by RNA-Seq.
The KEGG enrichment pathways in 7 dpa hearts.
The immune system disease genes enriched in 7 dpa hearts.
A number of ROS genes are up- or down-regulated in 7 dpa hearts.
Gene-specific primer sequences