Abstract
Objective: The wound healing response may be viewed as partially overlapping sets of two physiological processes, regeneration and wound repair with the former overrepresented in some lower species such as newts and the latter more typical of mammals. A robust and quantitative model of regenerative healing has been described in Murphy Roths Large (MRL) mice in which through-and-through ear hole wounds in the ear pinna leads to scarless healing and replacement of all tissue through blastema formation and including cartilage. Since these mice are naturally autoimmune and display many aspects of an enhanced inflammatory response, we chose to examine the inflammatory status during regenerative ear hole closure and observed that inflammation has a clear positive effect on regenerative healing.
Approach: The inflammatory gene expression patterns (Illumina microarrays) of early healing ear tissue from regenerative MRL and nonregenerative C57BL/6 (B6) strains are presented along with a survey of innate inflammatory cells found in this tissue type pre and postinjury. The role of inflammation on healing is tested using a COX-2 inhibitor.
Innovation and Conclusion: We conclude that (1) enhanced inflammation is consistent with, and probably necessary, for a full regenerative response and (2) the inflammatory gene expression and cell distribution patterns suggest a novel mast cell population with markers found in both immature and mature mast cells that may be a key component of regeneration.

Ellen Heber-Katz, PhD
Scope and Significance
The Murphy Roths Large (MRL) mouse strain is a unique “mammalian laboratory” in which to study the mechanistic details of spontaneous regeneration at all scales—from genes to whole tissues—and to compare this process to wound repair, the typical default healing response of mammals. In this review, we show that the innate immune system, metabolic differences, and tissue remodeling genes are differentially expressed between the MRL and the C57BL/6 strain, a well-characterized model of wound repair. We show differences in inflammatory cell types in the two strains pre and postinjury and identify a novel type of mast cell that may play a role in regeneration.
Translational Relevance
Although humans typically heal wounds via wound repair, the unexpected existence of a retained capacity for regeneration in mice offers the possibility that this capacity may be elicited in patients by the controlled modulation of inflammation.
Clinical Relevance
The observation that the approved Cox-2 inflammatory inhibitor meloxicam profoundly inhibits regeneration in a mouse wound model and should be considered for future clinical study of the effect of NSAIDs on wound healing. This may be especially relevant in the case of chronic wounds and postsurgical intervention in general.
Introduction
The MRL mouse originally generated to transfer the “cn” gene mutation for achondroplagia from the AK virus–susceptible, from Rockefeller mouse strain with a high leukemic background, to mice with a low background1 has been a “laboratory” for mammalian regeneration studies for the past 15 years in our hands and in other laboratories.2–5 A key feature that has allowed rapid progress along many parallel paths in mammalian regeneration has been the ear hole closure model that was first identified in rabbits.6–10 Standardized reproducible ear holes in mice are easily created, ear hole closure measurements are easily quantifiable, and ear punctures are considered to be relatively noninvasive as they have been a traditional life-long identification marker pioneered at the Jackson Laboratories.
This mouse ear hole model of regeneration also lends itself well to large genetic studies, and quantitative genetic trait loci have been mapped by multiple laboratories using both the MRL/MpJ mouse and its parental regenerative strain LG/J.11–19 Although the unusual healing property of the MRL mouse has been confirmed in many different organ systems including the heart,20–22 digit,23,24 articular cartilage and joint,25–27 cornea,28 muscle,29,30 skin grafts,31 and central nervous system and peripheral nervous system,32–37 the ear being an external appendage is easily followed in longitudinal studies of regenerative ear hole closure. Since the MRL mouse is especially prone to autoimmunity (characterized by chronic inflammation, it has for decades also been a workhorse model of systemic lupus erythematosis (SLE).38,39 We previously proposed that these same chronic inflammatory SLE properties were perhaps key to the regenerative response of MRL as well.40,41
In higher organisms, and mammals in particular, wound healing occurs via the wound repair process as opposed to the tissue/organ regeneration seen, for example, in newts and axolotls. This process involves a short and potent inflammatory response to infectious organisms and tissue trauma that lasts only a few days and engages both circulating cells of the innate immune system such as platelets, neutrophils, eosinophils, basophils, and monocytes in addition to stationary mast cells and plasma proteins of the blood. The cellular activation mechanisms of healing include the release of inflammatory factors that enhance cell migration through blood vessels via adhesion toward the affected site, and the resolution phase mediated by TGFb, apoptosis, extracellular matrix components, and specific inhibitory molecules such as resolvins. Hence, inflammation is a requirement for effective wound repair.42–44 However, the early inflammatory episode in healing can also take on a morbid character if excessive and prolonged, or chronic. This delicate balance of inflammation and healing outcomes is exemplified by the observation that inflammation converts the scarless healing response seen in embryos to scar formation in adults.45,46 The importance of the inflammatory response is further supported by the macrophage PU.1 knockout mouse that also displays enhanced wound healing,47 as do many other inflammatory gene knockout mice.42,48
On the other hand, there are also reports that inflammation has a totally different effect on the process of regeneration.49 It has been shown that inflammation enhances both neural protection and axonal regeneration.50,51 Pro-inflammatory mediators such as leukotriene B4 and lipoxin A4 have been shown to both positively and negatively regulate stem cell proliferation and differentiation52 and IL-4 has been found to enhance liver regeneration.53 The innate immune response (inflammatory cells) appears to also be an important factor in MRL mouse regeneration41 that brings cells to the injury site resulting in enhanced remodeling, including neutrophils and monocytes, which produce increased levels of matrix metalloproteinases.40 In the MRL mouse, prolonged inflammation is very apparent during blastema formation at the site of the ear hole injury, while in control mice, the inflammation is much less.
Further support comes from novel lines of mice that show that MRL ear hole closure is positively associated with inflammation. AIRmax and AIRmin mice were selectively bred for maximum and minimum acute inflammation, respectively.54 These two mouse lines display different neutrophil and macrophage migration phenotypes with AIRmax having enhanced ear hole closure. A genetic analysis showed a significant linkage of ear hole closure, neutrophil infiltration, and the AIRmax allele of the gene Slc11a1 or Nramp, a protein involved in endosomal ion transport in macrophages and neutrophils,55 reviewed in this issue.56
Thus, the contrarian view of a positive role of inflammation in regeneration, as opposed to wound healing, is well supported. In this article, we will discuss the cells, genes, and pathways that are differentially expressed in MRL and B6 ear hole wound tissue both at steady state and during ear hole closure to determine the possible functioning of inflammatory genes in regenerative as opposed to nonregenerative wound healing. Using meloxicam, an inhibitor of inflammation, which blocks the pro-inflammatory molecule COX-2, we show that this treatment blocks regenerative healing in the MRL mouse. We further suggest that the MRL has a unique mast cell, as noted previously57 and discussed in this issue,58 which may also be a major player in the MRL inflammatory and regenerative response. On another level, inflammation is also known to be dependent on glycolytic metabolism,59–61 and in fact we find that the basal metabolic state of the MRL mouse is indeed characterized by extensive aerobic glycolysis, reminiscent of the Warburg effect.62 In this review we will also explore genes involved in the glycolytic response, which could directly contribute to inflammation.
Discussion
Gene expression in MRL versus B6 mice
To understand the baseline molecular events associated with ear hole closure, we compared whole genome gene expression profiles between MRL and B6 mice at time 0 of the ear hole punch. Out of 18,761 expressed genes, 158 genes were significantly and differentially expressed at least two-fold between the two strains: 28 genes were upregulated in MRL/B6 and 130 genes were downregulated. An analysis of functions and processes overrepresented in the list of the 158 genes described above showed 13 enriched categories (Fig. 1), the majority [10 different gene ontology (GO) categories] were related to innate immune responses, metabolic differences, or tissue remodeling.
Figure. 1.
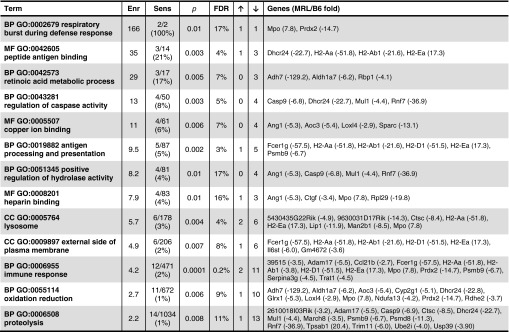
Gene ontology categories enriched among genes differentially expressed between Murphy Roths Large (MRL) and B6. RNA samples were prepared from ear holes from MRL/MpJ (Jackson Laboratories) and C57BL/6 (Taconic Laboratories) female mice in duplicate with each sample being from two ears from one mouse. RNA was extracted using TRIzol. RNA quality was assessed by gel electrophoresis and nanodrop analysis and then amplified according to Illumina. Raw intensities and detection p-values were extracted using Illumina Bead Studio v3.0. Arrays were then quantile normalized and probes were removed from further analysis if their intensity was low (detection p-value >0.05) relative to background in all samples. Differentially expressed genes were determined using SAM algorithm63 with significance level set to FDR <30% and fold change threshold of 2.0, calculated as a ratio between averages of two groups of replicates. Enrichment of gene ontology annotation terms were done using DAVID software64 and only enrichment results of more than two-fold and FDR <20% were reported. Enr, enrichment (fold over randomly expected); Sens, sensitivity K/N where K=number of genes in analyzed list and N=total number of genes within the enriched category; p, p-value of the enrichment; ↑/↓, number of genes with higher/lower expression in MRL; BP, biological process; MF, molecular function; CC, cellular component.
In terms of the inflammatory process, those categories and their definitions that are involved in the activation and recruitment phases include (1) heparin binding, which contains glycosaminoglycans mainly found as intracellular components of mast cells; (2) the immune response defined as a response to potential internal or invasive threat; (3) lysosomes, the cytoplasmic organelles containing a variety of hydrolases responsible for elimination of pathogens and the regulation of inflammation; (4) proteolysis or the ATP-dependent hydrolysis of proteins involved in defense responses; (5) external side of plasma membrane, which includes receptors for cell activation; and (6) respiratory burst during defense, defined as a phase of elevated metabolic activity during which oxygen consumption increases as part of a defense response leading to NADH-dependent production of hydrogen peroxide (H2O2), superoxide anions, and hydroxyl radicals.
The categories that may be involved in the resolution phase of the inflammatory response include (1) regulation of caspase activity using cysteine-type endopepetidase activity for apoptotic processes; (2) Cu ion-binding proteins that have anti-inflammatory effects; (3) retinoic acid metabolism being anti-inflammatory; and (4) oxidation reduction that could oppose or reverse glycolytic processes.
Inflammatory genes
With the analysis of the 158 differentially expressed between MRL and B6 genes on day 0, we found a number of genes that were significantly different (at least twofold with p<0.05) and involved in inflammation. We will discuss how these molecules might affect wound healing and could provide worthwhile candidates for further study. Two of those molecules were found to be upregulated in MRL tissue: Tpsab1 and Mpo. The rest were found to be downregulated in MRL including Fcer1g, Adam17, Aoc3, Ctsc, Ctgf, Ang1, Serpina3g, and Loxl4.
The first upregulated molecule, Tpsab1, encodes the mast cell protease 7 (MCP7)/tryptase alpha, which is 20-fold upregulated in the MRL mouse and is a potent inflammatory mediator. It is a tryptase that can regulate clot formation, fibrinogen/integrin interactions, and is pro-angiogenic.65,66 This has also been shown to be important in wound repair.67 The second upregulated gene, Mpo (7.8-fold up), myeloperoxidase, is a well-known marker of inflammation and mainly expressed by neutrophils and macrophages, which produces hypochlorous acid from H2O2 and chloride and is toxic to pathogens and tissue.68–72 Here, we show that MPO is upregulated in normal MRL tissue compared with B6 and is found in mast cells (Fig. 2). After wounding, it is upregulated in MRL neutrophils (data not shown). Thus, with increased mast cell numbers as well as increased MCP7 and MPO levels in the MRL mouse, this suggests a highly active MRL mast cell population.
Figure. 2.
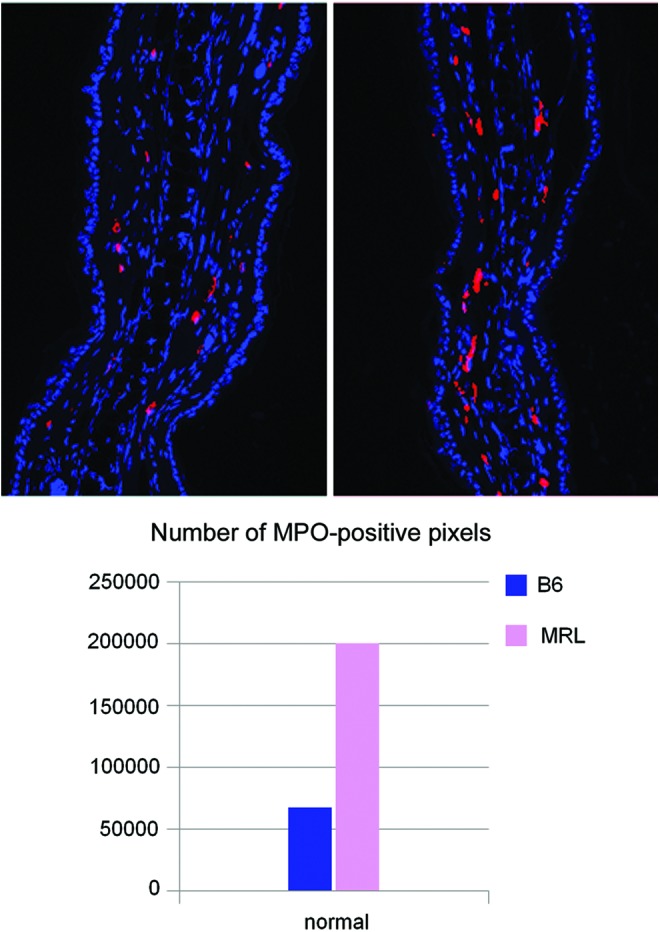
Normal ear tissue from B6 (Taconic; left) and MRL/MpJ (JAX; right) 8-week-old female mice were stained with anti-MPO antibody (red) and DAPI (blue). The staining pattern was consistent with toluidene blue staining for mast cells (data not shown). The level of staining was quantified and shown in the lower panel, indicating that MRL showed approximately threefold more staining than B6. Image analysis was carried out using an Olympus AX70 fluorescent microscope and SPOT camera with bounded software. The specific signal was measured by the ImageJ v1.46f software, provided by the NIH. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Several genes with antioxidant activity, ald1a7 (−6.2-fold), glrx1 or glutaredoxin (−5.3-fold), and prdx2 or peroxiredoxin (−14.7-fold) are all reduced in the MRL.73–75 Since these molecules reduce H2O2, this gene expression pattern could further potentiate MPO.
However, the mast cell receptor Fcer1g, which encodes the high affinity IgE receptor g chain, a key element in inflammation though involved with many different cell types, is 57-fold downregulated in the MRL. It has been reported that the Fcer1g-deficient mouse showed reduced infarct size76 and may be similarly advantageous to the healing response in MRL.
The downregulation of other inflammatory factors found in the MRL mouse include Adam17 or Tace (−5.5-fold), a key enzyme in notch signaling and energy homeostasis. It has been reported that Adam17-deficient mice are hyper-metabolic and lean,77 while Adam17 inhibition reduces tumor invasiveness under hypoxia.78 Low Adam17 level may be related to decreased L-selectin shedding,79 though L-selectin is also necessary for neutrophil accumulation.
Another molecule involved with cell adhesion and migration into inflammatory sites is Aoc3 (amine oxidase or vascular adhesion protein 1),80 which is 5.4-fold down in the MRL. This is a copper-containing molecule found on adipocyte and endothelial cell membranes and has been reported to be responsible for leukocyte extravasation.81 Likewise, Ctgf, or connective tissue growth factor, is associated with cell migration, adhesion, wound healing, and fibrosis. It is 3.4-fold down in the MRL.82,83 Also, Loxl4, or lysyl oxidase-like 4, which is involved in collagen cross-linking and potentiating cell migration from bone marrow-derived populations is 2.88-fold down and may indicate reduced recruitment of cells from circulation and scar formation.84 This suggests that the MRL has mechanisms that mitigate inflammatory cell accumulation.
On the other hand, Ang1 (−5.3-fold) or angiopoietin1 is important in angiogenesis and vascular development and potenitally reduces vascular leakage and maintains lymphatic integrity during inflammation.85 This would have an opposite effect of the above two molecules with increased vessel leakiness in the MRL.
Ctsc (cathepsin C) is a dipeptidyl peptidase I lysosomal peptidase, which is central to the activation of serine proteases in inflammatory cells86 and is 8.4-fold down in the MRL. The cathepsin inhibitor Serpina3g is a serine protease inhibitor found upregulated in activated macrophages stimulated with INFg and LPS and is found in the nucleolus where it can interact with individual cathepsins.87 It is regulated by HIF1a and hypoxia88 and could explain why it is 4.5-fold downregulated in MRL ear (see “Hypoxia-Related Genesis MRL Regeneration”). However, it has also been reported to be upregulated in the MRL/lpr mouse spleen by autoimmune antibodies through an Fc-independent mechanism.89
Finally the molecule Adh7 or alchohol dehydrogenase 7 is involved in the production of retinoic acid and is dramatically reduced (−129-fold) in the MRL. Adh7 levels have been shown to be inversely associated with the degree of inflammation in human gastric mucosa90 and the molecule BCMO1 involved in vitamin A synthesis when knocked out also leads to increased inflammation.91 Taken together, this suggests that an increased inflammatory response may be caused by decreased retinoic acid production.91
Hypoxia-related genes in MRL regeneration
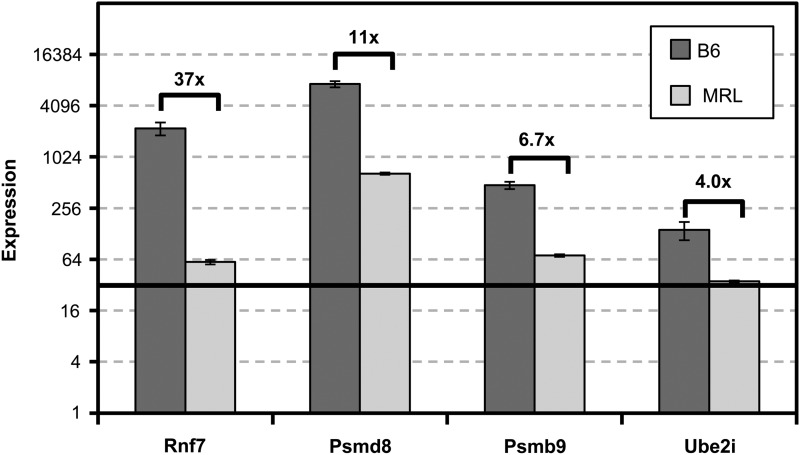
HIF1a is a major potentiator of the metabolic aerobic gylcolytic state, de-differentiation, cell migration, and specifically and importantly for this discussion, a key promotor of inflammation in response to hypoxia.59,60,92,93 One mechanism for the regulation of HIF1a, a protein that is constitutively made, is its breakdown through ubiquination and degradation processes.94 Though no differences in MRL/B6 Hif1a mRNA are seen, there are three differentially expressed genes involved in degradation of HIF1α protein: Rnf7, Psmd8, and Psmb9. These molecules are all significantly down-regulated in MRL tissue which might lead us to predict that HIF1a protein is not being degraded and is both elevated and stabilized in MRL. Most dramatic is the gene Rnf7, also known as Sag/Roc2/Rbx2, which is 37-fold lower in MRL and regulates HIF7 levels (Fig. 3). RNF7 targets HIF1α through the ubiquination/degradation pathway and has been previously described.95 RNF7 functions as a reactive oxygen species scavenger when acting alone, or with other E3 ubiquitin ligase components, promoting ubiquitination and degradation of a number of protein substrates.96 Suppressing Rnf7 mRNA using siRNA specifically stabilizes HIF1α levels.95 Furthermore, the importance of this gene comes from our recent studies mapping ear-hole closure genes16,18,19 in which we identified Rnf7 as a candidate healer gene at a locus on Ch 9.
Figure. 3.
Relative expression levels of genes related to HIF1a. Experimental details of microarray transcript studies are described in Figure 1.
The proteasomal protein Psmd8, which encodes a non-ATPase 19S subunit and Psmb9, which encodes a 20S core beta subunit are both downregulated (Fig. 3) and could prevent or reduce HIF1a degradation by proteasomes. Interestingly, proteasome inhibitors were reported to stimulate bone formation and hair growth.97
Another molecule, Ube2i or Ubc9, is also downregulated in the MRL (4.0-fold) and could influence HIF1α stability. Ube2i acts as a SUMO conjugase and being expressed in reduced levels could decrease HIF1α SUMOylation. The role of SUMOylation in HIF1α stability, however, is controversial98 being reported to target HIF1α for pVHL-dependent degradation or have an opposite effect by increasing its stability.99
In support of HIF1a's role in MRL regeneration, HIF1a target genes are in fact upregulated in MRL tissue. From the array results, we observed increases in Ldha (10%), Vegfc (74%), Hmox1 (12%), Met (7%), Nt5e (19%), Pkm2 (13%), and Gadph (20%). Examination of endothelial cell gene expression showed MRL/B6 ratios for Vegfc at 15×, hemoxygenase (Hmox1) at 2.9×, Met at 3.5×, and Nt5e at 5.8× and examination of neutrophil populations showed MRL/B6 ratios for Ldha at 7.3×, Pkm2 at 8.2×, Gadph at 6.2×, and Hmox1 at 32.6× (data not shown). Furthermore, HIF1a protein levels are indeed upregulated in normal MRL mice and expression levels increase during injury compared with nonregenerator mice suggesting an important role in the regenerative response (Zhang et al., submitted).
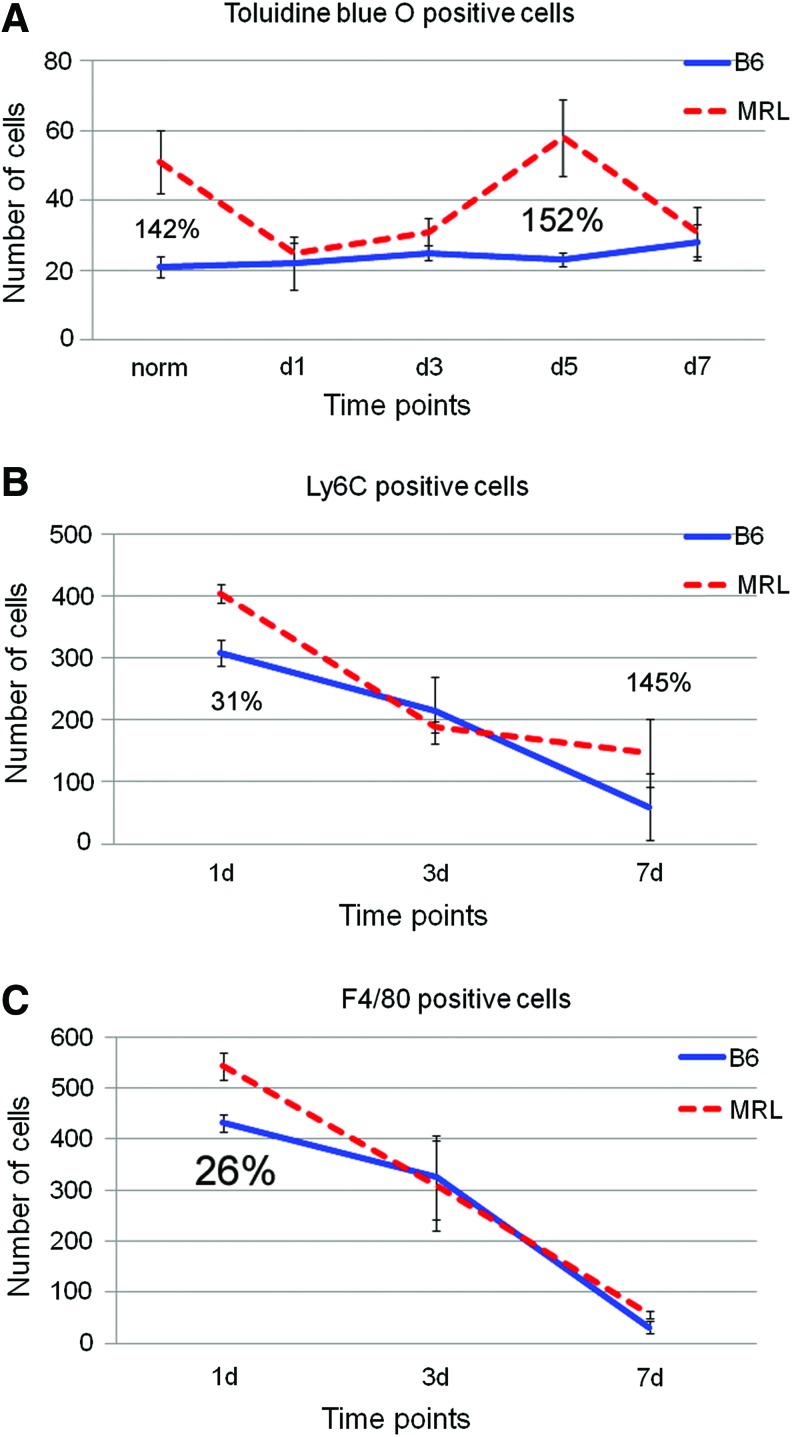
Increased inflammatory cell populations in the MRL regenerative versus C57BL/6 (B6) nonregenerative mouse ears during regenerative healing
As a preliminary study, we examined differences in the numbers of innate immune cells participating in the inflammatory response between MRL and B6 ear punch tissue. Early time points in MRL pre and post ear hole injury reveal differences in three inflammatory cell components during the early phases of the regenerative response (Fig. 4A–C). Mast cells stained with toluidine blue O are elevated in normal MRL ear tissue compared with B6. The number of cells drops after injury and then increases to uninjured levels by day 5. Mast cells are found in the dermis around blood vessels and act as a sentinel for injury. Upon injury, mast cell histamine release causes capillary dilation and leakiness, edema, and inflammatory cell infiltrates. Two of these infiltrate populations, neutrophils and macrophages, are significantly increased in MRL over B6 day 1 postinjury, during the activation phase. Along with the gene expression data, our analysis supports a pro-inflammatory regenerative environment with an increased number of pro-inflammatory cells.
Figure. 4.
Mouse ear tissue from female MRL/MpJ (JAX; red) and C57BL/6 mice (Taconic Labs; blue) show more mast cells (determined by toluidine blue O staining), sentinels of inflammation, in the MRL ear (A) on day 0, preinjury. The number of neutrophils (determined by anti-Ly-6C immunostaining; B) and macrophages (determined by anti-F4/80 immunostaining; C) are increased in the MRL ear, 1 day postinjury. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Inhibition of healing by an anti-inflammatory COX-2 inhibitor
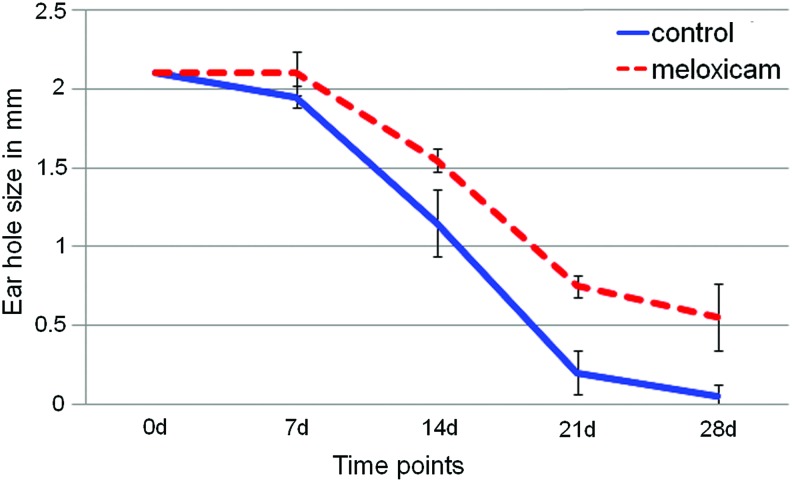
To more directly test whether the inhibition of inflammation would affect regenerative ear hole closure, we used meloxicam, a COX-2 inhibitor. Meloxicam is a nonsteroidal anti-inflammatory drug, which inhibits cyclooxygenase or COX, an enzyme that converts arachidonic acid into prostaglandin H2, a mediator of inflammation.100 Of the two COX species, meloxicam targets COX-2 over COX-1.101,102 Since inflammatory genes are upregulated in the MRL mouse and inflammation so prominently accompanies blastema formation, we proposed that blocking inflammation might also block the regenerative response. As seen in Fig. 5, meloxicam given for just the first 3 days significantly reduces the level of hole closure. It should be noted that a longer treatment period may yield a more complete inhibition given the increased levels of mast cells and neutrophils in the MRL on days 5 and 7 respectively (Fig. 4).
Figure. 5.
MRL/MpJ female mice, 6–8 weeks of age (Jackson Laboratories) were ear punched, intraperitoneally injected with PBS or Meloxicam (40 mg/kg) on days 0–2, and followed for 28 days for ear hole closure. For day 21, p=0.03 and for day 28, p=0.08. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
A unique population of mast cells
From our studies discussed above, we propose that the MRL mouse has a unique population of mast cells that may be important in the regenerative response. Mast cells, as mature cells, are found at the interface with the outside world such as blood vessels, skin, and mucosa, and are in the perfect position to be one of the first populations that will respond to insult or injury. They begin as myeloid progenitors in bone marrow without any specific mast cell receptors or granules, but do express CD34. Once they enter the vascular circulation, they become committed mast cell precursors, or essentially immature mast cells, still lacking most of the granules and specific markers of mature mast cells such as FCER1G and CD14/CDw17 but now do express CD34/ckit and MCP7 or mast cell protease 7 (Tpsab1).103 The final step in their maturation includes migration into different tissues where the cells become fully mature and express markers including Adam17 (TACE), which “processes” ckit, ANG1, CTGF, PKM2, LDHA, VEGF and HMOX1, and FCER1G, which allows the cells to bind IgE and to degranulate in response to antigen+C′, playing a role in anaphylaxis.104,105 Interestingly, all these cell markers are differentially expressed in MRL vs. B6 ear tissue. Since we did not actually isolate mature tissue mast cells, we will assume that the highly specific mast cell markers found by our microarrays on ear tissue are indeed represented on mast cells.
As mentioned above (Fig. 4), one noticeable difference in MRL vs. B6 tissue is the number of Toluidine Blue O-positive cells where MRL has 2.5-fold 150% more stationary mast cells full of granules, strong evidence of their mature state with increased levels of MCP7 and MPO (Fig. 2). Also PKM2, LDHA, VEGF, and HMOX1 (HO1), all HIF1a targets and markers of glycolysis and mature mast cells, are, as we would predict, higher in MRL tissue. HMOX1 in particular leads to less mast cell sensitivity to stimuli and more resistant to degranulation, and this is a property of tumor-associated mast cells.106,107
On the other hand, multiple mature mast cell markers including FCER1G, ADAM17 (TACE), ANG1, and CTGF are reduced in MRL over B6. Another interesting “deviation” in MRL tissue is a lower level of the molecule DAO1 (diamine oxidase),57 which is one of two major enzymes degrading histamine thus regulating its function. Though it is not expressed by mast cells, it is made by other cells and may affect mast cell function. This lower level might affect MRL cells when they degranulate with their histamine staying longer in the injury area.
This MRL gene expression pattern should lead to mast cells with unique properties. These cells may be less likely to degranulate due to antibody but we know that these cells will effectively degranulate due to injury and disappear from the injury site similar to B6 cells. However, MRL injured ear tissue shows a second wave of mast cells at the injury area by day 5, only to have them degranulate again by day 7. This phenomenon is completely absent in the control B6. These cells may be capable of releasing more cytokines and chemokines without degranulating. Also, MRL mast cells are probably more glycolytic and thus potentially more migratory108,109 and have a more stable membrane due to increased HMOX1 levels and have histamine that is longer acting.
These MRL mast cells are more abundant, express both mature and immature cell markers, display membrane stability characteristic of tumor-associated mast cells and produce longer acting histamine. Their unusual phenotype could be one of the major players in longer and more potent inflammation. This could prevent basement membrane formation allowing motile fibroblasts to expand the regenerative blastema and to fully close the ear hole. Thus, the MRL mast cell has characteristics that would place them somewhere between an immature and mature mast cell (Fig. 6). At an estimated density of 7,000 to 20,000 mast cells per cubic millimeter for skin,110 a rough extrapolation would indicate that mast cells comprise about one percent (i.e., one kilogram) of the human total whole body soft tissue mass. Thus, there is a significant physiological and evolutionary commitment to this well-distributed cell type whose primary physiological function is still somewhat obscure. Is it possible that the mast cell is a “regeneratiive remnant” in mammals that can yet be recruited for medicine?
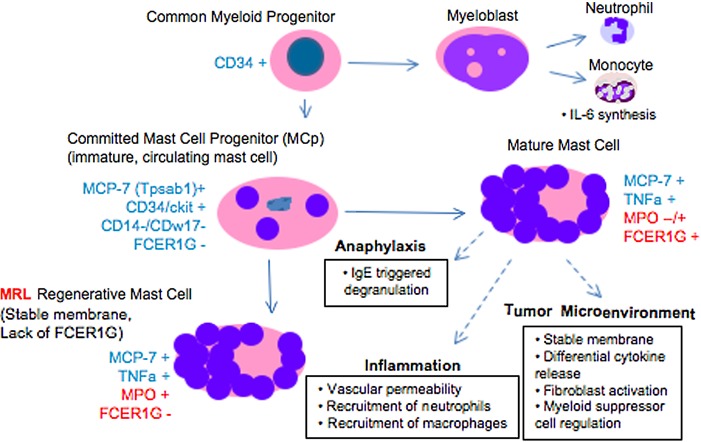
Figure. 6.
The maturation of mast cell populations is shown here. The myeloid progenitor begins as a CD34+ cell that leaves the bone marrow and matures into a myeloblast and then into neutrophils and monocytes or into a circulating mast cell progenitor, which now expresses MCP-7. This cell becomes a mature mast cell that weakly expresses TNFa, FCER1G, and MPO and is involved with degranulation upon exposure to antigen, is involved with the enhancement of inflammation, or can be active in the tumor microenvironment by producing multiple cytokines. In the MRL mouse, increased numbers of mast cells at the regenerative site show increased MPO levels, but the tissue is strikingly low in FCER1G, a marker of mast cell maturity, and high in MCP7. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Conclusion
The MRL mouse starkly illustrates the major biological differences between wound repair and regeneration in mammalian wound healing. This article has focused on the role of inflammation in regeneration as elucidated by MRL mouse gene expression. Globally, the MRL displays enhanced inflammatory gene expression at the basal level and in its response to wounding compared with control nonregenerating strains. Most studies in mammalian wound healing have historically focused on wound repair as this is the dominant wound healing process found in mammals in general and in humans. The role of inflammation except in the most delimited sense has been shown to be pathologic. Examples include chronic wounds, keloids, and autoimmunity following tissue damage or viral insult. Thus, enhanced inflammatory responses that are either mediating or necessary for regeneration is a contrarian view only because of the historical absence of a mammalian regeneration model.
The course of the inflammatory response is known to determine wound repair,43 which is so spectacular in embryos, where no inflammation is observed in the wound repair site.46 The MRL mouse is known to show immunologic abnormalities and several genes related to immune response have been already found to be differentially regulated in the MRL.49 The differential regulation of several genes involved in the immune response in the MRL may support the idea that modulation of the inflammatory response to the injury could explain reduced scarring.49 Finally, the MRL mouse may have inflammatory cell populations, which may make it quite unique.
Take-Home Messages.
• Ear hole closure is a model of mammalian regenerative healing as seen in the MRL mouse.
• An enhanced inflammatory response is required for regenerative healing (see also de Franco and colleagues, this issue).56
• At baseline, the genes differentially expressed between a regenerator and a nonregenerator are enriched for inflammatory responses, metabolic differences, and remodeling.
• As suggested from the genes expressed, the inflammatory response in the regenerator shows more remodeling, reduced scarring, with more stable cell populations that are less cytotoxic. There is increased vascular leakage but less cell migration, more angiogenesis, with an enhanced HIF1a response.
• A novel population of mast cells is proposed for the regenerator mouse, which is more immature than normal, is less sensitive to lysis, and may produce more factors, similar to what is seen for cancer-associated mast cells.
Abbreviations and Acronyms
- ADH7
alchohol dehydrogenase 7
- ANG1
angiopoietin1
- AOC3
amine oxidase
- B6
C57BL/6
- BP
biological process
- CC
cellular component
- COX
cyclooxygenase
- CTSC
cathepsin C
- CTGF
connective tissue growth factor
- DAO1
diamine oxidase
- FCER1G
Fc receptor for IgE (high affinity, gamma chain)
- GLRX1
glutaredoxin
- HIF1a
hypoxia-inducible factor 1a
- H2O2
hydrogen peroxide
- LOXL4
lysyl oxidase-like 4
- MCP7
mast cell protease 7
- MF
molecular function
- MPO
myeloperoxidase
- PRDX2
peroxiredoxin
- RNF7
ring finger protein 7
- SERPINa3g
serine protease inhibitor a3g
- SLE
systemic lupus erythematosis
Acknowledgments and Funding Sources
This research was supported funds from the National Institutes of Health (NIDCR and NIGMS) and the Department of Defense (DARPA).
Author Disclosure and Ghostwriting
The authors declare no competing interests in the studies discussed in this article. No ghostwriters were used to write this article.
About the Authors
The authors are members of the Molecular and Cellular Oncogenesis Group at the Wistar Institute. Andrew Kossenkov, PhD, is Director of the Bioinformatics Facility. Celia Chang is the Director of the Genomics Facility. Louise Showe, PhD, is Associate Director of the Center for Systems and Computational Biology. Dmitri Gourevitch, MD, Yong Zhang, PhD, Lise Clark, PhD, DVM, and Ellen Heber-Katz, PhD, have been studying regeneration biology and immunology for up to the past 15 and 30+ years, respectively.
References
- 1.Murphy ED: Lymphoproliferation (lpr) and other single-locus models for murine lupus. In Immunologic Defects in Laboratory Animals, edited by Gershwin ME. and Merchant B. New York: Plenum Press, 1981, p. 143 [Google Scholar]
- 2.Clark LD, Clark RK, and Heber-Katz E: A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol 1998; 88:35. [DOI] [PubMed] [Google Scholar]
- 3.Kench JA, Russell DM, Fadok VA, et al.: Aberrant wound healing and TGF-beta production in the autoimmune-prone MRL/+mouse. Clin Immunol 1999; 92:300. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Mohan S, Gu W, Miyakoshi N, and Baylink DJ: Differential protein profile in the ear-punched tissue of regeneration and non-regeneration strains of mice: a novel approach to explore the candidate genes for soft-tissue regeneration. Biochim Biophys Acta 2000; 1524:102. [DOI] [PubMed] [Google Scholar]
- 5.Rajnoch C, Ferguson S, Metcalfe AD, Herrick SE, Willis HS, and Ferguson MWJ: Regeneration of the ear after wounding in different mouse strains is dependent on the severity of wound trauma. Dev Dyn 2003; 226:388. [DOI] [PubMed] [Google Scholar]
- 6.Joseph J. and Dyson M: Tissue replacement in the rabbit's ear. Br J Surg 1966; 53:372. [DOI] [PubMed] [Google Scholar]
- 7.Goss RJ. and Grimes LN: Tissue interactions in the regeneration of rabbit ear holes. Am Zool 1975; 12:151 [Google Scholar]
- 8.ten Koppel PGJ, van Osch GJVM, Verwoerd CDA, and Verwoerd-Verhoef HL. A new in vivo model for testing cartilage grafts and biomaterials: the ‘rabbit pinna punch-hole’ model. Biomaterials 2001; 22:1407. [DOI] [PubMed] [Google Scholar]
- 9.Labandeira-Garcia J. and Guerra-Seijas M: Intracellular lipids in rabbit ear cartilage during tissue regeneration. Acta Anat 1986; 127:249. [DOI] [PubMed] [Google Scholar]
- 10.Edwards RG: From embryonic stem cells to blastema and MRL mice. Reprod Biomed Online 2008; 16:425. [DOI] [PubMed] [Google Scholar]
- 11.McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, and Heber-Katz E: Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci USA 1998; 95:11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masinde GL, Li X, Gu W, Davidson H, Mohan S, and Baylink DJ: Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res 2001; 11:2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankenhorn EP, Troutman S, Clark LD, Zhang XM, Chen P, and Heber-Katz E: Sexually dimorphic genes regulate healing and regeneration in MRL mice. Mamm Genome 2003; 14:250. [DOI] [PubMed] [Google Scholar]
- 14.Heber-Katz E, Chen P, Dvm LC, Zhang X-M, Troutman S, and Blankenhorn EP: Regeneration in MRL mice: further genetic loci controlling the ear hole closure trait using MRL and M. m. castaneus mice. Wound Repair Regen 2004; 12:384. [DOI] [PubMed] [Google Scholar]
- 15.Yu H, Mohan S, Masinde G, and Baylink D: Mapping the dominant wound healing and soft tissue regeneration QTL in MRL x CAST. Mamm Genome 2005; 16:918. [DOI] [PubMed] [Google Scholar]
- 16.Blankenhorn E, Bryan G, Kossenkov A, et al.: Genetic loci that regulate healing and regeneration in LG/J and SM/J mice. Mamm Genome 2009; 20:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li XM, Gu WK, Masinde G, et al.: Genetic control of the rate of wound healing in mice. Heredity 2010; 86:668. [DOI] [PubMed] [Google Scholar]
- 18.Cheverud JM, Lawson HA, Funk R, Zhou J, Blankenhorn EP, and Heber-Katz E: 2012. Healing Quantitative Trait Loci in a Combined Cross Analysis using Related Mouse Strain Crosses. Heredity 108: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheverud JM, Lawson HA, Bouckaert K, et al.: 2014. Fine-mapping quantitative trait loci affecting murine external ear tissue regeneration in the LG/J by SM/J advanced intercross line. Heredity [Epub ahead of print]; DOI: 10.1038/hdy.2013.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leferovich JM, Bedelbaeva K, Samulewicz S, et al.: Heart regeneration in adult MRL mice. Proc Natl Acad Sci USA 2001; 98:9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haris Naseem R, Meeson AP, Michael DiMaio J, et al.: Reparative myocardial mechanisms in adult C57BL/6 and MRL mice following injury. Physiol Genomics 2007; 30:44. [DOI] [PubMed] [Google Scholar]
- 22.Alfaro MP, Pagni M, Vincent A, et al.: The Wnt modulator sFRP2 enhances mesenchymal stem cell engraftment, granulation tissue formation and myocardial repair. Proc Natl Acad Sci USA 2008; 105:18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chadwick RB, Bu L, Yu H, et al.: Digit tip regrowth and differential gene expression in MRL/MpJ, DBA/2, and C57BL/6 mice. Wound Repair Regen 2007; 15:275. [DOI] [PubMed] [Google Scholar]
- 24.Gourevitch DL, Clark L, Bedelbaeva K, Leferovich J, and Heber-Katz E: Dynamic changes after murine digit amputation: The MRL mouse digit shows waves of tissue remodeling, growth, and apoptosis. Wound Repair Regen 2009; 17:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fitzgerald J, Rich C, Burkhardt D, Allen J, Herzka AS, and Little CB: Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthritis Cartilage 2008; 16:1319. [DOI] [PubMed] [Google Scholar]
- 26.Rai MF, Hashimoto S, Johnson EE, et al.: Heritability of articular cartilage regeneration and its association with ear-wound healing. Arthritis Rheum 2012; 64:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward BD, Furman BD, Huebner JL, Kraus VB, Guilak F, and Olson SA: Absence of posttraumatic arthritis following intra-articular fracture in the MRL/MpJ mouse. Arthritis Rheum 2008; 58:744. [DOI] [PubMed] [Google Scholar]
- 28.Ueno M, Lyons BL, Burzenski LM, et al.: Accelerated wound healing of alkali-burned corneas in MRL mice Is associated with a reduced inflammatory signature. Invest Ophthalmol Vis Sci 2005; 46:4097. [DOI] [PubMed] [Google Scholar]
- 29.Buhimschi CS, Zhao G, Sora N, Madri JA, and Buhimschi IA: Myometrial wound healing post-Cesarean delivery in the MRL/MpJ mouse model of uterine scarring. Am J Pathol 2010; 177:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heydemann A, Swaggart KA, Kim GH, Holley-Cuthrell J, Hadhazy M, and McNally EM: The superhealing MRL background improves muscular dystrophy. Skelet Muscle 2012; 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolba RH, Schildberg FA, Decker D, et al.: Mechanisms of improved wound healing in Murphy Roths Large (MRL) mice after skin transplantation. Wound Repair Regen 2010; 18:662. [DOI] [PubMed] [Google Scholar]
- 32.Hampton DW, Seitz A, Chen P, Heber-Katz E, and Fawcett JW: Altered CNS response to injury in the MRL/MpJ mouse. Neuroscience 2004; 127:821. [DOI] [PubMed] [Google Scholar]
- 33.Baker KL, Daniels SB, Lennington JB, et al.: Neuroblast protuberances in the subventricular zone of the regenerative MRL/MpJ mouse. J Comp Neurol 2006; 498:747. [DOI] [PubMed] [Google Scholar]
- 34.Balu DT, Hodes GE, Anderson BT, and Lucki I: Enhanced sensitivity of the MRL/MpJ mouse to the neuroplastic and behavioral effects of chronic antidepressant treatments. Neuropsychopharmacology 2009; 34:1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thuret S, Toni N, Aigner S, Yeo GW, and Gage FH: Hippocampus-dependent learning is associated with adult neurogenesis in MRL/MpJ mice. Hippocampus 2009; 19:658. [DOI] [PubMed] [Google Scholar]
- 36.Thuret S, Thallmair M, Horky LL, and Gage FH: Enhanced functional recovery in MRL/MpJ mice after spinal cord dorsal hemisection. PLoS One 2012; 7:e30904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buckley G, Metcalfe AD, and Ferguson MWJ: Peripheral nerve regeneration in the MRL/MpJ ear wound model. J Anat 2011; 218:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen PL. and Eisenberg RA: Lpr and gld: single gene models of systemic autoimmunity and lymphoproliferative disease. Annu Rev Immunol 1991; 9:243. [DOI] [PubMed] [Google Scholar]
- 39.Kono DH. and Theofilopoulos AN: Genes and genetics of murine lupus. In: The Molecular Pathology of Autoimmune Diseases, edited by Theofilopoulos AN. and Bona CA. New York: Taylor and Francis, 2002, p. 353 [Google Scholar]
- 40.Gourevitch D, Clark L, Chen P, Seitz A, Samulewicz SJ, and Heber-Katz E: Matrix metalloproteinase activity correlates with blastema formation in the regenerating MRL mouse ear hole model. Dev Dyn 2003; 226:377. [DOI] [PubMed] [Google Scholar]
- 41.Heber-Katz E. and Gourevitch D: The relationship between inflammation and regeneration in the MRL mouse: potential relevance for putative human regenerative (scarless wound healing) capacities? Ann NY Acad Sci 2009; 1172:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin P. and Leibovich SJ: Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol 2005; 15:599. [DOI] [PubMed] [Google Scholar]
- 43.Eming SA, Krieg T, and Davidson JM: Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol 2007; 127:514. [DOI] [PubMed] [Google Scholar]
- 44.Koh TJ. and Luisa Ann DiPietro LA: Inflammation and wound healing: The role of the macrophage. Expert Rev Mol Med 2013; 13:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson MWJ, Whitby DJ, Shah M, Armstrong J, Siebert JW, and Longaker MT: Scar formation: the spectral nature of fetal and adult wound repair. Plast Reconstr Surg 1996; 97:854. [DOI] [PubMed] [Google Scholar]
- 46.Redd MJ, Cooper L, Wood W, Stramer B, and Martin P: Wound healing and inflammation: embryos reveal the way to perfect repair. Philos Trans R Soc Lond B Biol Sci 2004; 359:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin P, D'Souza D, Martin J, et al.: Wound Healing in the PU.1 Null mouse—tissue repair is not dependent on inflammatory cells. Curr Biol 2003; 13: 1122. [DOI] [PubMed] [Google Scholar]
- 48.Grose R. and Werner S. Wound healing studies in transgenic and knockout mice. Mol Biotechnol 2004; 28: 147. [DOI] [PubMed] [Google Scholar]
- 49.Harty M, Neff AW, King MW, and Mescher AL: Regeneration or scarring: an immunologic perspective. Dev Dyn 2003; 226:268. [DOI] [PubMed] [Google Scholar]
- 50.Yin Y, Henzl MT, Lorber B, et al.: Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat Neurosci 2006; 9:843. [DOI] [PubMed] [Google Scholar]
- 51.Kyritsis N, Kizil C, Zocher S, et al.: Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 2012; 338:1353. [DOI] [PubMed] [Google Scholar]
- 52.Wada K, Arita M, Nakajima A, et al.: Leukotriene B4 and lipoxin A4 are regulatory signals for neural stem cell proliferation and differentiation. FASEB J 2006; 20:1785. [DOI] [PubMed] [Google Scholar]
- 53.Goh YP, Henderson NC, Heredia JE, et al.: Eosinophils secrete IL-4 to facilitate liver regeneration. Proc Natl Acad Sci USA 2013; 110:9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Araujo LMM, Ribeiro OG, Siqueira M, et al.: Innate resistance to infection by intracellular bacterial pathogens differs in mice selected for maximal or minimal acute inflammatory response. Eur J Immunol 1998; 28:2913. [DOI] [PubMed] [Google Scholar]
- 55.De Franco M, Carneiro P, Peters L, et al.: Slc11a1 (Nramp1) alleles interact with acute inflammation loci to modulate wound-healing traits in mice. Mamm Genome 2007; 18:263. [DOI] [PubMed] [Google Scholar]
- 56.Canhamero T, Valino Garcia L, and De Franco M: Acute inflammation loci are involved in wound healing in the mouse ear punch model. Adv Wound Care 2014January31 [Epub ahead of print]; DOI: 10.1089/wound.2013.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furukawa F, Yoshimasu T, Yamamoto Y, Kanazawa N, and Tachibana T: Mast cells and histamine metabolism in skin lesions from MRL/MP-lpr/lpr mice. Autoimmun Rev 2009; 8:495. [DOI] [PubMed] [Google Scholar]
- 58.Morales K, Rowehl L, Smith J, et al.: Mapping novel subcutaneous angiogenesis quantitative trait loci in [B6×MRL]F2 mice. Adv Wound Care 2014April21 [Epub ahead of print]; DOI: 10.1089/wound.2013.0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cramer T, Yamanishi Y, Clausen BE, et al.: HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell 2003; 112:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zarember KA. and Malech HL: HIF-1alpha: a master regulator of innate host defenses? J Clin Invest 2005; 115:1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Everts B, Amiel E, van der Windt GJ, et al.: Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood 2012; 120:1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naviaux RK, Le TP, Bedelbaeva K, et al.: Retained features of embryonic metabolism in the adult MRL mouse. Mol Genet Metab 2009; 96:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S: A comprehensive evaluation of SAM, the SAM R-package and a simple modification to improve its performance. BMC Bioinform 2007; 8:23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang DW, Sherman BT, and Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44. [DOI] [PubMed] [Google Scholar]
- 65.Coussens L, Raymond W, Bergers G, et al.: Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev 1999; 13:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang C, Wong G, Ghildyal N, et al.: The tryptase, mouse mast cell protease 7, exhibits anticoagulant activity in vivo and in vitro due to its ability to degrade fibrinogen in the presence of the diverse array of protease inhibitors in plasma. J Biol Chem 1997; 272:31885. [DOI] [PubMed] [Google Scholar]
- 67.Somasundaram P, Ren G, Nagar H, et al.: Mast cell tryptase may modulate endothelial cell phenotype in healing myocardial infarcts. J Pathol 2005; 205:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sinclair RD. and Ryan TJ: Proteolytic enzymes in wound healing: the role of enzymatic debridement. Australas J Dermatol 1994; 35:35. [DOI] [PubMed] [Google Scholar]
- 69.Zhang R, Brennan ML, Shen Z, et al.: Myeloperoxidase functions as a major enzymatic catalyst for initiation of lipid peroxidation at sites of inflammation. J Biol Chem 2002; 277:46116. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Rosen H, Madtes DK, et al.: Myeloperoxidase inactivates TIMP-1 by oxidizing its N-terminal cysteine residue: an oxidative mechanism for regulating proteolysis during inflammation. J Biol Chem 2007; 282:31826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu X, Kassim SY, Parks WC, and Heinecke JW: Hypochlorous acid generated by myeloperoxidase modifies adjacent tryptophan and glycine residues in the catalytic domain of matrix metalloproteinase-7 (matrilysin): an oxidative mechanism for restraining proteolytic activity during inflammation. J Biol Chem 2003; 278:28403. [DOI] [PubMed] [Google Scholar]
- 72.Yager DR. and Nwomeh BC: The proteolytic environment of chronic wounds. Wound Repair Regen 1999; 7:433. [DOI] [PubMed] [Google Scholar]
- 73.Leonard MO, Kieran NE, Howell K, et al.: Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J 2006; 20:2624. [DOI] [PubMed] [Google Scholar]
- 74.Porras P, Pedrajas JR, Martínez-Galisteo E, et al.: Glutaredoxins catalyze the reduction of glutathione by dihydrolipoamide with high efficiency. Biochem Biophys Res Commun 2002; 295:1046. [DOI] [PubMed] [Google Scholar]
- 75.Shen C. and Nathan C: Nonredundant antioxidant defense by multiple two-cysteine peroxiredoxins in human prostate cancer cells. Mol Med 2002; 8:95. [PMC free article] [PubMed] [Google Scholar]
- 76.Takaya N, Katoh Y, Iwabuchi K, et al.: Platelets activated by collagen through the immunoreceptor tyrosine-based activation motif in the Fc receptor gamma-chain play a pivotal role in the development of myocardial ischemia-reperfusion injury. J Mol Cell Cardiol 2005; 39:856. [DOI] [PubMed] [Google Scholar]
- 77.Gelling RW, Yan W, Al-Noori S, et al.: Deficiency of TNFalpha converting enzyme (TACE/ADAM17) causes a lean, hypermetabolic phenotype in mice. Endocrinology 2008; 149:6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng X, Jiang F, Katakowski M, et al.: Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer Sci 2007; 98:674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hafezi-Moghadam A, Thomas KL, Prorock AJ, Huo Y, and Ley K: L-selectin shedding regulates leukocyte recruitment. J Exp Med 2001; 193:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith DJ, Salmi M, Bono P, Hellman J, Leu T, and Jalkanen S: Cloning of vascular adhesion protein 1 reveals a novel multifunctional adhesion molecule. J Exp Med 1998; 188:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stolen CM, Marttila-Ichihara F, Koskinen K, et al.: Absence of the endothelial oxidase AOC3 leads to abnormal leukocyte traffic in vivo. Immunity 2005; 22:105. [DOI] [PubMed] [Google Scholar]
- 82.Babic AM, Chen CC, and Lau LF: Fisp12/mouse connective tissue growth factor mediates endothelial cell adhesion and migration through integrin alphavbeta3, promotes endothelial cell survival, and induces angiogenesis in vivo. Mol Cell Biol 1999; 19:2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sonnylal S, Shi-Wen X, Leoni P, et al.: Selective expression of connective tissue growth factor in fibroblasts in vivo promotes systemic tissue fibrosis. Arthritis Rheum 2010; 62:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wong CC, Gilkes DM, Zhang H, et al.: Hypoxia-inducible factor 1 is a master regulator of breast cancer metastatic niche formation. Proc Natl Acad Sci USA 2011; 108:16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kajiya K, Kidoya H, Sawane M, et al.: Promotion of lymphatic integrity by angiopoietin-1/Tie2 signaling during inflammation. Am J Pathol 2012; 180:1273. [DOI] [PubMed] [Google Scholar]
- 86.Turk D, Janjić V, Stern I, et al.: Structure of human dipeptidyl peptidase I (cathepsin C): Exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J 2001; 20:6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Konjar S, Yin F, Bogyo M, Turk B, and Kopitar-Jerala N: Increased nucleolar localization of SpiA3G in classically but not alternatively activated macrophages. FEBS Lett 2010; 584:2201. [DOI] [PubMed] [Google Scholar]
- 88.Zhao J, Yan Y, Salnikow K, Kluz T, and Costa M: Nickel-induced down-regulation of serpin by hypoxic signaling. Toxicol Appl Pharmacol 2004; 194:60. [DOI] [PubMed] [Google Scholar]
- 89.Qing X, Zavadil J, Crosby MB, et al.: Nephritogenic anti-DNA antibodies regulate gene expression in MRL/lpr mouse glomerular mesangial cells. Arthritis Rheum 2006; 54:2198. [DOI] [PubMed] [Google Scholar]
- 90.Matsumoto M, Yokoyama H, Suzuki H, Shiraishi-Yokoyama H, and Hibi T: Retinoic acid formation from retinol in the human gastric mucosa: role of class IV alcohol dehydrogenase and its relevance to morphological changes. Am J Physiol Gastrointest Liver Physiol 2005; 289:G429. [DOI] [PubMed] [Google Scholar]
- 91.van Helden YG, Heil SG, van Schooten FJ, et al.: Knockout of the Bcmo1 gene results in an inflammatory response in female lung, which is suppressed by dietary beta-carotene. Cell Mol Life Sci 2010; 67:2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kominsky DJ, Campbell EL, and Colgan SP: Metabolic shifts in immunity and inflammation. J Immunol 2010; 184:4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Semenza GL: Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harbor Symp Quant Biol 2011; 76:347. [DOI] [PubMed] [Google Scholar]
- 94.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al.: HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 2001; 292:464. [DOI] [PubMed] [Google Scholar]
- 95.Tan M, Gu Q, He H, Pamarthy D, Semenza GL, and Sun Y: SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1[alpha] ubiquitination and degradation. Oncogene 2008; 27:1404. [DOI] [PubMed] [Google Scholar]
- 96.Sun Y, Tan M, Duan H, and Swaroop M: SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal 2001; 3:635. [DOI] [PubMed] [Google Scholar]
- 97.Mundy G, Gutierrez G, Garrett R, et al.: Proteasome inhibitors stimulate both bone formation and hair growth by similar mechanisms. Ann NY Acad Sci 2007; 1117:298. [DOI] [PubMed] [Google Scholar]
- 98.Liu B. and Shuai K: Summon SUMO to wrestle with inflammation. Mol Cell 2009; 35:731. [DOI] [PubMed] [Google Scholar]
- 99.Bae SH, Jeong JW, Park JA, et al.: Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun 2004; 324:394. [DOI] [PubMed] [Google Scholar]
- 100.Hawkey CJ: COX-1 and COX-2 inhibitors. Best Pract Res Clin Gastroenterol 2001; 15:801. [DOI] [PubMed] [Google Scholar]
- 101.Hoshino T, Tabuchi K, Hirose Y, et al.: The non-steroidal anti-inflammatory drugs protect mouse cochlea against acoustic injury. Tohoku J Exp Med 2008; 216:53. [DOI] [PubMed] [Google Scholar]
- 102.Dudhgaonkar S, Tandan SK, Kumar D, Arunadevi R, and Prakash V: Synergistic interaction between meloxicam and aminoguanidine in formalin-induced nociception in mice. Eur J Pain 2008; 12:321. [DOI] [PubMed] [Google Scholar]
- 103.Walczak-Drzewiecka A, Salkowska A, Ratajewski M, and Dastych J: Epigenetic regulation of CD34 and HIF1A expression during the differentiation of human mast cells. Immunogenetics 2013; 65:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pennock JL. and Grencis RK: In vivo exit of c-kit+/CD49d(hi)/beta7+mucosal mast cell precursors from the bone marrow following infection with the intestinal nematode Trichinella spiralis. Blood 2004; 103:2655. [DOI] [PubMed] [Google Scholar]
- 105.Kawaguchi M, Mitsuhashi Y, and Kondo S: Overexpression of tumour necrosis factor-alpha-converting enzyme in psoriasis. Br J Dermatol 2005; 152:915. [DOI] [PubMed] [Google Scholar]
- 106.Xia Z-W, Xu L-Q, Zhong W-W, et al.: Heme oxygenase-1 attenuates ovalbumin-induced airway inflammation by up-regulation of Foxp3 T-regulatory cells, interleukin-10, and membrane-bound transforming growth factor-β1. Am J Pathol 2007; 171:1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Theoharis C: Theoharides, Pio Conti the JEKYLL and HYDE of tumor growth. Trends Immunol 2004; 25:235. [DOI] [PubMed] [Google Scholar]
- 108.Nathan C: Immunology: Oxygen and the inflammatory cell. Nature 2003; 422:675. [DOI] [PubMed] [Google Scholar]
- 109.O'Neill LAJ. and Hardie DG: Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 2013; 493:346. [DOI] [PubMed] [Google Scholar]
- 110.Eady RA, Cowen T, Marshall TF, Plummer V, and Greaves MW: Mast cell population density, blood vessel density and histamine content in normal human skin. Br J Dermatol 1979; 100:623. [DOI] [PubMed] [Google Scholar]