Dear Editor,
Drosophila tudor is a maternal effect gene required for germ cell formation and abdominal segmentation during oogenesis1,2. It encodes a large protein, Tudor (Tud), of 2 515 amino acids, containing 11 copies of a ∼60-residue sequence motif, termed the tudor domain. Tudor domains are best characterized by their methyllysine and methylarginine binding abilities3,4,5. Biochemically, Tud interacts with Aubergine (Aub), a Piwi family protein, in a manner dependent on symmetrically dimethylated arginine (sDMA) residues located at the N-terminal end of Aub6,7,8. The sDMA-dependent interaction between Tud and Aub is part of a broad range of phenomena involving tudor domain and Piwi family proteins, in species ranging from fruit flies to mammals3. We and others have shown previously that domains 7-11 of Tud (Tud7-11) were necessary and sufficient for germ cell formation and interaction with arginine-methylated Aub9,10,11. The structure of Tud11 in complex with sDMA-Aub peptides, together with the structure of human SND1 (TDRD11) bound to sDMA peptides from PIWIL1, uncovered the composition of the sDMA-binding pocket, which consists of a cage of four aromatic residues and an asparagine10,12. Interestingly, the sDMA-binding tudor domains are rigidly embedded in a structural module termed the extended Tudor domain (eTud), which comprises an additional juxtaposed OB-fold domain13,14. It is likely that all tudor domains in Tud7-11 have an eTud fold (Supplementary information, Figure S1A)10. However, apart from Tud11, the structures and sDMA binding properties of tudor domains are unknown. More importantly, a mechanistic understanding of Tud's molecular functions in germ plasm requires the knowledge of the spatial organization of the tandem tudor domains as a whole.
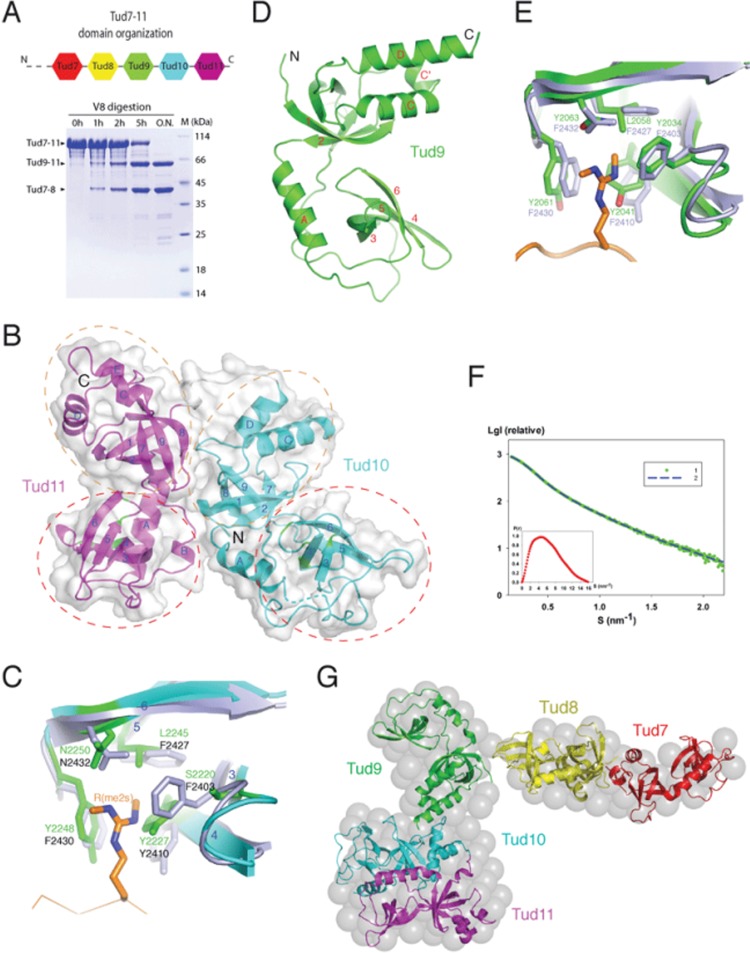
To learn the overall structure of Tud7-11 and the features of individual structural units, we expressed Tud7-11 (aa 1 617-2 515) in E. coli and probed its domain organization by limited proteolysis. Digestion of Tud7-11 with V8 (Glu-C) protease produced two smaller, stable bands corresponding to Tud9-11 and Tud7-8, as determined by N-terminal sequencing (Figure 1A). However, crystallization attempts of recombinant Tud7-11 and Tud9-11 were unsuccessful despite many efforts. We did crystallize a recombinant Tud10-11 fragment (aa 2 163-2 515) and solved a 3.0 Å structure (see Supplementary information, Table S1). The structure shows that, like Tud11, Tud10 is also embedded in a Tudor-SN/p300-like OB-fold scaffold, forming an eTud unit (Figure 1B). The two eTud modules have a similar overall fold, with a root-mean-squared (r.m.s.) deviation of 1.81 Å. Comparison of the present Tud11 structure with the previously determined peptide-bound Tud11 structure (Tud11-pep) gives rise to an r.m.s. deviation of 1.29 Å (Supplementary information, Figure S1B). The aromatic cage involved in binding sDMA in Tud11 is not conserved in Tud10: Ser2220 and Leu2245 in Tud10 take the place of Phe2403 and Phe2427 in Tud11, respectively (Figure 1C and Supplementary information, Figure S1A). Superposition of the Tud10 and Tud11-pep structures reveals two deleterious effects of Ser2220 for sDMA binding: 1) it lacks cation-π interactions with sDMA; 2) its small size allows Tyr2227 to move closer, making the pseudo sDMA-binding pocket too small to accommodate sDMA (Figure 1C). It appears that Leu2245 is less harmful for sDMA binding, as it can maintain hydrophobic interaction with the methyl group. Consistent with the structural observation, we detected no interaction between recombinant Tud10 and methylated Aub peptides by ITC.
Figure 1.
Structure of Tud7-11. (A) Domain organization of Tud7-11. Top, a schematic diagram showing distribution of eTud modules based on sequence alignment and secondary structure analyses. The domains and their spacing were approximately drawn in scale. Bottom: time course of limited proteolysis with V8 protease generating two stable fragments corresponding to Tud7-8 and Tud9-11. (B) Overall structure of Tud10-11 shown as a ribbon model superimposed with a semi-transparent surface representation. Regions spanning the canonical tudor and OB-fold domains are enclosed in red and brown dashed ovals, respectively. Residues at the putative sDMA-binding pockets are indicated in green. (C) Superposition of the putative sDMA-binding site of Tud10 with that of Tud11 in the Tud11-pep complex. The involved residues are shown in a stick model (light blue for Tud11 and green for Tud10). The Aub peptide bound to Tud11 is shown as a reference, with the sDMA residue shown in a stick model (brown). (D) Overall structure of Tud9. (E) Comparison of the sDMA-binding pockets of Tud9 and Tud11. The Aub peptide bound to Tud11 is shown in brown, with the sDMA residue shown in a stick model. (F) Experimental SAXS curve from Tud7-11 in solution. Green dots represent the experimental SAXS data points. Blue dashed line represents the scattering curve computed from the ab initio envelope. The inset at left bottom corner shows the distance distribution functions p(r). (G) Molecular envelope of Tud7-11. The ab initio low-resolution SAXS-derived envelope is shown in a sphere representation. The crystal structures of Tud9, Tud10-11, and the simulated models of Tud7 and Tud8, all shown in a ribbon representation, were docked into the envelope as rigid bodies.
The Tud10-11 structure revealed for the first time the packing mode of methylarginine-binding tandem eTud domains. In the structure, the two eTud modules are rigidly juxtaposed in a head-to-head manner (Figure 1B). The packing buries a total surface area of 1 007 Å2, which indicates a moderate binding affinity between the two eTud modules. A majority of the sidechain contacts are between residues located in the OB-fold domains, and they are mostly polar interactions. In particular, Glu2305 and Tyr2326 each forms a hydrogen bond with Arg2454, and Arg2323 forms two hydrogen bonds with Asp2369 (Supplementary information, Figure S1C). An E2305R/R2323E/Y2326A triple mutation results in an altered spatial organization between Tud10 and Tud11, as evidenced by the change of sedimentation coefficients in analytic ultracentrifugation experiments (Supplementary information, Figure S1D). These features are consistent with the properties of two independently folded, moderately associated eTud modules that can be separately expressed and purified, even crystallized in the case of Tud1110.
We also crystallized and solved a 2.5 Å structure of Tud9 (aa 1 978-2 160), which shows that it also forms an eTud unit (Figure 1D and Supplementary information, Table S1). Superposition of Tud9 with Tud11 of the Tud11-pep complex yielded an r.m.s. deviation of 1.9 Å. Major differences between Tud9 and Tud11 include: 1) the conformation of the loop connecting αA and β3 in the tudor domain; 2) the presence of an additional short helix, αC', in the OB-fold domain of Tud9; and 3) an extended αD in Tud9 (Supplementary information, Figure S1E). Despite these differences, the putative sDMA-binding pocket in Tud9 is relatively well conserved (Figure 1E). One main difference is between Leu2058 of Tud9 and Phe2427 in Tud11. ITC measurements of the binding of the wild-type and the L2058F mutant of Tud9 to sDMA peptides of Aub revealed dissociation constants (Kd) in the sub-millimolar range for both proteins (Supplementary information, Figure S1F). The weak but robust interactions indicate that: 1) the putative sDMA-binding pocket is an authentic one; 2) the insensitivity to the flanking sequences of sDMA peptide suggests that either Tud9 is a promiscuous sDMA binder or that Aub is not its true binding partner; and 3) an aromatic residue at the position occupied by Leu2058 is not necessary for sDMA binding — a hydrophobic residue is most likely sufficient. Both Tud7 and Tud8 have intact sets of sDMA-binding residues, and ITC analyses indicate that Tud7-8 can simultaneously bind two ariginine-methylated Aub peptides with a Kd value of 10 μM (Supplementary information, Figure S1G). In comparison, an isolated Tud8 binds Aub peptides with Kd values ranging from 14 to 32 μM, depending on the location of the sDMA residue within the Aub peptide (Supplementary information, Figure S1G).
To obtain an overall understanding of the domain organization in Tud7-11, we performed solution small angle X-ray scattering (SAXS) experiments to determine the protein envelope. Ab initio modeling using the SAXS data yielded an L-shaped envelope of Tud7-11 (Figure 1F and 1G). Three lobes of the envelope are clearly visible, one elongated lobe connected via a thin neck to a bilobal, calabash-shaped body. Based on the proteolysis result and the structural information, the globular, calabash-shaped body can be unambiguously assigned to Tud9-11, with Tud9 being the smaller lobe, connected to the larger lobe of Tud10-11 (Figure 1B, 1D and 1G). The size of the elongated lobe is capable of accommodating two eTud modules. Crystallization of Tud7-8 or Tud7 and Tud8 separately was not successful despite many efforts. We then generated the structures of Tud7 and Tud8 by homology modeling using the Tud9 structure as the template. The crystal structures of Tud9, Tud10-11, and the modeled Tud7 and Tud8 structures were then placed into the SAXS-generated envelope using a combined approach of computational docking and manual adjustment (Figure 1G and Supplementary information, Figure S1H). The low-resolution, approximate model of Tud7-11 conforms to the molecular envelopes well, and it can account for known biochemical properties of Tud7-11. The Tud7-11 model structure shows that all sDMA-binding sites are well spaced and accessible, with the sDMA-binding sites of Tud7, Tud9 and Tud11 located on one side of the protein surface (Supplementary information, Figure S1H).
The domain organization of Tud7-11 provided first evidence that certain eTud modules may pack into a compact structure. This finding is biologically significant, as the packing creates asymmetry among individual Tud units that may affect their ligand binding properties, as exampled in methyllysine-binding tandem tudor domains. A different domain organization was observed for the four-tudor domain fragment of murine TDRD1, which showed a flexible, extended conformation from a SAXS study15. The extended vs the more compact conformation of the tandem tudor domains would place the binding partners of individual tudor units at different geometric arrangements, constraining the interactions of binding partners with the tudor domain proteins, as well as those between themselves and with downstream targets, hence determining the biological properties of the tudor domain proteins. In Tud7-11, the bulky Tud9-11 is connected to the elongated Tud7-8 via a thin hinge, suggesting that the two structural ensembles have a flexible relative positioning, in agreement with the domain mapping result by proteolysis. The model also indicates that all sDMA-binding sites are accessible, suggesting that Tud7-11 can bind multiple arginine-methylated proteins simultaneously. Our previous study showed that mutations compromising aromatic cages in each of the five tudor domains, except Tud8, resulted in defective germ cell formation10. While it can be argued that ligand binding by Tud8 is not essential for germ cell formation, it is surprising that mutation in Tud10 has a detrimental effect, as its tudor domain does not have a complete set of aromatic residues needed for binding sDMA. In fact, localization of the Tud10 mutant to the germ plasm was unaffected, pointing to the requirement of a separate set of intermolecular interactions involving unmethylated binding partners for its function in germ cell formation. Coincidently, a highly conserved residue, Arg2228, required for stabilizing the putative ligand-binding site in Tud10, is also needed for Tud function in germ cell formation9.
In summary, the overall spatial organization of the tandem eTud modules of Tud7-11, together with the knowledge of structures and sDMA binding properties of individual eTud units, provided new mechanistic insights into ligand binding properties of Tud in germ cell formation, and the information should be useful to guide further efforts in identifying new interaction partners and aid the understanding of the assembly of polar granules.
Acknowledgments
We would like to thank Yuanyuan Chen for assistance with ITC experiments, Xiaoxia Yu, Ting Yao and Ping Chen for help with analytic ultracentrifugation, beamline scientists at BSRF, DESY and SSRF for help with data collection, Ying Huang at Shanghai Institute of Biochemistry and Cell Biology for participation at early stage of the work. The work is supported by the “Key New Drug Creation and Manufacturing Program” of China (2014ZX09507-002), and grants from the National Natural Science Foundation of China (31370734, 91219307 and 31210103914), and the Strategic Priority Research Program (XDB08010100) and the Key Research Program (KJZD-EW-L05) of Chinese Academy of Sciences. RL is an HHMI investigator.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Structural, biochemical and biophysical characterizations of Tud7-11.
References
- Boswell RE, Mahowald AP. Cell. 1985. pp. 97–104. [DOI] [PubMed]
- Ephrussi A, Lehmann R. Nature. 1992. pp. 387–392. [DOI] [PubMed]
- Pek JW, Anand A, Kai T. Development. 2012. pp. 2255–2266. [DOI] [PubMed]
- Lu R, Wang GG. Trends Biochem Sci. 2013. pp. 546–555. [DOI] [PMC free article] [PubMed]
- Thomson T, Lasko P. Cell Res. 2005. pp. 281–291. [DOI] [PubMed]
- Kirino Y, Kim N, de Planell-Saguer M, et al. Nat Cell Biol. 2009. pp. 652–658. [DOI] [PMC free article] [PubMed]
- Nishida KM, Okada TN, Kawamura T, et al. EMBO J. 2009. pp. 3820–3831. [DOI] [PMC free article] [PubMed]
- Kirino Y, Vourekas A, Sayed N, et al. RNA. 2010. pp. 70–78. [DOI] [PMC free article] [PubMed]
- Arkov AL, Wang JY, Ramos A, et al. Development. 2006. pp. 4053–4062. [DOI] [PubMed]
- Liu H, Wang JY, Huang Y, et al. Genes Dev. 2010. pp. 1876–1881. [DOI] [PMC free article] [PubMed]
- Anne J. PLoS One. 2010. p. e14378. [DOI] [PMC free article] [PubMed]
- Liu K, Chen C, Guo Y, et al. Proc Natl Acad Sci USA. 2010. pp. 18398–18403. [DOI] [PMC free article] [PubMed]
- Shaw N, Zhao M, Cheng C, et al. Nat Struct Mol Biol. 2007. pp. 779–784. [DOI] [PubMed]
- Friberg A, Corsini L, Mourao A, et al. J Mol Biol. 2009. pp. 921–934. [DOI] [PubMed]
- Mathioudakis N, Palencia A, Kadlec J, et al. RNA. 2012. pp. 2056–2072. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural, biochemical and biophysical characterizations of Tud7-11.