Abstract
Objective: MRL/MpJ mice are known for enhanced healing, but mechanistic details or how specific aspects of wounding (e.g., angiogenesis) contribute to healing are unknown. While previous studies investigated the systemic effects of immunity in MRL/MpJ healing, few have focused on tissue-intrinsic effects.
Approach: Ex vivo skin biopsies from MRL/MpJ and C57BL/6J mice were cultured in ex vivo conditions that favor endothelial cell growth to compare their angiogenic potential. We localized enhanced angiogenesis quantitative trait loci (QTL) in an F2 intercross. We then performed an expression analysis in cultured skin biopsies from MRL/MpJ and C57BL/6J mice to determine the pathways that are associated with the capacity for differential growth.
Results: MRL/MpJ biopsies have a two- to threefold greater growth potential than C57BL/6J mice, supporting the hypothesis that angiogenesis may contribute to enhanced healing in MRL/MpJ skin. We mapped two QTLs that are unique from previously mapped MRL/MpJ wound healing QTLs and detected interactions between wound healing QTLs and loci in this cross. Additionally, we found that pathways previously implicated in MRL/MpJ healing are also enriched in skin biopsies.
Innovation: We have developed a novel approach to determine how specific aspects of tissue development contribute to wound healing that will ultimately lead to the discovery of unidentified genes that contribute to enhanced healing.
Conclusion: We have shown that, consistent with previous studies following wound closure in MRL/MpJ mice, vessel growth during healing is also influenced by genetic background. Our ongoing work will identify the genetic factors that should be useful biomarkers or as therapeutic targets for enhanced wound healing.

Bruce J. Herron, PhD
Introduction
Genetic control of wound healing is well known in humans and model systems1,2 and many pathways have been implicated in the healing process.3 Despite this knowledge, nonhealing wounds are a prevalent problem, especially among the elderly and in conditions like diabetes.4 Both genetic polymorphisms and environmental factors contribute to wound healing; however, analyzing these variables in humans is difficult due to confounding comorbidities and genetic heterogeneity within the population. Alternatively, preclinical models that follow wound closure and resolution are known to be strongly influenced by genetic background and these differences can be mapped to multiple quantitative trait loci (QTLs).2,5,6
Early stages of tissue healing include a granular appearance of the tissue stroma that is principally due to the presence of newly formed capillaries.7 Neovessels are critical for the delivery of nutrients to the developing tissue and also provide a way for immune cells to enter and stimulate wound repair. Whereas it has been established that differential angiogenesis between individuals is, in part, mediated by genetic factors, the precise genes that are involved in this process, particularly in the early stages of wound healing are largely unexplored.
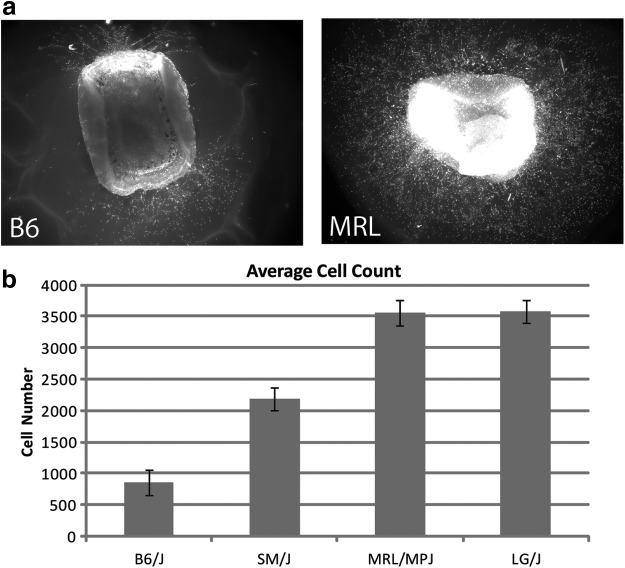
The Murphy Roth's large (MRL/MpJ, abbreviated MRL) inbred mouse strain is well known for its unique abilities to achieve scarless healing in juvenile and adult mice.8 This complex trait is shared with the large (LG/J, abbreviated LG) mouse strain and many QTLs have been associated with healing in these strains; however, none of these QTLs have been identified.2,5 Wound healing can be broken down into temporal phases that include, hemostasis, inflammation, proliferation, and remodeling.8 The genetic factors that influence a wound healing study are dependent on the timing during which data are collected. For example, MRL and LG mice are known to remove and reestablish basement membrane in the resolution phase of healing, whereas other strains do not.9 Alternatively, we and others have illustrated divergent outcomes among inbred mouse strains with respect to vessel growth at early stages of wound healing (Fig. 1b),10,11 suggesting that multiple polymorphisms in mammals contribute to healing at different points in the process.
Figure 1.
Examples of differential vessel growth between B6 and MRL mice. (a) Representative 20×images of B6 (right) and MRL (left) skin biopsies grown for 7 days using the ex vivo skin assay. (b) Average angiogenic responses of different inbred mouse strains. Four inbred strains (n=13/strain) were quantified as described in the accompanying text and the average cell number growing from each strain were compared. There was a greater than threefold difference in the total cell number between B6 and MRL after 7 days of culture.
We report here the mapping of two novel QTLs that mediate early stages of angiogenesis between MRL and C57BL/6J (B6) mice using an ex vivo subcutaneous assay. To compare and contrast this approach with other models of wound healing, we performed genomewide expression analysis of skin biopsies subjected to our culture model. Our results show that genetic factors that modify angiogenesis interact with previously reported Heal and Sth QTLs known to modify later stages of wound healing in mice, demonstrating cooperation between vessel growth and wound healing in this model.
Clinical Problem Addressed
The etiology of poorly healing wounds is complex and can be influenced by environmental and genetic factors. By developing preclinical models of healing, we can dissect this process into discrete phenomena that are amenable to genetic analyses. The discovery of the underlying genetic mechanisms that mediate healing will be useful in clinical settings as biomarkers for identifying poor healers and as potential targets for therapeutic intervention.
Materials and Methods
Animals
Mice were purchased from Jackson Laboratories or were bred at the Wadsworth Center. Animals were maintained on a 7am–7pm light–dark cycle with food and water available ad libitum. All procedures were approved by the Wadsworth Center Institutional Animal Care and Use Committee.
Ex vivo angiogenesis assays
Vessel growth assays were performed as previously described.10 Briefly, euthanized 3-day mice were sanitized and transferred to the Eagle's minimal essential medium containing 1× antibiotic–antimycotic (Gibco). A 2 cm×2 cm full-thickness skin segment was removed from the dorsal flank between the scapulae and four 2 mm circular biopsies were isolated with a dermal punch (Miltex). Biopsies were embedded, dermis side down, in Matrigel (BD Biosciences) and warmed at 37°C in a tissue culture incubator for 1 h. The biopsies were then fed the endothelial cell basal medium 2 (Lonza) containing 20 ng/mL recombinant human vascular endothelial growth factor (VEGF; R&D Systems), 2% fetal bovine serum (Sigma), 1× antibiotic–antimycotic, and 12.5 μg/μL chloramphenicol (Sigma). Cultures were maintained for 7 days, replacing the media ∼72 h after the start of culture.
Vessel quantification
After 7 days of culture, Matrigel-embedded explants were fixed for 1 h at 25°C in 1× phosphate-buffered saline (PBS) containing 4% paraformaldehyde. Fixed samples were rinsed three times in 1× PBS and stained with 10 μg/mL Hoechst 33342 stain (Sigma) for 1 h. Biopsies were visualized with a light microscope (Nikon TE2000), equipped for epifluorescence (2×0.1 NA) at an excitation of 340 to 380 nm with UV fluorescence collected at 435 to 485 nm. ImagePro 6.2 (Media Cybernetics) was used to collect a Z-series (70 μm step) with a CCD camera (Roper HQ). The upper and lower imaging focal limits were determined manually by visualization of the uppermost and lowermost stained nuclei in each biopsy during image collection. A maximum local contrast projection was generated from each image stack by ImagePro 6.2.
Composite images with growth irregularities were excluded, including biopsies that produced a monolayer on the surface of the gel plug, or where the original biopsy was lost from the culture. Fewer than 4% of the images were rejected in this manner. Images of the skin biopsy were cropped from the composite image using Photoshop CS3 (Adobe Systems). The cropped 2D grayscale projections were converted to black and white images and filtered with a Watershed routine to distinguish overlapping nuclei. The nuclei were then counted, using a size exclusion paradigm designed to count objects of similar size (ImagePro 6.2; Media Cybernetics). Specific settings are available on request.
Intercross
C57BL6/J mice were crossed with MRL/MpJ mice to produce F1 progeny that were intercrossed to produce F2 mice used in subsequent mapping analyses. The age of mice at the time of the assay (day 3) was based on day 1 being the first day pups were seen in the cage. The weight of F2 animals was recorded at the time of sacrifice and did not significantly influence the outcome of the assay (data not shown).
Mapping
Liver or skin samples were used for genomic DNA isolation with the Purogene DNA isolation kit as per the manufacturer's protocol (Qiagen). Genomic DNA was then submitted to Geneseek (www.geneseek.com) for mapping analysis with a custom SNP panel designed to be informative between B6 and MRL mice in 150 genetic loci across the mouse genome. An additional 22 mouse simple sequence length polymorphisms were mapped based on their close proximity to previously identified wound healing QTLs in MRL mice.2,5,6 The variations between groups of cultured skin biopsies were normalized by subtracting the average growth rate of one batch of samples (6–15 F2 progeny per batch) from the average growth score of the individual (3–4 biopsies per animal).*
Data analysis
Mean normalized growth scores from F2 progeny, sex, and genotype information were determined for mapping analysis with R/qtl version 1.012 using the J/qtl interface version 1.3.3.13 Single interval QTL mapping was performed using either the EM algorithm, or a Haley Knott regression treating the data distribution as normal. Both algorithms produced similar results. Genomewide significance levels were determined by permutation of the entire dataset using 2000 replicates. A p-value of<0.05 was considered to be significant, while p>0.63 was considered suggestive. The 95% confidence interval of each peak was determined using confidence interval function in J/qtl. The percentage of total trait variance attributable (effect size) to each locus was determined using the Fit QTL function provided within the J/qtl program. The effect size is defined as the percentage of phenotypic variation explained by a given locus. QTLs reaching a significant threshold value were named in accordance with the International Committee on Standard Genetic Nomenclature for Mice (www.informatics.jax.org/mgihome/nomen/strains.shtml).
Expression analysis
Skin biopsies from B6 mice and MRL mice were cultured as described above for 5 days and rinsed twice in PBS at room temperature. Eight biopsies per RNA sample were scraped into 1 mL of Qiazol (Qiagen) and immediately homogenized for RNA extraction, as per the manufacturer's protocol. The aqueous phase was transferred to RNeasy mini columns and processed according to the manufacturer's protocol (Qiagen). RNA integrity was confirmed with the Agilent Bioanalyzer, which gave a RIN value of >8 for all samples.
One hundred micrograms of total RNA from six independent samples (three each from B6 and MRL strains) was submitted for microarray analysis. Affymetrix mouse gene ST 1.0 arrays were probed by the Wadsworth Center Advanced Genomics Technologies Core.
Gene expression normalization and analysis was carried out using Genespring GX Ver 10.0 (Agilent Technologies) with the GCRMA algorithm. Data were filtered to focus analysis on samples with a present or marginal flag in at least one sample. Statistical analysis was achieved using an unpaired t test (by strain), and false positives were minimized with a Benjamini-Hochberg multiple testing correction factor (cutoff p<0.05).
Results
Whereas many molecular processes are known to contribute to wound healing and several QTLs are known to modify healing abilities in MRL and LG mice, the genetic basis of these traits remain partially characterized. Our goal in this study was to focus on a specific aspect of healing (i.e., the early stages of angiogenesis) to reduce the complexity of the experimental wounding model. To accomplish this goal, we utilized a novel ex vivo culture system that has been described elsewhere.10 This method focuses on a short window of vessel growth from full-thickness skin biopsies cultured over 7 days. Whereas the process of biopsy isolation is presumed to activate the wound response, our approach should limit this response to tissue intrinsic differences that are present in the skin. Finally, we drive vascular growth through the addition of VEGF to the culture media and minimize environmental variance in our model by performing multiple assays per individual (four per proband).
The differences between inbred mouse strains in endothelial outgrowth are evident under fluorescent microscopy after 7 days of culture (Fig. 1a), but we needed a more quantitative means to measure this variance in an unbiased fashion. We developed a semiautomated method to count the total number of cells in outgrowth by staining them with a fluorescent dye, followed by counting individual cell nuclei in the outgrowth with image analysis software (see Materials and Methods). As shown in Fig. 1b, highly significant differences (p<0.00001) in the total number of cells between B6 and SM/J, MRL or LG are present when making pairwise comparisons. In contrast, the ancestrally related MRL and LG strains are not significantly different from one another (Fig. 1b) indicating that they likely share common alleles for subcutaneous angiogenesis QTLs.
To localize these differences to specific QTL, we utilized a [B6×MRL]F1 intercross to generate 178 F2 animals that were analyzed for vessel growth with our ex vivo method. These data showed significant diversity with average endothelial cell counts per F2 animal ranging from an average of 121 cells to 3204 cells. In contrast to the ear hole closure phenotypes previously reported for crosses between B6 and MRL mice, we only observed minimal differences between sexes in the F2 angiogenic response (p=0.0909, data not shown).
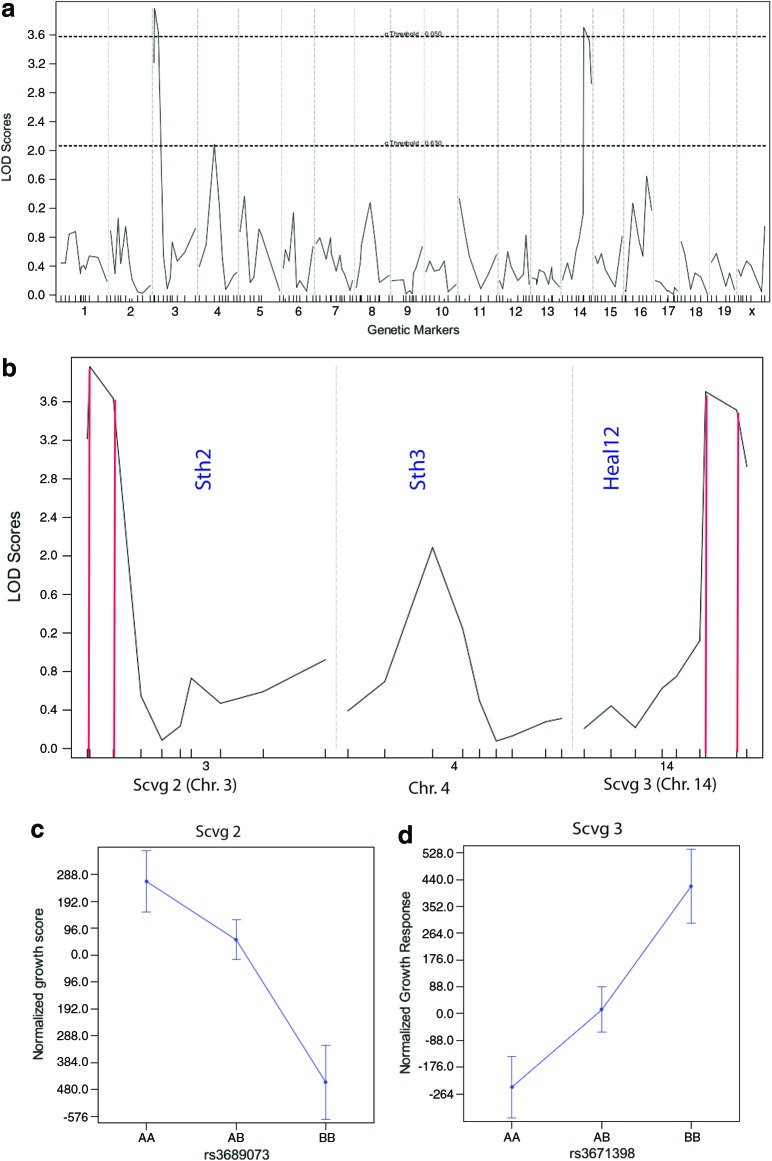
We genotyped each animal from the F2 cohort with a genomewide panel of 172 informative markers and these data were used to perform linkage analysis in R/qtl using the normalized growth response for individuals as our phenotypic trait (Fig. 2a). We detected significant associations on chromosomes 3 (Chr. 3) and 14 and a suggestive QTL was detected on Chr. 4 based on 2000 permutations of this data. The two significant QTLs were designated subcutaneous vessel growth QTL 2 (Scvg2) (Chr. 3, logarithm of the odds [LOD] 4.090) and Scvg3 (Chr. 14, LOD 3.909) based on the magnitude of their LOD scores.
Figure 2.
Mapping analysis for angiogenesis QTL. (a) Genomewide mapping analysis of the normalized quantity of endothelial cells growing from F2 biopsies. The y-axis represents the LOD score and the x-axis represents mouse chromosomes with each hash mark showing the position of the markers tested. Horizontal dashed lines represent the significant (p=0.05 upper line) and suggestive (p=0.63, lower line) cutoff based on 2000 permutations of the data. (b) Expanded view of significant and suggestive QTL detected in this cross. For significant QTL, vertical red lines delineate the 95% confidence intervals of the QTL. Vertical blue text indicates the approximate position of the closest previously mapped wound healing QTL for each chromosome. Effect plots for (c) Scvg2 and (d) Scvg3 QTLs. The x-axis lists the genotype at each locus (AA, B6/B6; AB, B6/MRL; BB, MRL/MRL) and the y-axis indicates the average effect on growth for those genotypes. LOD, logarithm of the odds; QTL, quantitative trait loci. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/wound
Scvg2 is the strongest loci detected in this cross (12% of the total phenotypic variance) by a small fraction over Scvg3 (11.5% of the variance). The growth effect of Scvg2 is transgressive, with the allele from the lower growth B6 parental strain providing a positive influence on vessel growth in our F2 cross (Fig. 2c). Scvg3 behaves in the opposite fashion with the MRL allele conveying positive growth in our model (Fig. 2d). The effects of both of these loci are additive in nature, with two copies providing roughly twice the influence on growth (data not shown).
We compared the position of our newly discovered QTLs to previous studies investigating wound healing in MRL and LG mice. Based on the previously reported positions for Sth loci14 and Heal Loci,5 we were unable to colocalize any of the previously mapped QTL with either Scvg2 or 3. Investigating QTL of lower significance, the suggestive enhanced angiogenesis peak on Chr. 4 was in the vicinity of Sth314 indicating that these QTLs may share polymorphisms. However, this hypothesis awaits the identification of the underlying mutation(s). The bayesian confidence intervals, which define the most likely limits for the QTL, were calculated in J/qtl.13 The Scvg2 interval spans from 7384755 to 26195496 base pair on Chr. 3, a region containing 215 genes (Fig. 2b). The Scvg3 confidence interval spans from 105307202 to 117038577 base pair on Chr. 14, a region containing 54 genes (Fig. 2b).
Interactions with other enhanced healing QTLs
A pairwise mapping analysis was done to determine if there were interactions between QTLs that influenced vessel growth. None of these interactions attained a p-value less than 0.05 since our cross was relatively small, but two notable interactions were detected between previously identified wound healing QTLs and markers in our study. The suggestive QTL detected on Chr. 4 was shown to interact with a locus on Chr. 6 at rs13478872 (37 cM Fig. 3a), which was previously shown to also contain wound healing QTL Sth5.14 This interaction was only detected in mice that carried a homozygous B6 genotype at the Chr. 4 locus. The Chr. 15 interaction is detected between markers rs13482752 (58 cM) and D8Mit211, markers previously used to detect Heal1 on Chr. 8.6 This interaction also requires homozygous B6 alleles at D8Mit11 to positively influence vessel growth in the presence of homozygous B6 alleles at rs13478872. Additional data will need to be analyzed to confirm these preliminary interactions and their significance.
Figure 3.
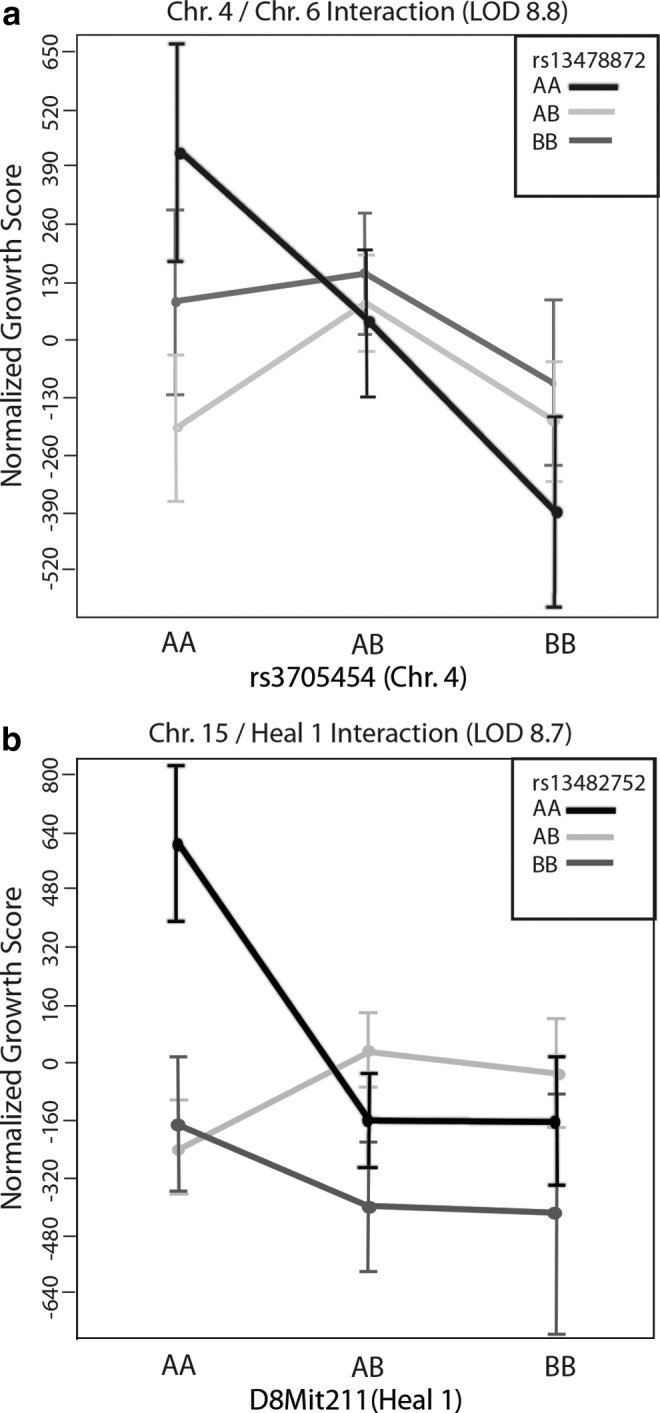
Pairwise interactions between Chrs. 4 and 6 (a) and Chrs. 8 and 15 (b). Effect plots for the two interactions with the highest LOD scores. The x-axis designates the genotype at the first locus, while the black, light gray, and dark gray lines show the effect of B6, heterozygous, and MRL alleles, respectively, at the interacting loci (inset). Note that the B6/B6 genotype (black line) for both interactions is required to elicit an interaction with the second locus.
Expression analysis of B6 and MRL ex vivo cultures
To elucidate the mechanisms that may be associated with differential angiogenesis in B6 and MRL mice, we performed global gene expression analysis on RNA transcripts from skin biopsies cultured for 5 days from these parental strains. After correction for multiple tests, we uncovered 1019 differentially expressed genes out of a total of 28,815 genes detected as expressed on the expression arrays. Based on the relative fold change between strains, the highest expressed transcripts in MRL biopsies were associated with eosinophils and mast cells (Ear11, Tpsab1, and Hal) (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/wound), while those highly expressed in B6 biopsies included MHC genes and skin differentiation genes (H2-D1|H2-L, H2-Q6|0610037M15Rik, Lce1i, Lce3c and others) (Supplementary Table S1). Tryptase alpha/beta 1 is notable among these transcripts since it is primarily expressed in mast cells and its activity has been associated with angiogenesis in multiple studies.15–17
We used functional annotation clustering of gene expression in the database for annotation, visualization, and integrated discovery (DAVID) Bioinformatics Resources 6.7 to connect expression differences with putative functional pathways.18 The complete list of significant differences used in this analysis was further refined by selecting only transcripts with greater than twofold expression differences between strains and then splitting them into those increased in B6 or MRL. These gene lists were submitted to the functional annotation clustering tool on the DAVID website to generate clusters of related transcripts significantly overrepresented in gene ontology categories, KEGG pathways, or other classifications.
The top categories in functional classifications were associated with terms like “Glycoprotein” and “transmembrane region” that were less informative with respect to function due to the large number of genes within these groups (data not shown). However, other categories were present (Table 1) that provided clues on the processes that were differentially regulated in our ex vivo assay. For example, significant increases in genes previously associated with lysosomes were seen in this data. The lysosome is implicated in invasion and angiogenesis in tumor cells,19 and lysosomes are more abundant in certain inflammatory cells.20 Wound response genes were also well represented in this analysis supporting the hypothesis that healing genes are activated in our assay and may contribute to the differences we see in angiogenesis. A group of induced immune response genes was also detected by the DAVID algorithm (Table 1); however, several of the targets increased in MRL like Il6 are not thought to be proinflammatory in skin, or are cytokines likely to impact vessel growth (e.g., Cxcl14). Finally, whereas the number genes in the GO term “angiogenesis” that were deregulated in this model did not achieve statistical significance, other classes of genes, including “cell projection morphogenesis,” “cell motion,” and “Wnt signaling,” were present, supporting the hypothesis that transcripts associated with vessel growth are differentially expressed between B6 and MRL skin.
Table 1.
Functional classes overrepresented in MRL and B6 expression data
| Term | % Total | p-Value | Increased in MRL | Increased in B6 |
|---|---|---|---|---|
| GO:0005764 Lysosome |
1.9 | 0.003 | Lgmn, Ctsf, Ctsk, Ctss, Sort1, Laptm5, Fam176a, Rab27a, Hyal1, Acpp, Ids | Arsb, Arsb, Entpd4, Naga, Ctsw, Idua, Hgsnat, Glb1 |
| GO:0009611 Response to wounding |
2.9 | 0.004 | Sgk3, Fcgr3, Fcer1g, Casp14, Igfbp3, Bcl11b, Foxc1, Clu, Glo1, Malt1, Lgals12, Bcl2l11, Nudt2, Tgfbr1, Dhcr24, Runx3, Il6, Niacr1, Bcl2l13, Cidec,Ercc1, Apoe, Rab27a, Gpx1 | Phlda3, Pnp, Pnp2, Cd74, Pcsk9, Spp1, Uaca, Rnf7, Timp1 |
| GO:0006928 Cell motion |
3.06 | 0.004 | Plxna2, Fcgr3, Fcer1g, Srgap1, B3gnt2, Fut8, Foxc1, Fgf10, Robo1, Alcam, Robo2, Efna5, Lbp, Tgfbr1, Runx3, Sema3a, Cxcl2, Gpx1, Plxna3 | Arid5b, Etv1, Cdh2, Emx2, Schip1, Slit2, Itga11 |
| GO:0006955 Immune response |
4.14 | 0.009 | Il18r1, Ccl20, Fcgr2b, Fcgr3, Fcer1g, Cr1l, Gp49a, Lilrb4, Osm, Csf3, Cd300lh, Cxcl14, Lcp1, Il7r, Enpp2, Myo1f, Tap2, Malt1, Lbp, Masp2, Il6, Cxcl2, Ercc1, Rab27a, Myd88 | Ly96, Irgm1, Ccl5, H2-Ab1, H2-D1, H2-Q8, H2-Q6, H2-K1, Cd74, Gbp7, Ddx58, Tnfrsf1b, Oasl2 |
| GO:0016055 Wnt signaling |
1.37 | 0.014 | Fzd5, Rac2, Senp2, Fzd8, Tcf7l2, Vangl1, Ppp2r1b | |
| GO:0048858 Cell projection morphogenesis |
1.58 | 0.072 | B3gnt2, Bcl11b, Clu, Slitrk6, Robo1, Alcam, Robo2, Efna5, Runx3, Sema3a, Plxna3 | Als2, Etv1, Stmn1, Stmn1, Slit2 |
| GO:0001525 Angiogenesis |
0.70 | 0.7407 | Fzd5, Zc3h12a, Gpx1, C1galt1, Itgav, Fgf2, Ccbe1 | Ccbe1, Itgav, Fgf2 |
Wnt, wingless-int.
We then focused our attention on the genes that reside under the 95% confidence intervals for the two QTLs discovered in our mapping study. For Scvg2, three genes were differentially expressed, including carbonic anhydrase 2, carboxypeptidase A3, and the ribonuclease P RNA-like 2 gene (Table 2). Genes deregulated under the Scvg3 locus included the sprouty homolog 2, SLIT- and NTRK-like family member 6, and a citrate lyase beta-like protein (Table 2). Whereas expression differences could potentially account for the divergent vessel growth, no clear connection between these transcripts and our observed phenotype is obvious at this time, although additional experiments are underway to follow up on specific targets.
Table 2.
Candidate genes differentially expressed between B6 and MRL skin near Scvg2 and Scvg3
| Gene Symbol | Description | p-Value | Fold Change | Average B6 | Average MRL | Location (GRCm38) |
|---|---|---|---|---|---|---|
| Car2 | Carbonic anhydrase 2 | 8.80E‐04 | −2.13 | 84.49 | 373.22 | Chr. 3: 14,866,426 |
| Cpa3 | Carboxypeptidase A3 | 0.00482 | −1.76 | 128.67 | 440.87 | Chr. 3: 20,215,617 |
| Rprl2 | ribonuclease P RNA‐like 2 | 0.033065 | 1.51 | 705.03 | 466.25 | Chr. 3: 22,251,374 |
| Spry2 | Sprouty homolog 2 | 0.02308 | −1.62 | 405.77 | 656.89 | Chr. 14: 105,891,949 |
| Slitrk6 | SLIT and NTRK‐like family, member 6 | 0.02856 | −1.41 | 58.71 | 82.78 | Chr. 14: 110,748,578 |
| Clybl | Citrate lyase beta like | 0.01678 | 2.38 | 1096.90 | 457.32 | Chr. 14: 122,181,694 |
Chr., chromosome.
Discussion
The impetus for this work was to establish an ex vivo wounding paradigm that can model early events in healing that are associated with vessel growth and to use this model to uncover genetic factors that mediate angiogenesis. Vessel growth is a critical initial step to wound healing that is tightly regulated, and deviations from this process can impact and ultimately prevent proper tissue regeneration.
The complexity of healing is modified by many factors that can be further exacerbated by insufficient angiogenesis in the elderly and in diseases like diabetes, and atherosclerosis. These same conditions are associated with poorly healing wounds supporting the idea that discovery of novel factors impacting angiogenesis will ultimately lead to better treatments for wound therapy related to these disorders.
The MRL mouse model has been studied extensively due to its regenerative capacity and many QTLs associated with healing have been reported in this strain.2,5,6,9,14 The most common phenotype to follow in these studies has been some measure of wound closure like ear hole diameter postwounding, which can be influenced by many factors. We show here that angiogenesis is also divergent in MRL mice and the number of QTLs uncovered is reduced when compared with wound closure mapping studies, indicating that there may be reduced genetic complexity in our model. Additional refinements, like performing the phenotypic analysis on skin from young animals, are intended to eliminate confounds like sex effects that are observed in wounding studies performed on adult animals.5
The positive impact of angiogenesis on wound healing in MRL skin has been observed by others.21 Using a transplant model of wound healing between haplotypically identical MRL and B10.BR mice, it was shown that MRL transplant recipients had better tissue engraftment that included enhanced VEGF expression, increased microcirculation, and increased STAT 3 activities at the site of transplantation. A reduction in neutrophil infiltration to thermal injuries was also previously correlated with enhanced angiogenesis in MRL mice.22 Our model extends these observations to show that early aspects of vessel growth like endothelial cell sprouting and proliferation are enhanced in MRL skin. VEGF is an important mediator of the proliferative phase of wound healing in humans23 and angiogenic sprouting into the wound clot is one of the earliest events in the healing process.24 Whereas multiple studies have investigated effects of polymorphisms within VEGF as a surrogate for differential angiogenesis, far less is known about the downstream responses to VEGF and the potential for genetic heterogeneity in those outcomes.
Whereas the QTLs discovered here did not map to previous wound healing QTL positions, we did observe interactions between two loci in this study with previously discovered healing QTLs. In each case, we found that homozygous B6 alleles were required at one locus to show the effect at the second allele. Gene by gene interactions are common among complex traits and these interactions can be useful when selecting candidate genes for further study because they indicate an interdependency between the two loci such as a receptor–ligand interaction.
Our expression studies revealed both unique and previously discovered transcripts and pathways that may be implicated in the pathology of differential angiogenesis between B6 and MRL mice. One of the highest expressed transcripts in MRL skin was associated with Mast cells, suggesting that these cells may be in greater numbers in MRL skin at the time of biopsy isolation. Additionally, we saw increases in the expression of lysosomal genes in our pathway analysis that could be connected to increased Mast cell activity in MRL skin.25 Immune cells provide many cytokines in the local wound environment that stimulate angiogenesis, and their abnormal activity in MRL has been demonstrated by others.21
Among the other pathway clusters discovered in our gene ontology analysis, many are potentially associated with enhanced vessel growth in the MRL background with a notable presence of several wingless-int (Wnt) receptor pathway genes increased in MRL skin. The Wnt pathway is critical to both embryonic vasculogenesis and postnatal angiogenesis.26–28 Deregulated transcripts associated with Wnt signaling have been previously detected in MRL mice undergoing digit healing, supporting the possibility that this pathway contributes to their differential healing.29 Whereas none of the Wnt genes differentially regulated are present under the QTL confidence intervals reported here, it is possible that a novel transcript alteration or polymorphism in the Wnt pathway is, in part, responsible for the differences we see in vessel growth in MRL wounds.
Looking forward, we will focus our studies on the two QTL regions discovered here to further refine the intervals that carry these differential angiogenesis QTLs. This will be accomplished using a combination of breeding congenic animals derived from the two parental strains and through the use of the increasing public sequence datasets available for inbred strains.30
In conclusion, we have identified two QTLs that mediate differential angiogenesis between MRL and B6 mice using a novel ex vivo angiogenesis assay. Based on these preliminary studies, we are currently moving forward to better clarify the basis for differential angiogenesis in MRL and B6 mice using a combination of genetic and genomic approaches. These studies may hold promise for discovery of genes that can potentially be targeted for treatment of nonhealing wounds in the future.
Innovation
The genetic pathways that modify wound healing in humans are poorly characterized. Defining the genetic basis of differential wound healing in preclinical models can benefit humans by providing candidate genes to evaluate in clinical populations. This work uses a novel ex vivo approach to determine how angiogenesis contributes to enhanced wound healing in MRL mice. The QTLs discovered here will ultimately be cloned and lead to development of biomarkers and therapeutic targets for treating poorly healing wounds in humans.
Key Findings.
• There are significant variations in subcutaneous vessel growth between B6 and MRL inbred mouse strains that are due to differences in their genetic background.
• Two novel QTLs were discovered that influence angiogenesis in subcutaneous vessels.
• Interactions between the angiogenesis phenotype shown here and previously reported wound closure QTLs indicate these phenotypes are interdependent.
• The molecular signatures of full-thickness MRL and B6 skin biopsies cultured in our ex vivo model are similar to previous studies following wounds in vivo.
Supplementary Material
Abbreviations and Acronyms
- AA
B6/B6 genotype
- AB
B6/MRL genotype
- B6
C57BL/6J mouse strain
- BB
MRL/MRL genotype
- DAVID
database for annotation, visualization, and integrated discovery
- Lg
large mouse strain
- LOD
logarithm of the odds
- MRL
Murphy Roth's large mouse strain
- PBS
phosphate-buffered saline
- QTL
quantitative trait loci
- Scvg
designation for angiogenesis QTL described here
- VEGF
vascular endothelial growth factor
- Wnt
wingless-int
Acknowledgments and Funding Sources
We thank the Wadsworth Center Advanced Light Microscopy core, the Advanced Genomics Technologies core, and the Wadsworth Center veterinary sciences program for their support in the completion of this research. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number R01AR054828 (B.J.H.).
Author Disclosure and Ghostwriting
No competing financial interests exist. The content of the article was expressly written by the authors listed. No ghostwriters were used to write this article.
About the Authors
Bruce J. Herron, PhD, is a research Scientist at the Wadsworth Center; the basic research arm of the New York State Department of Health, whose research focuses on mouse genetics impacting skin development. He is also an Assistant Professor at the University at Albany in the School of Public Health in the departments of Biomedical Sciences and Environmental Health Sciences. The coprimary authors Krista Morales, BS, and Leahana Rowehl, BS, were visiting undergraduates in Dr. Herron's laboratory who performed this work, in part, through funding by an ARRA Administrative supplement for students and science educators from NAIMS. Ms. Morales is currently obtaining an MD from the Albany Medical College and Ms. Rowehl is a research technician at the Stony Brook University Medical Center working on identification of genetic and environmental factors of inflammatory bowel disease. Jason Smith, BA, and Barbara Beyer, BS, are technicians at the Wadsworth Center. Richard Cole, MA, is director of the Wadsworth Center advanced image analysis core. Fang Liu, PhD, was a graduate student in Dr. Herron's lab who is currently completing a postdoctoral fellowship in the Department of Dermatology at the University of Pennsylvania.
Smith J. et al.: Angiogenesis QTL on mouse Chr. 8 co-localizes with differential beta defensin expression. Wadsworth Center, Albany, NY, 2013. (Unpublished data). Manuscript submitted to Mammal Genome.
References
- 1.Kathju S, Gallo PH, and Satish L: Scarless integumentary wound healing in the mammalian fetus: molecular basis and therapeutic implications. Birth Defects Res C Embryo Today 2012; 96:223. [DOI] [PubMed] [Google Scholar]
- 2.Cheverud JM, Lawson HA, Funk R, Zhou J, Blankenhorn EP, and Heber-Katz E: Healing quantitative trait loci in a combined cross analysis using related mouse strain crosses. Heredity (Edinb) 2012; 108:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynn TA. and Ramalingam TR: Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 2012; 18:1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falanga V: Wound healing and its impairment in the diabetic foot. Lancet 2005; 366:1736. [DOI] [PubMed] [Google Scholar]
- 5.Heber-Katz E, Chen P, Clark L, Zhang XM, Troutman S, and Blankenhorn EP: Regeneration in MRL mice: further genetic loci controlling the ear hole closure trait using MRL and M.m. Castaneus mice. Wound Repair Regen 2004; 12:384. [DOI] [PubMed] [Google Scholar]
- 6.Masinde GL, Li X, Gu W, Davidson H, Mohan S, and Baylink DJ: Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res 2001; 11:2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt TK: Disorders of wound healing. World J Surg 1980; 4:271. [DOI] [PubMed] [Google Scholar]
- 8.Heydemann A: The super super-healing MRL mouse strain. Front Biol 2012; 7:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gourevitch D, Clark L, Chen P, Seitz A, Samulewicz SJ, and Heber-Katz E: Matrix metalloproteinase activity correlates with blastema formation in the regenerating MRL mouse ear hole model. Dev Dyn 2003; 226:377. [DOI] [PubMed] [Google Scholar]
- 10.Liu F, Smith J, Zhang Z, Cole R, and Herron BJ: Genetic heterogeneity of skin microvasculature. Dev Biol 2010; 340:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohan RM, Fernandez A, Udagawa T, Yuan J, and D'Amato RJ: Genetic heterogeneity of angiogenesis in mice. FASEB J 2000; 14:871. [DOI] [PubMed] [Google Scholar]
- 12.Arends D, Prins P, Jansen RC, and Broman KW: R/qtl: high-throughput multiple QTL mapping. Bioinformatics 2010; 26:2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith R, Sheppard K, DiPetrillo K, and Churchill G: Quantitative trait locus analysis using J/qtl. Methods Mol Biol 2009; 573:175. [DOI] [PubMed] [Google Scholar]
- 14.Yu H, Mohan S, Masinde GL, and Baylink DJ: Mapping the dominant wound healing and soft tissue regeneration QTL in MRL x CAST. Mamm Genome 2005; 16:918. [DOI] [PubMed] [Google Scholar]
- 15.Ranieri G, Ammendola M, Patruno R, et al. : Tryptase-positive mast cells correlate with angiogenesis in early breast cancer patients. Int J Oncol 2009; 35:115. [DOI] [PubMed] [Google Scholar]
- 16.Cho S, Shin MH, Kim YK, et al. : Effects of infrared radiation and heat on human skin aging in vivo. J Investig Dermatol Symp Proc 2009; 14:15. [DOI] [PubMed] [Google Scholar]
- 17.Hiromatsu Y. and Toda S: Mast cells and angiogenesis. Microsc Res Tech 2003; 60:64. [DOI] [PubMed] [Google Scholar]
- 18.Huang DA W, Sherman BT, and Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44. [DOI] [PubMed] [Google Scholar]
- 19.Joyce JA, Baruch A, Chehade K, et al. : Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell 2004; 5:443. [DOI] [PubMed] [Google Scholar]
- 20.Benado A, Nasagi-Atiya Y, and Sagi-Eisenberg R: Protein trafficking in immune cells. Immunobiology 2009; 214:507. [DOI] [PubMed] [Google Scholar]
- 21.Tolba RH, Schildberg FA, Decker D, et al. : Mechanisms of improved wound healing in Murphy Roths Large (MRL) mice after skin transplantation. Wound Repair Regen 2010; 18:662. [DOI] [PubMed] [Google Scholar]
- 22.Davis TA, Amare M, Naik S, Kovalchuk AL, and Tadaki D: Differential cutaneous wound healing in thermally injured MRL/MPJ mice. Wound Repair Regen 2007; 15:577. [DOI] [PubMed] [Google Scholar]
- 23.Nissen NN, Polverini PJ, Koch AE, Volin MV, Gamelli RL, and DiPietro LA: Vascular endothelial growth factor mediates angiogenic activity during the proliferative phase of wound healing. Am J Pathol 1998; 152:1445. [PMC free article] [PubMed] [Google Scholar]
- 24.Tonnesen MG, Feng X, and Clark RAF: Angiogenesis in wound healing. J Investig Dermatol Symp Proc 2000; 5:40. [DOI] [PubMed] [Google Scholar]
- 25.Furukawa F, Yoshimasu T, Yamamoto Y, Kanazawa N, and Tachibana T: Mast cells and histamine metabolism in skin lesions from MRL/MP-lpr/lpr mice. Autoimmun Rev 2009; 8:495. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin AM. and D'Amore PA: Wnt signaling in the vasculature. Angiogenesis 2002; 5:1. [DOI] [PubMed] [Google Scholar]
- 27.Robitaille J, MacDonald ML, Kaykas A, et al. : Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat Genet 2002; 32:326. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa T, Tamai Y, Zorn AM, et al. : Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development 2001; 128:25. [DOI] [PubMed] [Google Scholar]
- 29.Chadwick RB, Bu L, Yu H, et al. : Digit tip regrowth and differential gene expression in MRL/Mpj, DBA/2, and C57BL/6 mice. Wound Repair Regen 2007; 15:275. [DOI] [PubMed] [Google Scholar]
- 30.Maddatu TP, Grubb SC, Bult CJ, and Bogue MA: Mouse Phenome Database (MPD). Nucleic Acids Res 2012; 40:D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.